Abstract
With the increased popularity of zebrafish (Danio rerio) for mutagenesis studies, efficient methods for manipulation of its genome are needed. One approach is the use of a transposable element as a vector for gene transfer in this species. We report here the transformation of zebrafish and germ-line transmission of the mariner element from Drosophila mauritiana. The mariner element was selected because its transposition is independent of host-specific factors. One- to two-cell-stage zebrafish embryos were coinjected with a supercoiled plasmid carrying the nonautonomous mariner element peach and mRNA encoding the transposase. Surviving larvae were reared to adulthood, and the transmission of peach to the F1 generation was tested by PCR. Four of the 12 founders, following plasmid injections on 2 different days, transmitted the element to their progeny. Inheritance of the transgene from the F1 to the F2 generation showed a Mendelian pattern. No plasmid sequences were detected by PCR or Southern blot analysis, indicating transposition of peach rather than random integration of the plasmid DNA. These data provide evidence of transformation of a vertebrate with a transposable element and support the host-independent mechanism for transposition of the mariner element. We suggest this system could be used for insertional mutagenesis or for identifying active regions of the genome in the zebrafish.
Transposons are naturally occurring genetic elements capable of moving from one chromosomal location to another (1). The fundamental components of transposable elements are a gene encoding the enzyme necessary for transposition, a transposase, and flanking sequences required for recognition by the transposase. Autonomous elements encode functional copies of the transposase and possess the recognition sequences to mediate transposition. Nonautonomous elements often carry mutated copies of the transposase but retain the recognition sequences; thus, they can be mobilized by transposase expressed from another source (1, 2).
In many eukaryotes, transposons are powerful tools for genetic research (3–5). This finding is convincingly illustrated by the widespread use of the P-element transposon in Drosophila melanogaster for mutating genes and for transferring foreign DNA sequences into the genome (3, 6–9). Sequence homology has identified many remnants of transposable elements in vertebrates, although the frequency of active transposition is comparatively low (10), and no functional transposase has been isolated as yet in vivo (11). Attempts to use the P-element for transformation in vertebrates failed, apparently because of the requirement of additional host-specific factors necessary for transposition (12–14). To overcome these difficulties, we have investigated the utility of the mariner element isolated from Drosophila mauritiana as a transformation vector for vertebrates (15).
The original, nonautonomous, mariner element was isolated from an unstable eye color allele, white-peach, in D. mauritiana (15). The Mos1 factor, an autonomous copy of mariner, was identified by its ability to promote high frequency excision of the mariner element, peach, from its position near the white gene (16, 17). The mariner element is ≈1.3 kb in length, encodes a single polypeptide chain of 345 amino acids, and is flanked by 28-bp inverted repeats. mariner-like elements (MLEs) have been identified in a wide range of host species including humans (18–21). Recent findings that Mos1 can transform the parasitic protozoan Leishmania (22) and a MLE from horn fly can transpose in vitro provide evidence that transposition is independent of host-specific factors (23). Thus, mariner has been proposed as a vector for transformation of a wide range of eukaryotes (22–24).
In this report, we present evidence for transposition in the zebrafish of mariner from D. mauritiana. Zebrafish embryos were injected with a plasmid carrying the nonautonomous element, peach, and mRNA encoding the Mos1 transposase. The strategy was to rely upon the host’s cellular machinery to translate the mRNA into functional Mos1 transposase. The transposase should recognize and then transpose the peach element from the plasmid vector into the host genome. A transposition event in cells giving rise to the germ line would be detected by inheritance of the peach element in the F1 progeny. Although random integration of plasmid DNA is plausible, distinguishing random integration from transposition is possible by screening for co-inheritance of peach and plasmid vector sequences (e.g., the β-lactamase gene and regions adjacent to the multicloning site). We propose that by manipulating the presence of the transposase, a general transformation system for mutagenesis and gene tagging can be developed in a vertebrate model.
MATERIALS AND METHODS
Fish Stocks and DNA Constructs.
The Ekwill strain of zebrafish, originally obtained from Ekwill Fish Farm (Gibsonton, FL), was maintained as an inbred population in the Harvard Zebrafish Facility.
The construct pAD31 consists of an immobile version of Mos1 cloned into the multicloning site of pBluescript II (24). The 1,450-bp insert lacks the 5′-inverted repeat necessary for recognition by the transposase and is flanked by 200 bp of Drosophila simulans genomic DNA. pAD31 was linearized 3′ of the insert with XhoI or HindIII, digested with proteinase K, and phenol/chloroform extracted. mRNA was generated by in vitro transcription by using T7 RNA polymerase and the mCAP Capping Kit according to the manufacturer’s instructions (Strategene). The mRNA was brought to a concentration of 1 μg/μl with water, and the purity and sizes of the transcript were examined by agarose gel electrophoresis. The pWE1 construct (provided by Daniel De Aguiar, Harvard University) was generated by ligating a PCR amplification product of the original 1.3-kb peach element (15) generated with sequence-specific primers flanked by EcoRI restriction sites into the unique EcoRI site of pBluescript II (Fig. 1Ñ). Prior to injection, plasmid DNA was purified on Wizard miniprep columns (Promega), digested with proteinase K, phenol/chloroform extracted, and precipitated with ethanol. The DNA was dissolved in diethyl-pyrocarbonate-treated water at a concentration of 0.1 μg/μl. Gel electrophoresis demonstrated the preparation to consist primarily of supercoiled plasmid.
One- to two-cell-stage zebrafish embryos from timed, pairwise matings were micro-injected just below the cell cytoplasm with an estimated 1–4 nl of a solution containing 45 ng/μl pWE1 and 50 ng/μl mRNA or 45 ng/μl pWE1 plasmid DNA alone. Surviving embryos were reared to breeding stage.
DNA Isolation and PCR.
Founder fish were mated pairwise or outcrossed to the original Ekwill line. DNA was prepared from pools of 20–30 embryos by digestion at 52°C in 100 μl of 10 mM Tris (pH 8.0) containing 200 μg/ml proteinase K, 0.2% SDS, and 20 mM NaCl. The reactions were stopped by heating to 95°C. The digests were diluted 1:5 with water and centrifuged to remove suspended material. Two microliters of the digest were used as template in 35 or 50 μl PCRs. Individual embryos were digested in 25 μl digestion buffer and processed as above. Individual zebrafish older than 21 days were screened by isolating DNA from caudal fin clippings. Fish were anesthetized in tricaine (25), and either the dorsal or ventral region of the caudal fin was removed with a razor. The tissue was digested as above for individual embryos.
The PCRs contained 10 mM Tris⋅HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 2.25 mM MgCl2, 20 mM each dNTP, 25 pmol each primer, and 1 unit Taq DNA polymerase (Promega). The reactions were initially denatured at 94°C for 3 min, followed by 35 cycles of 94°C for 30 sec and 60°C for 1 min 30 sec, and a final 72°C elongation for 15 min to 2 hr. For most samples, the presence of suitable template DNA in the preparations was confirmed by amplification in separate reactions of a 250-bp fragment of the zebrafish homeobox gene ZF21 (26). The primer pairs for the peach element were sense primer JMF31 (GAA GTG TCA ACC TTG ACT GTC) and antisense primer JMF36 (CTC ATG TGG CAC CCA TCT AC), which yield an amplification product of 450 bp. These primers demonstrate no known homology to endogenous zebrafish genomic sequences. Primers complementary to plasmid DNA were JMF32 (GCG GCC AAC TTA CTT CTG AC) and JMF33 (CTG ACG CTC AGT GGA ACG AA), which amplify a 650-bp fragment of the β-lactamase gene, and JMF6 (GTT TTC CCA GTC ACG ACG TTG TAA) against M13 forward sequence, which in combination with JMF36 yields a 680-bp product. The primer pair used for amplification of the ZF21 gene was JMF21 (GCG TTT TGT CCA CAT TAT TCA G) and JMF22 (GTA GCC AGA CTC ATG CTC TTC AG). Amplification products were identified by agarose gel electrophoresis.
Southern Blot Analysis.
Adult zebrafish were ground into powder in liquid nitrogen, resuspended in digestion buffer (10 mM Tris, pH 8.0/25 mM EDTA/0.5% SDS/10 mM NaCl/100 μg/ml proteinsase K), and incubated overnight at 50°C. Preparations were repeatedly extracted with phenol, followed by phenol/chloroform, and precipitated with ethanol. Ten to 15 μg of DNA was digested to completion with the BamHI, EcoRI, or SacI (Life Technologies, Gaithersburg, MD) in appropriate buffer. Samples were precipitated with ethanol and resuspended in TE prior to electrophoresis on 0.8% agarose gels in TAE. Following electrophoresis, the DNA was blotted to GeneScreenPlus (DuPont/NEN) by capillary transfer and UV cross-linked to the membrane. Prehybridization and hybridized were conducted at 65°C according the manufacturers recommendations (DuPont/NEN) by using prehybridization solution (Life Technologies). The probe, labeled with [32P]dCTP, was generated from the 1,300-bp EcoRI fragment of the pWE1 vector by random priming.
RESULTS
The plasmid pWE1 carries a single copy of the nonautonomous mariner element, peach, cloned into a unique EcoRI restriction site. mRNA encoding the transposase was transcribed from a plasmid containing the Mos1 factor, an autonomous copy of mariner. On two separate days, 64 1- to 2-cell-stage zebrafish embryos were injected with pWE1 and Mos1 mRNA. The surviving larvae (13 and 33, respectively) were reared to breeding stage and mated in pairs. DNA was isolated from pools of 20–30 embryos and screened by PCR for the presence of the peach element. If a pool tested positive for the transgene, clutches of embryos from the same founding pair were raised to breeding stage. DNA isolated from caudal fin clips was screened to identify transgenic F1 individuals. The frequency of transmission of the transgene to the F2 progeny was determined.
Pools of embryos from 4 pairs of founders tested positive for the transgene (Fig. 2). This result represents the identification of a minimum of 4 independent integration events in the 12 founders successfully mated, a frequency of 33% for germ-line transmitting founders. From 3 of these, the transgene was inherited by 1 or 2 (3–14%) of the individual F1 fish, indicating mosaicism of the germ line. In the fourth case, the transgene was not identified in any of the 22 available F1 fish. This low frequency of transmission is consistent with the frequency observed in the pooled DNA samples in which possibly less than 2% of the progeny inherited this copy of the transgene.
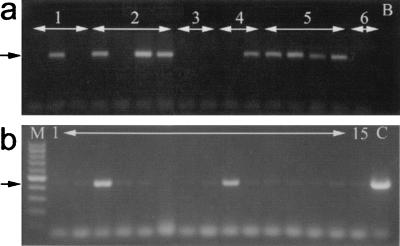
Figure 2.
PCR amplification of the mariner element in F1 progeny. (a) Genomic DNA was isolated from pools of 20–30 F1 generation larvae from six pairwise matings of founder fish (numbered 1–6) and used as template for amplification of mariner sequences. Nine of the pools of larvae from four different crosses tested positive for the transgene as indicated by a product at 450 bp (arrow). (b) Analysis of DNA samples isolated from fin clippings of individual F1 offspring from mating of founder pair number 5. Genomic DNA from 2 of the 15 individuals demonstrated the presence of mariner sequences (arrow) (lane M, 100-bp marker; Promega; lane B, no DNA control; lane C, positive control containing 1 ng pWE1 plasmid).
From four F1 fish identified in the remaining three independent lines, the frequency of transmission of the transgene to their F2 progeny was examined. F1 fish 1060.07, 1061.03, and 1085.26 transmitted the transgene to 50–56% (7/12, 10/18, and 9/18, respectively) of the F2 generation, consistent with a Mendelian pattern of segregation of a single heterozygous locus. F1 fish 1061.09 transmitted the transgene to 68% and 70% (11/16 and 7/10) of F2 progeny in two clutches, respectively (Fig. 3). This result is consistent with the inheritance of two independent insertions. Because the F1 fish in question was reared from a clutch derived from mating two putative founder fish, the two potential inserts could have been inherited from a single founder or as separate inserts from each parent. Our data cannot rule out either possibility. Overall, the data do demonstrate isolation in the F1 generation of at least four independent inserts from the original five possible.
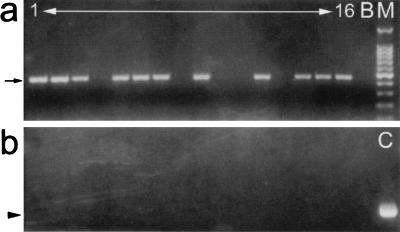
Figure 3.
PCR demonstration of inheritance of the mariner element without plasmid vector sequences. (a) Genomic DNA isolated from individual F2 larvae was tested for the presence of the mariner element and flanking plasmid sequences. Amplification of mariner sequences identified 11 of 16 (68%) larvae as positive for the transgene (arrow) (lane M, 100-bp marker, Promega; lane B, no DNA control). (b) Amplification with one primer specific for mariner and one for the M13 reverse sequence flanking the insert in the pWE1 plasmid revealed none possessed this plasmid DNA. The control lane (C), containing 1 ng pWE1 plasmid, demonstrated the expected amplification product of 680 bp (arrowhead).
DNA preparations from individual F2 fish from each of the F1s were also tested for the presence of plasmid DNA sequences. Two sets of primer pairs were used: primers specific to the β-lactamase gene and a primer pair with one specific to peach sequences and one specific to the plasmid M13 sequences flanking the insert (Fig. 1). If the transgenic lines were the result of random integration of plasmid DNA into the fish genome, we would expect to find plasmid sequences inherited with the peach sequences. However, in all cases tested, the DNA did not support amplification of either the ampicillin resistance gene or M13 sequences (Fig. 3). Lowering the annealing temperature in PCR conditions gave a similar result. These data suggest that the transgenic lines result from transposition of peach into the zebrafish genome.
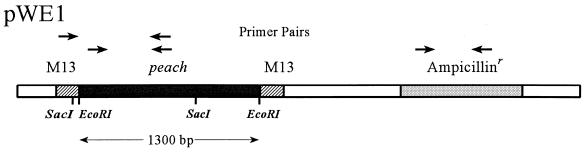
Figure 1.
Diagrammatic representation of the mariner-containing vector pWEI. A 1,300-bp PCR amplification product derived from the nonautonomous element, peach, was ligated into the unique EcoRI site of pBluescript II. The locations of the M13 sequences flanking the polylinker site and the ampicillin resistance gene are represented by the hatched and stippled boxes, respectively. The locations of primer pairs used to screen for mariner and plasmid sequence in genomic DNA preparations are indicated.
Genomic DNA isolated from three of the lines was subjected to Southern blot analysis to provide further evidence for transposition. If the transgene had integrated as plasmid DNA, the sequences of the multicloning site of the pWE1 vector should also be inherited. Digestion with EcoRI should then liberate the peach element from the integrated plasmid, yielding a 1,300-bp band on a Southern blot. The same argument implies that digestion with SacI, which would cut once within the element and once within the multicloning site, would generate one fragment of 800 bp in each line and a second of variable size. However, such results were not observed. Digestion with EcoRI resulted in bands of 10, 7, and 9 kb in the three lines tested, and digestion with SacI produced two fragments of high molecular weight in each line (Fig. 4). These data demonstrate that the peach element inserted into the zebrafish genome without the flanking plasmid sequences, providing evidence that transgenesis was the result of transposition and not random integration into the host genome. In our study, injection of the plasmid alone yielded no transgenic animals (data not shown) indicating that integration of the transgene was dependent upon the presence of the transposase.
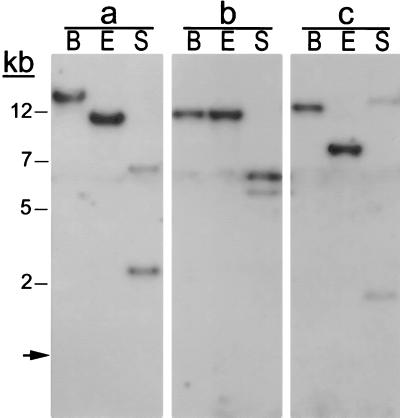
Figure 4.
Evidence for transposition of the peach element by Southern blot analysis. Genomic DNA from lines 1085 (a), 1060 (b) and 1061 (c) were digested with BamHI (B), EcoRI (E), and SacI (S), and filters were probed with the 1.3-kb peach element from pWE1. If the peach element had integrated with flanking plasmid sequences, then digestion with EcoRI should generate a single 1.3-kb band, and digestion with SacI, which recognizes one site in the peach element and one in the multicloning site of pWE1, should produce a fragment of 800 bp and a second fragment with a size dependent upon the site of integration. However, the hybridization patterns for the three samples differ, and the fragments are much larger than expected. These findings indicate that integration of the peach element occurred without flanking plasmid sequences and that the genomic sites of integration are different. (Molecular weight standards are shown to the left. The arrow indicates the location of the intact 1.3-kb peach element from pWE1 following digestion with EcoRI.)
DISCUSSION
We have demonstrated that the mariner element from D. mauritiana can be used for generation of stable transgenic lines of zebrafish. This result was achieved by microinjection, into 1- and 2-cell-stage zebrafish embryos, a plasmid carrying a nonautonomous element and mRNA encoding the transposase. This strategy is similar to that originally used for transformation of D. melanogaster, which relied upon microinjection of a helper plasmid encoding the transposase with the DNA containing the transposon (6, 7). While this work was under review, transformation of zebrafish with the Caenorhabditis elegans transposable element, Tc3 was achieved (27). Tc1-related elements are known to occur in teleosts including zebrafish (10, 11). Although no functional transposase has been isolated in vivo, a low frequency of transposition has been demonstrated (10), and a functional transposase based upon these sequences has been engineered in vitro (11); thus, it may be that the whole mariner/Tc1 superfamily of transposons is functional in this genome. These results suggest the possibility of replicating in zebrafish, a vertebrate model, the success observed with transposable elements in D. melanogaster (6–9).
Although transformation of zebrafish and germ-line transmission of a transgene was first demonstrated a decade ago (28), the technology is not well developed. The most common method relies on random integration of DNA microinjected into a 1- to 2-cell-stage embryo. The efficiency of transmission through the germ line is usually low (0–20% in different reports), and founder fish demonstrate varying degrees of mosaicism (28–31). Rearrangements and concatomerization of the transgene are common (28–30). Expression of the transgene after passage through the germ line is unpredictable and may decrease in subsequent generations (29, 30, 32).
A major advance in zebrafish transgenics occurred with the application of a pseudotype retrovirus vector for insertional mutagenesis (33–35). First developed as a vector for gene therapy and genetic studies (36), the engineered virus can infect a wide range of organisms and it efficiently integrates into the genome. In the zebrafish, transformation rates are approaching 100%, with most founders transmitting on average 10 proviral inserts to their progeny (37). This vector has been successfully employed for the mutating and cloning of several genes involved in zebrafish embryogenesis (34, 35). Based upon a frequency of isolating 1 embryonic lethal mutation per 70 insertions, it has been proposed that 250–350 mutations can be recovered over a period of several years. However, the application of the retrovirus to modestly sized mutagenesis screens or as a general transformation vector has not been demonstrated.
The mariner/Tc1 elements could be used as vectors for insertional mutagenesis. One strategy would be similar to that using the pseudotype retrovirus (34–36): generate many transposon inserts, breed these to homozygosity, and screen the larvae for mutant phenotypes. An alternative strategy would be an “enhancer-trap” screen of heterozygous insertions looking for specific patterns of expression of a reporter gene (8, 38–40). Expression of the reporter gene is regulated by host enhancer sequences near the site of insertion. Once a mutant phenotype or interesting pattern of reporter gene expression are identified, the transposon acts as a molecular tag and facilitates cloning of the gene. If large numbers of inserts can be efficiently mobilized to new chromosomal loci in each generation, it would be possible to undertake complementation analysis to clone genes previously identified in chemical mutagenesis screens. A limitation of any such mutant screen is the ability to generate the large number of insertions needed to approach saturation of the potential insertion sites. Steps are underway in our laboratories to improve this transformation frequency.
Acknowledgments
We thank Stephen M. Beverly for reviewing this manuscript and for helpful comments. We also thank Bill McCarthey and Amy Chin of the Harvard University Fish Facility for maintenance of zebrafish, Daniel De Aguiar for plasmid constructs, and Robert Hagan for assistance in the laboratory. This work was supported by grants awarded by the National Institutes of Health (EY00811 to J.E.D. and EY06554 and EY11774 to J.M.F.).
ABBREVIATION
- MLE
mariner-like element
References
- 1.Howe M, Berg D, editors. Mobile DNA. Washington, DC: Am. Soc. Microbiol.; 1989. [Google Scholar]
- 2.Hartl D L, Lozovskaya E R, Lawrence J. Genetica. 1992;86:47–53. doi: 10.1007/BF00133710. [DOI] [PubMed] [Google Scholar]
- 3.Engels W R. In: Mobile DNA. Howe M, Berg D, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 437–484. [Google Scholar]
- 4.Moerman D G, Waterston R H. In: in Mobile DNA. Howe M, Berg D, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 537–556. [Google Scholar]
- 5.Osborne B I, Baker B. Curr Opin Cell Biol. 1995;7:406–413. doi: 10.1016/0955-0674(95)80097-2. [DOI] [PubMed] [Google Scholar]
- 6.Spradling A C, Rubin G M. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 7.Rubin G M, Spradling A C. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 8.O’Kane C J, Gehring W J. Proc Natl Acad Sci USA. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spradling A C, Stern D M, Kiss I, Roote J, Laverty T, Rubin G M. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam W L, Lee T S, Gilbert W. Proc Natl Acad Sci USA. 1996;93:10870–10875. doi: 10.1073/pnas.93.20.10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ivics Z, Hackett P B, Plasterk R H, Izsvak Z. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 12.Laski F A, Rio D C, Rubin G M. Cell. 1986;44:7–19. doi: 10.1016/0092-8674(86)90480-0. [DOI] [PubMed] [Google Scholar]
- 13.Rio D C, Rubin G M. Proc Natl Acad Sci USA. 1988;85:8929–8933. doi: 10.1073/pnas.85.23.8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beall E, L, Admon A, Rio D C. Proc Natl Acad Sci USA. 1994;91:12681–12685. doi: 10.1073/pnas.91.26.12681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobson J W, Medhora M M, Hartl D L. Proc Natl Acad Sci USA. 1986;83:8684–8688. doi: 10.1073/pnas.83.22.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bryan G J, Jacobson J W, Hartl D L. Science. 1987;235:1636–1638. doi: 10.1126/science.3029874. [DOI] [PubMed] [Google Scholar]
- 17.Medhora M M, MacPeek A H, Hartl D L. EMBO J. 1988;7:2185–2189. doi: 10.1002/j.1460-2075.1988.tb03057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson H M. Nature (London) 1993;362:241–245. doi: 10.1038/362241a0. [DOI] [PubMed] [Google Scholar]
- 19.Auge-Gouillou C, Bigot Y, Pollet N, Hamelin M H, Meunier-Rotival M, Periquet G. FEBS Lett. 1995;24:541–546. doi: 10.1016/0014-5793(95)00735-r. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Fernandez J, Bayascas-Ramirez J R, Marfany G, Munoz-Marmol A M, Casali A, Baguna J, Salo E. Mol Biol Evol. 1995;12:421–431. doi: 10.1093/oxfordjournals.molbev.a040217. [DOI] [PubMed] [Google Scholar]
- 21.Robertson H M, Zumpano K L, Lohe A R, Hartl D L. Nat Genet. 1996;12:360–361. doi: 10.1038/ng0496-360. [DOI] [PubMed] [Google Scholar]
- 22.Gueiros-Fiho F J, Beverley S M. Science. 1997;276:1716–1719. doi: 10.1126/science.276.5319.1716. [DOI] [PubMed] [Google Scholar]
- 23.Lampe D J, Churchill M E A, Robertson H M. EMBO J. 1996;15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- 24.Lohe A R, Lidholm D-A, Hartl D L. Genetics. 1995;140:183–192. doi: 10.1093/genetics/140.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Westerfield M. The Zebrafish Book. 3rd Ed. Eugene: Univ. of Oregon Press; 1995. [Google Scholar]
- 26.Njolstad P R, Molven A, Fjose A. FEBS Lett. 1988;230:25–30. doi: 10.1016/0014-5793(88)80634-3. [DOI] [PubMed] [Google Scholar]
- 27.Raz E, van Luenen H G A M, Schaerringer B, Plasterk R H A, Driever W. Curr Biol. 1998;8:82–88. doi: 10.1016/s0960-9822(98)70038-7. [DOI] [PubMed] [Google Scholar]
- 28.Stuart G W, McMurray J V, Westerfield M. Development (Cambridge, UK) 1988;103:403–412. doi: 10.1242/dev.103.2.403. [DOI] [PubMed] [Google Scholar]
- 29.Stuart G W, Vielkind J R, McMurray J V, Westerfield M. Development (Cambridge, UK) 1990;109:577–584. doi: 10.1242/dev.109.3.577. [DOI] [PubMed] [Google Scholar]
- 30.Culp P, Nusslein-Volhard C, Hopkins N. Proc Natl Acad Sci USA. 1991;88:7953–7957. doi: 10.1073/pnas.88.18.7953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin S, Yang S, Hopkins N. Dev Biol. 1994;161:77–83. doi: 10.1006/dbio.1994.1009. [DOI] [PubMed] [Google Scholar]
- 32.Gibbs P D L, Peek A, Thorgaard G. Mol Mar Biol Biotech. 1994;3:307–316. [PubMed] [Google Scholar]
- 33.Lin S, Gaiano N, Culp P, Burns J, C, Friedmann T, Yee J-K, Hopkins N. Science. 1994;265:666–669. doi: 10.1126/science.8036514. [DOI] [PubMed] [Google Scholar]
- 34.Gaiano N, Amsterdam A, Kawakami K, Allende M, Becker T, Hopkins N. Nature (London) 1996;383:829–832. doi: 10.1038/383829a0. [DOI] [PubMed] [Google Scholar]
- 35.Allende M L, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Genes Dev. 1996;10:3141–3155. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- 36.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J-K. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaiano N, Allende M, Amsterdam A, Kawakami K, Hopkins N. Proc Natl Acad Sci USA. 1996;93:7777–7783. doi: 10.1073/pnas.93.15.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allen N D, Cran D G, Barton S C, Hettle S, Reik W, Surani M A. Nature (London) 1988;333:852–855. doi: 10.1038/333852a0. [DOI] [PubMed] [Google Scholar]
- 39.Gossler A, Joyner A L, Rossant J, Skarnes W C. Science. 1989;244:463–465. doi: 10.1126/science.2497519. [DOI] [PubMed] [Google Scholar]
- 40.Bayer T A, Campos-Ortega J A. Development (Cambridge, UK) 1992;115:421–426. doi: 10.1242/dev.115.2.421. [DOI] [PubMed] [Google Scholar]