Abstract
Background
The matrix protein of the influenza A virus and the matrix and capsid proteins of the human immunodeficiency virus (HIV) share striking structural similarities which may have evolutionary and biological significance. These similarities led us to hypothesize the existence of cross-reactivity between HLA-A2-restricted FLU-M1:58–66 and HIV-1 p17 GAG:77–85 epitopes.
Methods
The hypothesis that these two epitopes are cross-reactive was tested by determining the presence and extent of FLU/GAG immune cross-reactivity in lymphocytes from HIV-seropositive and seronegative HLA-A2+ donors by cytotoxicity assays and tetramer analyses. Moreover, the molecular basis for FLU/GAG cross-reactivity in HIV-seropositive and seronegative donors was studied by comparing lymphocyte-derived cDNA sequences corresponding to the TCR-β variable regions, in order to determine whether stimulation of lymphocytes with either peptide results in the expansion of identical T-cell clonotypes.
Results
Here, we report evidence of cross-reactivity between FLU-M1:58–66 and HIV-1 p17 GAG:77–85 epitopes following in vitro stimulation of PBMC derived from either HIV-seropositive or seronegative HLA-A2+ donors as determined by cytotoxicity assays, tetramer analyses, and molecular clonotyping.
Conclusion
These results suggest that immunity to the matrix protein of the influenza virus may drive a specific immune response to an HLA-A2-restricted HIV gag epitope in HIV-infected and uninfected donors vaccinated against influenza.
Background
The matrix protein of the influenza A virus and the matrix and capsid proteins of human immunodeficiency virus 1 (HIV-1) share striking structural similarities [1]. When the influenza and HIV matrix proteins were compared by aligning selected stretches of their amino acid chains (from areas that produced similar three-dimensional protein structure, rather than shared amino acid sequence homology), the HLA-A2-restricted FLU-M1:58–66 peptide (GILGFVFTL) [2] was almost perfectly aligned with the sequence SLYNTIAVL at the level of the Helix 4 domain. This sequence is a variant of the HIV-1 clade B p17 GAG:77–85 epitope consensus sequence (SLYNTVATL) [3], widely utilized as an HLA-A2-restricted HIV matrix epitope. This coincidence led us to investigate possible cross-reactivity between FLU-M1:58–66 and GAG:77–85 epitopes in cytotoxicity experiments following in vitro stimulation of PBMC derived from either HIV-seropositive or seronegative HLA-A2+ donors.
We report in the present study that PBMC from different HLA-A2+, HIV-infected donors, stimulated in vitro with FLU-M1:58–66 or GAG:77–85 peptide in the presence of interleukin-2 (IL-2), were capable of HLA-restricted killing of target cells loaded with either FLU-M1:58–66 or GAG:77–85 peptide. These cytotoxic activities correlated with binding of stimulated CD8+ T lymphocytes with FLU-M1:58–66 and GAG:77–85 HLA-A2 tetramers [4]. These results are consistent with the concept that immunity to structural components of the influenza virus may result in a cross-reactive response to structural components of HIV in infected patients.
In vitro stimulation of PBMC from HLA-A2+, seronegative donors with FLU-M1:58–66 or GAG:77–85 peptide also generated measurable cross-reactive responses to each epitope as measured by cytotoxicity studies. Analysis of the β-chain gene of the T-cell receptor (TCR) in lymphocytes from one of the seronegative donors strongly suggests that cross-recognition of FLU-M1:58–66 and GAG:77–85 epitopes results from in vitro expansion of influenza-specific memory T cells.
Methods
Human Subjects and derivation of PBMC
Blood donors were enrolled according to a protocol approved by the Institutional Review Board (HR# 7748) at the Medical University of South Carolina and were tested for both HIV antigen and antibody immediately before leukapheresis (the designation "infected" donors refers only to their HIV status; the designation "uninfected" donors refers only to their HIV seronegativity). To obtain PBMC, leukapheresis samples from donor individuals were centrifuged through a ficoll gradient (Lymphocyte Separation Medium, Organon Teknika, Durham, NC, USA) according to manufacturer's instructions. HlV-seropositive donors were receiving highly-active anti-retroviral treatment and had a blood CD4 count of at least 300/mm3.
Peptides
FLU-M1:58–66 (GILGFVFTL) [2], HIV-1 p17 GAG:77–85 (SLYNTVATL) [3], and GP100:209–217 (ITDQVPFSV) [5] peptides were synthesized by Cell Essentials, Inc. (Boston, MA, USA) and purified to >95%. Stock solutions of peptides were prepared in dimethyl sulfoxide (Mallinckrodt Baker, Phillipsburg, NJ, USA).
In vitro stimulation of PBMC
PBMC derived from leukapheresis of donors were stimulated in vitro for two-weeks in Iscove's media (Life Technologies) supplemented with 10% human AB serum (Sigma, St. Louis, MO, USA), 300 IU/mL IL-2 (Proleukin, Chiron Corporation, Emeryville, CA, USA), plus antigenic peptides (1 μg/mL FLU-M1:58–66 peptide, 5 μg/mL GAG:77–85 or GP100:209–217 peptides).
Cytotoxicity assays
Effector cells were derived from stimulation of donor PBMC as described above. T2 target cells [6] were loaded overnight at 26°C with 100 μCi Cr51 (as sodium chromate, Amersham Pharmacia, Piscataway, NJ, USA) plus 5 μg/mL of the appropriate peptide (cells not loaded with peptide were used as control targets). Effector and target cells were incubated together for four hours at 37°C at indicated ratios. Maximum and minimum Cr51-release were calculated by incubating aliquots of target cells (loaded with peptides or control) in sodium dodecyl sulfate or medium, respectively. Cr51-release was measured by gamma counter. Percent lysis reflects the average of triplicate samples.
Tetramer analyses
PE-labeled FLU-M1:58–66/HLA-A2.1 and GAG:77–85/HLA-A2.1 tetramers [4] were purchased from Immunomics (Beckman Coulter, Fullerton, CA, USA). PBMC stimulated in vitro as described above were subjected to immunofluorescence cell surface staining with tetramers plus FITC-labeled anti-CD8 mAb (Beckman Coulter). To exclude dead cells, the dye 7-amino-actinomycin D (7-AAD; BD PharMingen, San Jose, CA, USA) was added before fixation, according to manufacturer's instructions. Stained cells were fixed with 2% formaldehyde and subjected to flow cytometry analysis using a Becton-Dickinson cell analyzer (Franklin Lakes, NJ, USA).
TCR clonotype mapping
PBMC from HIV-seronegative donor 1 were subjected to in vitro stimulation with FLU-M1:58–66 or GAG:77–85 peptides as described above. Cell separation was then performed by a modification of the procedure as previously described [7]. Briefly, 100 μL of streptavidin microbeads (Miltenyi Biotec, Auburn, CA, USA) were conjugated to 20 pmol biotinylated HLA-A2/FLU-M1:58–66 monomers (synthesized according to standard protocols for tetramer construction found on the NIAID Tetramer Facility website) by incubating the mixture for 15 min at room temperature and removing the unbound monomers by spinning the beads and washing twice. HLA-A2/FLU:58–66-conjugated beads were then used to label cells stimulated with either FLU-M1:58–66 or GAG:77–85 peptide, followed by separation of the bead-bound cells by passing through a MACS MS separation column (Miltenyi Biotec), all according to the manufacturer's instructions. Total RNA was purified from each cell type using a Perfect RNA kit (Eppendorf Scientific, Westbury, NY, USA) and cDNA synthesized by the ProStar First-Strand RT-PCR kit (Stratagene, La Jolla, CA, USA) using an oligo(dT) primer.
PCR amplification of TCR BV regions for each cell type was performed using primer sequences (all BV primers plus the GC-clamped BCseq2 primer) and protocols as previously reported [8], taking appropriate precautions to prevent cross-contamination of samples. All primers were synthesized by the Great American Gene Company (Ramona, CA, USA) and Taq 2000 DNA polymerase (Stratagene) was used for PCR. PCR products were analyzed by electrophoresis through a 1.8% agarose gel in 1X TBE and stained with ethidium bromide. Some samples were also analyzed by denaturing gradient gel electrophoresis as described [7]. Briefly, samples were run in a 6% polyacrylamide gel containing a gradient of urea and formamide from 20–80% (160 V, 4.5 hr, 58°C) and stained with ethidium bromide. For DNA sequencing, bands on the DGGE polyacrylamide gel were excised under UV light, and DNA extracted from the gel slices was re-amplified by PCR using appropriate BV and BC primers. The resultant PCR products were then purified by separation on an agarose gel, followed by excision of each band and DNA extraction. The purified DNAs were then sent to the MUSC Biotechnology Core for sequencing in both forward (BV primer) and reverse (BC primer) directions.
Results
Cross-recognition of influenza- and HIV-derived epitopes in HIV-seropositive donors
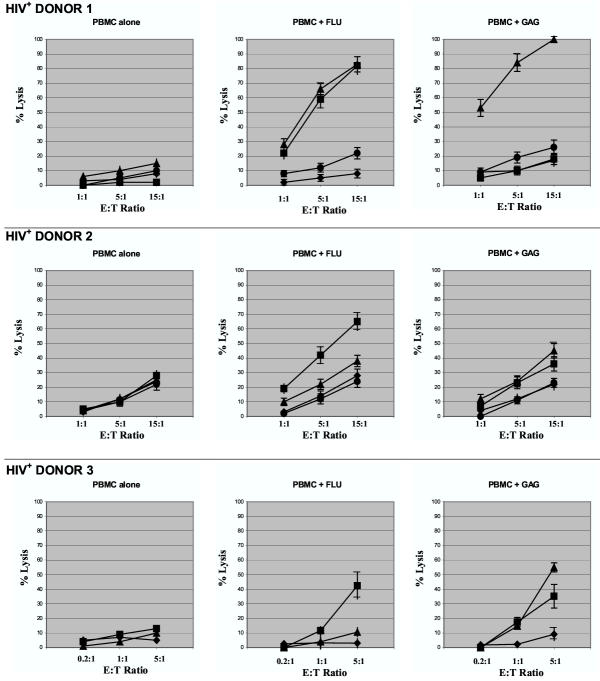
Three HLA-A2+, HIV-seropositive donors (under highly-active anti-retroviral treatment and with a blood CD4 count of at least 300/mm3) underwent leukapheresis, and their PBMC were purified by centrifugation through a ficoll gradient. Following a two-week in vitro stimulation of PBMC with either influenza A-derived FLU-M1:58–66 or HIV-1-derived p17 GAG:77–85 peptide in the presence of IL-2, effector cells were tested in 51Cr-release assays against the following targets: 1) unloaded HLA-A2+ T2 cells (no peptide); 2) T2 cells loaded with FLU-M1:58–66 peptide; 3) T2 cells loaded with GAG:77–85 peptide; and 4) T2 cells loaded with the unrelated, melanoma-associated GP100:209–217 [5] peptide. The results of these cytotoxicity experiments (Fig. 1) show that for HIV+ Donor 1 and to a lesser extent HIV+ Donor 2, effector cells stimulated with the FLU-M1:58–66 peptide lysed not only target cells presenting FLU-M1:58–66 peptide, but also target cells presenting the GAG:77–85 peptide. Conversely, effector cells from HIV+ Donor 3 (and to a lesser extent HIV+ Donor 2) stimulated with the GAG:77–85 peptide were able to lyse T2 cells presenting either the GAG:77–85 or FLU-M1:58–66 peptides. It is worth noting that lymphocytes from Donor 3 demonstrated very strong cytolytic activity after stimulation with antigenic peptides, hence the lower effector:target ratios shown in Fig. 1. No cytotoxicity above background was demonstrated against target cells presenting the HLA-A2-restricted, melanoma-associated GP100:209–217 peptide or when effector cells were stimulated with IL-2 in the absence of peptide (PBMC alone), suggesting that the observed cross-reactivity is dependent on the in vitro expansion of memory T cells in the presence of their cognate epitopes.
Figure 1.
Cytotoxic responses by lymphocytes from HIV-seropositive donors. Results are shown of representative cytotoxicity assays (Cr51-release) using stimulated lymphocytes derived from three HLA-A2+, HIV-seropositive donors. Effector cells were mixed with T2 target cells presenting HLA-A2-restricted influenza A-derived FLU-M1:58–66 (■) or HIV-1-derived p17 GAG:77–85 (▲) peptide. T2 cells loaded with GP100:209–217 (●, Donors 1 and 2 only) or no peptide (◆) were used as controls as shown. Effector cells were tested after in vitro stimulation of PBMC with IL-2 and: no peptide (PBMC alone); FLU-M1:58–66; or GAG:77–85 peptide. Data points refer to percent lysis at indicated effector:target ratios.
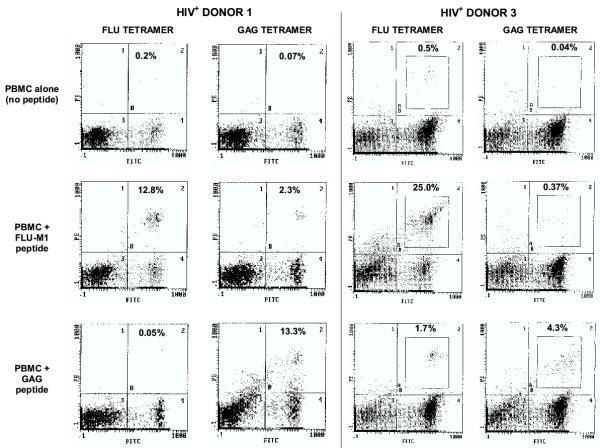
In parallel, the phenotypes of peptide-stimulated effector cell populations from these patients were also analyzed using HLA tetrameric complexes (Fig. 2). The results of these flow cytometry studies indicate that, after stimulation with FLU-M1:58–66 peptide in the presence of IL-2, CD8+ T lymphocytes from HIV+ Donor 1 recognized both FLU-M1:58–66 and GAG:77–85 HLA-A2 tetramers. In addition, GAG:77–85-stimulated CD8+ T lymphocytes from HIV+ Donor 3 recognized both GAG:77–85 and FLU-M1:58–66 tetramers. GAG:77–85 CD8+ T lymphocytes from HIV+ Donor 2 also recognized FLU-M1:58–66 tetramers, but to a much lesser extent than HIV+ Donor 3 (data not shown). It is not surprising that, despite the fact that the GAG:77–85 peptide has been determined to be the immunodominant p17-GAG antigenic epitope in HLA-A2+ individuals, the percentage of GAG-specific CD8+ cells in the donor PBMC population is too low to be seen with either tetramer staining or cytotoxicity assays unless reactive clones have first been specifically-expanded by in vitro peptide stimulation (this is also true of the FLU-M1:58–66 epitope in most donor samples).
Figure 2.
Tetramer analyses of lymphocytes from HIV-seropositive donors. After in vitro stimulation with IL-2 plus either FLU-M1:58–66 or HIV p17 GAG:77–85 peptide (or no peptide as background), lymphocytes from HLA-A2+, HIV-seropositive Donors 1 and 3 were stained with FITC-labeled anti-CD8 mAb and either HLA-A2/FLU-M1:58–66 tetramer or HLA-A2/GAG:77–85 tetramer (both PE-labeled), as indicated, and viable cells were analyzed by flow cytometry (nonviable cells positive for staining with the dye 7-AAD were excluded). Percentages of CD8+ cells that stain with tetramer appear in the upper-right quadrant of the histograms.
Lymphocytes from three additional HLA-A2+, HIV-seropositive donors were also tested by cytotoxicity assays and tetramer analyses. Effector cells from these three patients failed to respond to either FLU-M1:58–66 or GAG:77–85 peptide stimulation; therefore, no cross-reactivity was observed, indicating that their T-cell-mediated recall response was severely compromised. This is not surprising, considering that two of these patients had previously exhibited a nadir in CD4+ cell counts of 66 and 80 cells/mm3, and the third donor was experiencing a variety of immunocompromising ailments. These observations are consistent with our previously published report showing that stimulation of HIV-1 envelope-specific cytotoxic T lymphocytes (CTL) was observed in three out of six HIV-infected donors tested for their recall responses against viral antigens [9]. Therefore, in vitro stimulation seems unable to rescue a measurable CTL response from subjects who may have lost a considerable portion of their memory T-cell repertoire.
Control experiments to demonstrate HLA-restriction performed with PBMC derived from an HLA-A2-negative, HIV-infected donor showed that FLU-M1:58–66 or GAG:77–85 HLA-A2-restricted peptides were unable to stimulate either specific lysis of FLU-M1:58–66 or GAG:77–85 loaded target cells, or recognition of FLU-M1:58–66 and GAG:77–85 HLA-A2 tetramers by this patient's lymphocytes (data not shown).
Cross-recognition of influenza- and HIV-derived epitopes in seronegative donors
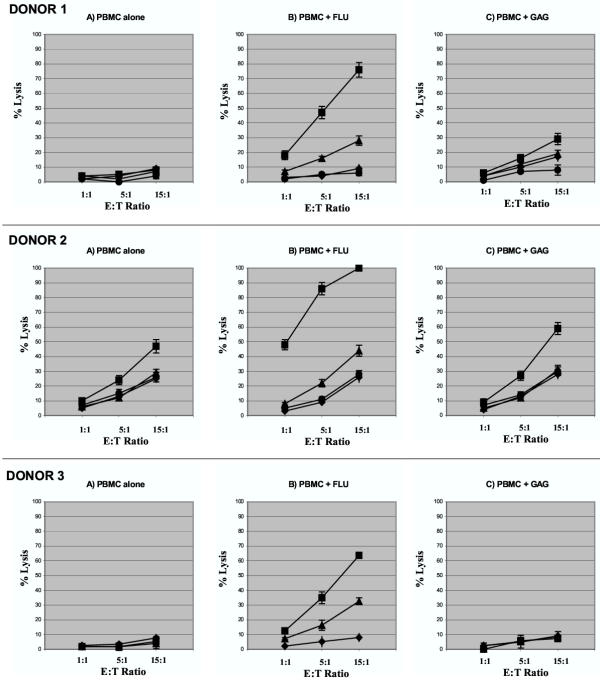
The results of the cytotoxicity and tetramer studies on the PBMC from the HLA-A2+, HlV-seropositive donors led us to investigate the possibility that HLA-A2-restricted cross-reactivity between FLU-M1:58–66 and GAG:77–85 epitopes might also be observed in lymphocytes from uninfected individuals. Therefore, we obtained PBMC from three HLA-A2+, seronegative donors. Following stimulation of PBMC with FLU-M1:58–66 or GAG:77–85 peptide in the presence of IL-2, effector cells were tested in cytotoxicity experiments as described for the HIV-seropositive donor cells above. For all three donors, effector cells stimulated with FLU-M1:58–66 peptide were able to specifically lyse target cells loaded with either FLU-M1:58–66 or GAG:77–85, indicating that it is possible to induce a primary in vitro response to an HIV-derived epitope in lymphocytes from seronegative subjects because of cross-reactivity between FLU-M1:58–66 and GAG:77–85 epitopes (Fig. 3). Stimulation of PBMC from Donors 1 and 2 with GAG:77–85 peptide resulted in some lysis of FLU-M1:58–66-loaded target cells, but no significant lysis of GAG:77–85-loaded target cells.
Figure 3.
Cytotoxic responses by lymphocytes from HIV-seronegative donors. Results are shown of representative cytotoxicity assays (Cr51-release) using stimulated lymphocytes derived from three HLA-A2+, HIV-seronegative donors. Effector cells were mixed with T2 target cells presenting HLA-A2-restricted influenza A-derived FLU-M1:58–66 (■) or HIV-1-derived p17 GAG:77–85 (▲) peptide. T2 cells loaded with GP100:209–217 (●, Donors 1 and 2 only) or no peptide (◆) were used as controls as shown. Effector cells were tested after in vitro stimulation of PBMC with IL-2 and: no peptide (PBMC alone); FLU-M1:58–66; or GAG:77–85 peptide. Data points refer to percent lysis at indicated effector:target ratios.
These cytotoxicity results suggest that stimulation of PBMC with FLU-M1:58–66 peptide induces the expansion of cognate memory T cells and the expansion of CTL that lyse not only target cells presenting FLU-M1:58–66, but also target cells presenting GAG:77–85. On the other hand, the absence in these seronegative subjects of GAG:77–85-specific memory T cells explains why PBMC stimulation with this peptide does not generate any measurable GAG:77–85-specific CTL activity. Interestingly, however, in seronegative donors 1 and 2, stimulation with GAG:77–85 results in the induction of a small but measurable CTL activity against FLU-M1:58–66-loaded target cells, consistent with the possibility that the GAG:77–85 peptide was able to stimulate the expansion of some FLU-M1:58–66 memory T cells because of cross-reactivity. Despite these repeatable results in multiple cytotoxicity assays, we could not definitively demonstrate the existence of GAG-specific, CD8+ lymphocytes in uninfected individuals by tetramer analyses (data not shown).
PBMC from two additional HLA-A2+, HlV-seronegative donors were tested that demonstrated a very weak or absent response to FLU-M1:58–66 or any other epitope examined in both cytotoxicity assays and tetramer analyses, suggesting that cross-reactivity can only be observed in subjects exhibiting a strong recall response to influenza.
Clonotype analysis of cross-reactive T cells
Because of the cross-reactivity between FLU-M1:58–66 and GAG:77–85 epitopes, we investigated whether in vitro stimulation of PBMC from seronegative Donor 1 with FLU-M1:58–66 and GAG:77–85 epitopes results in expansion of any identical T-cell clonotypes. Therefore, we performed TCR clonotype mapping by RT-PCR and denaturing gradient gel electrophoresis (DGGE) [7,8]. This procedure can be used to quickly detect clonally expanded T cells in a mixed T-cell population based on the differences in rearrangement of TCR genes from clone to clone. In most studies reported to date, this type of analysis has demonstrated that antigen-specific T cells are usually oligoclonal subpopulations of occasionally dozens of individual clones [7,8].
For this procedure, PBMC from this seronegative donor were subjected to in vitro stimulation with either FLU-M1:58–66 or GAG:77–85 peptides in the presence of IL-2, and the resulting lymphocytes purified by binding the cells to FLU-M1:58–66/HLA-A2 monomers conjugated to magnetic microbeads [7]. By attaching FLU-M1:58–66/HLA-A2 molecules to the microbeads, FLU-M1:58–66-specific T cell populations can be identified by separating bead-bound cells in a magnetic field. We observed that over ten-fold more FLU-M1:58–66-stimulated cells than GAG:77–85-stimulated cells bound to the magnetic microbeads conjugated with FLU-M1:58–66/HLA-A2.
To determine if these FLU-M1:58–66-specific T cells isolated from both FLU-M1:58–66-or GAG:77–85-stimulated cell populations shared any of the same rearrangements of the TCR-β chain gene, RNA was purified from each of these two cell populations and used as templates for making cDNA by reverse transcription. RT-PCR was then used to amplify possible transcripts for each of the 24 TCR β-variable gene (BV) regions or families (each region is amplified by the use of a common BC primer that binds in the TCR-β constant region and specific BV primer that binds in the TCR BV region; each primer pair will produce PCR products in a limited size range of about 150 to 300 bp in length; some BV families require two different BV primers to encompass all members of that BV family). To simplify analysis of the large number of samples, we first analyzed PCR products by electrophoresis in a 1.8% agarose gel. In this gel, all clones within a BV family were resolved based on their size. For the PCR products from FLU-M1:58–66-stimulated cells, clonotypes from at least 15 BV families were represented; for the PCR products from GAG:77–85-stimulated cells, clonotypes from at least 14 BV families were represented (data not shown). The results of this gel clearly demonstrated that FLU-M1:58–66-specific T cells from this donor are oligoclonal and belong to several different TCR BV families.
For 12 BV families that appeared, by agarose gel electrophoresis, to share clones between FLU-M1:58–66- and GAG:77–85-stimulated cells, PCR products were then resolved by DGGE to determine if any clones were indeed identical. This procedure can identify the presence of individual clones within a BV family by gel electrophoresis through a gradient of denaturants, during which different DNA molecules will partially melt at different locations based on their nucleotide composition, and their migration will be retarded. As a result of this gel-shift phenomenon, distinct bands will be seen on the gel even if DNA species of identical length differ in only a single base pair of their sequence [10].
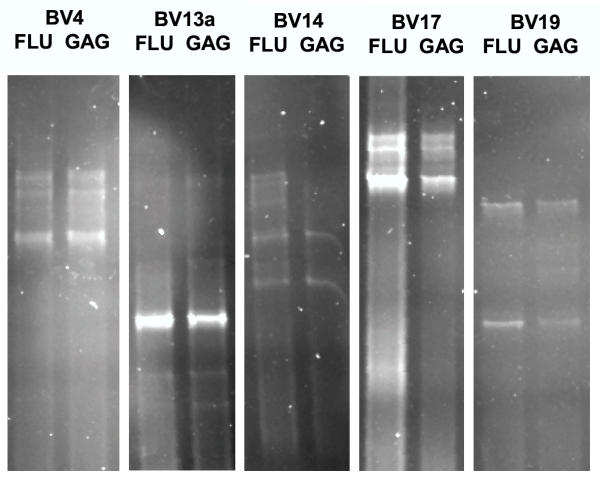
This analysis clearly showed that most bands of identical size on the agarose gel actually represented species of different nucleotide composition, as multiple bands resolved in most lanes of the DGGE. Moreover, some identical clonotypes between FLU-M1:58–66- and GAG:77–85-stimulated cells were seen in 5 of the 12 families BV families that shared clones (Fig. 4), consistent with the concept that stimulation of PBMC from this seronegative donor by either FLU-M1:58–66 or GAG:77–85 peptide results in expansion of some of the same FLU-M1:58–66-specific T-cell clonotypes, which the cross-recognition observed in cytotoxicity experiments and tetramer analyses. (For the sake of clarity, figure 4 was constructed to highlight the evidence of cross-reactivity, but as expected with two different peptides, many clones expanded by in vitro stimulation were, in fact, species of different nucleotide composition. Cross-reactive peptides would only be expected to induce some of the same FLU-M1:58–66-specific clonotypes).
Figure 4.
Clonotype analysis of FLU- and GAG-stimulated lymphocytes. Some identical FLU-specific clonotypes between FLU- and GAG-stimulated T cells were observed in 5 BV families analyzed by denaturing gradient gel electrophoresis (DGGE). PBMC from a HIV-seronegative Donor 1 were stimulated in vitro with FLU-M1:58–66 or GAG:77–85 peptides, and the resulting FLU-M1:58–66-specific cells purified by binding cells with the cognate TCR to HLA-A2/FLU-M1:58–66 peptide monomers conjugated to magnetic microbeads. RT-PCR was performed on RNA derived from each cell type (FLU-M1:58–66- or GAG:77–85-stimulated) using primers specific for all 24 TCR-β (BV) families. Each band resolved on a DGGE gel represents different clonotypes separated according to their nucleotide composition. The results of five different primers are shown here (BV13a is one of the two variable primers needed to amplify all possible members of the BV13 family).
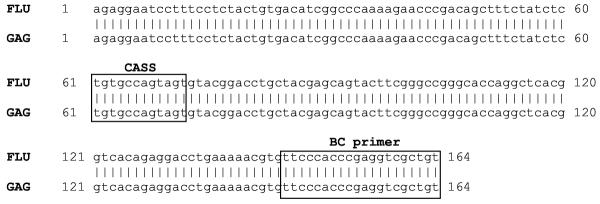
To further verify that in vitro stimulation of PBMC (from HIV-seronegative donor 1) with either FLU-M1:58–66 or GAG:77–85 epitopes results in the expansion of some identical T-cell clonotypes, two identically migrating, easily accessible bands on the DGGE polyacrylamide gel were excised, purified, and sequenced. The results of DNA sequencing confirmed that these distinct bands exhibiting identical migration in DGGE are indeed identical in sequence (Fig. 5).
Figure 5.
DNA sequencing of identical TCR-β clonotypes. 100% sequence identity of the TCR-β-variable gene region of FLU- or GAG-stimulated lymphocytes was obtained using a BV17 primer. This sequence includes the CDR3 hypervariable D/J region between the 'CASS' coding sequence and the BC region (both in gray), which greatly influences the HLA/peptide binding specificity of the TCR molecule.
Future experiments will hopefully determine the proportion of T-cell clones that are cross-reactive by simultaneously staining lymphocyte populations with both FLU and GAG tetramers conjugated to different fluorochromes. It may also be possible to isolate cross-reactive clonotypes using flow cytometry.
Discussion
In their seminal work on CTL response to cross-reactive viral epitopes, Selin, Welsh, and their collaborators emphasized that T-cell-mediated cross-reactivity is a far more widespread phenomenon than anticipated and may provide an explanation for the natural resistance to subsequent viral infections. The expansion of cross-reactive memory T lymphocytes following low-affinity cross-reactive interactions may be facilitated by the expression in these cells of ancillary molecules that lower the threshold of activation and enhance the killing of cognate target cells [11]. The pre-existing cross-reactive T-cell immunity can enhance clearance of a second, unrelated viral infection [12,13], although the sequence of viral infections is important and cross-protection is not necessarily reciprocal [14]. There may be a positive selection in favor of cross-reactive memory T cells compared to non-cross-reactive cells, which would be lost over time; such selection would contribute to a more efficient use and better homeostasis of the immune system [15]. Interestingly, little or no sequence homology between epitopes may be required for cross-recognition [16,17], consistent with the molecular mimicry model of cross-reactivity [18,19]. This may explain how the very large, but not unlimited repertoire of T-cell receptor specificities can accommodate a virtually endless variety of possible antigenic epitopes. The existence of a possible immunodominance hierarchy among cross-reactive epitopes [20] may also explain the non-symmetrical, non-reciprocal character of cross-reactivity, as we have also observed between FLU-M1:58–66 and GAG:77–85; although both epitopes are immunodominant, FLU-M1:58–66 appears to be the stronger of the two.
In other recent studies, Schlom and coworkers have developed a carcinoembryonic antigen (CEA) transgenic mouse model in which the human CEA transgene is expressed in fetal tissue and normal colonic mucosa at levels virtually identical to that found in humans [21]. They have demonstrated that these mice are tolerant to CEA as an immunogen when CEA protein in adjuvant is used as a vaccine. However, when the CEA gene is inserted into a vaccinia virus vector (rV-CEA), or into a recombinant avipox vector (avi-CEA/rF-CEA), and used as an immunogen, these CEA transgenic mice now mount a T-cell response directed against CEA [22]. Additional studies have shown that the T-cell response in CEA transgenic mice vaccinated with rV-CEA is much higher and sustained than the T-cell response in transgenic mice vaccinated with CEA protein alone [James Gulley, personal communication]. It is tempting to speculate that the role of the viral component of the rV-CEA vaccine may, at least in part, be the result of cross-reactivity, as the viral epitopes are able to mobilize large numbers of memory cells, some of which also recognize CEA-derived epitopes.
The cross-reactivity between FLU-M1:58–66 and GAG:77–85 is of particular interest since it involves two of the most intensively studied immunodominant epitopes. Cross-reactivity between FLU-M1:58–66 and a rotavirus epitope was previously reported [23] and recently, CTL-mediated cross-reactivity between hepatitis C virus (HCV) and influenza A virus has also been reported. This cross-reactivity appears to be the consequence of extensive sequence homology between the epitopes of the two viruses and is observed in 55–60% of tested individuals, either HCV-infected or uninfected [24].
Although a T-cell-mediated immune response against GAG:77–85 is readily measurable in most HLA-A2+, chronic HIV carriers, this epitope does not appear to be recognized following acute infection or vaccination [[25-27]; Kent Weinhold, personal communication]. Therefore, it seems interesting to observe a specific cytotoxic response against GAG:77–85 in three out of five seronegative donors following in vitro stimulation of their PBMC. The results of the cytotoxicity studies and tetramer analyses support the notion that the process of in vitro stimulation in the presence of FLU-M1:58–66 peptide and IL-2 activates FLU-M1:58–66-specific memory T cells, which are able to cross-recognize and lyse target cells presenting GAG:77–85. Therefore, the primary in vitro response against the GAG:77–85 epitope in seronegative subjects would be the result of FLU-M1:58–66/GAG:77–85 cross-reactivity. The small number of donors analyzed in this study prevents us from assessing the prevalence of the FLU-M1:58–66/GAG:77–85 cross-reactivity among HLA-A2+ individuals, and future studies would be necessary to fully characterize the relationship between the magnitude of the recall response to FLU-M1:58–66 and the extent of cross-reactivity involving GAG:77–85, including any possible correlations to flu vaccination history.
Conclusions
In a recently published study on clonality of HIV-specific T-cell response, it has been proposed that epitopes capable of engaging a multiplicity of T-cell clonotypes may help control viral escape [28]. Consistent with these observations, it is tempting to speculate that immunodominant epitopes are capable of engaging a greater variety of T-cell clonotypes, and they would exhibit a greater likelihood of cross-reactivity with different epitopes that engage some of the same clonotypes. Therefore, our experimental observations on the cross-recognition between the influenza A-derived, HLA-A2-restricted FLU-M1:58–66 epitope and the HIV-1-derived, HLA-A2-restricted HIV p17 GAG:77–85 epitope may have practical applications not only for immunotherapy of HIV-infected patients, but also for the prevention of HIV infection in healthy subjects, raising the possibility of inducing a protective immune response against HIV by identifying appropriate combinations and formulations of influenza- and HIV-derived antigens to be used as vaccines.
Competing interests
Competing interests: none declared.
Authors' contributions
P.M.A. participated in the design of the study, carried out most lymphocyte stimulations, cytotoxicity assays, and tetramer analyses, and assisted in the preparation of the manuscript. D.A.N. participated in the design of the study, performed the clonotype analyses, and helped draft the manuscript. E.A.B. participated in the coordination of the study and enrollment of volunteer blood donors. L.A.M. participated in the coordination of the study and enrollment of leukapheresis donors. J.E.B. participated in the design of the study and completion of clonotype analyses. S.G-C. conceived of the study, participated in its design and coordination, and helped draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
The authors would like to thank Drs. Gene Shearer, Jay Berzofsky, and Veffa Franchini for helpful comments and criticism. We also thank Ms. Anne Cope, Human Studies Coordinator. This work was supported by the MUSC Foundation for Research Development through a contract with HybriVax, Inc.
Contributor Information
Paula M Acierno, Email: pacierno@bidmc.harvard.edu.
Danforth A Newton, Email: newtond@musc.edu.
Edwin A Brown, Email: brownea@musc.edu.
Lou Anne Maes, Email: maesl@musc.edu.
John E Baatz, Email: baatzje@musc.edu.
Sebastiano Gattoni-Celli, Email: gattoni@radonc.musc.edu.
References
- Harris A, Sha B, Luo M. Structural similarities between influenza virus matrix protein M1 and human immunodeficiency virus matrix and capsid proteins: an evolutionary link between negative-stranded RNA viruses and retroviruses. J Gen Virol. 1999;80:863–869. doi: 10.1099/0022-1317-80-4-863. [DOI] [PubMed] [Google Scholar]
- Bodmer H, Ogg G, Gotch F, McMichael A. Anti-HLA-A2 antibody-enhancement of peptide association with HLA-A2 as detected by cytotoxic T lymphocytes. Nature. 1989;342:443–446. doi: 10.1038/342443a0. [DOI] [PubMed] [Google Scholar]
- Tsomides TJ, Aldovini A, Johnson RP, Walker BD, Young RA, Eisen HN. Naturally processed viral peptides recognized by cytotoxic T lymphocytes on cells chronically infected by human immunodeficiency virus type 1. J Exp Med. 1994;180:1283–1293. doi: 10.1084/jem.180.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman JD, Moss PA, Goulder PJ, Barouch DH, McHeyzer-Williams MG, Bell JI, McMichael AJ, Davis MM. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. doi: 10.1126/science.274.5284.94. [DOI] [PubMed] [Google Scholar]
- Salgaller ML, Marincola FM, Cormier JN, Rosenberg SA. Immunization against epitopes in the human melanoma antigen gpl00 following patient immunization with synthetic peptides. Cancer Res. 1996;56:4749–4757. [PubMed] [Google Scholar]
- Salter RD, Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell 879 hybrid. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrama D, Andersen MH, Terheyden P, Schrøder L, Østergaard Pedersen L, thor Straten P, Becker JC. Oligoclonal T-cell receptor usage of melanocyte differentiation antigen-reactive T cells in stage IV melanoma patients. Cancer Res. 2001;61:493–496. [PubMed] [Google Scholar]
- thor Straten P, Barfoed A, Seremet T, Saeterdal I, Zeuthen J, Guldberg P. Detection and characterization of αβ-T-cell clonality by denaturing gradient gel electrophoresis (DGGE) BioTech. 1998;25:244–250. doi: 10.2144/98252st05. [DOI] [PubMed] [Google Scholar]
- Grene E, Newton DA, Brown EA, Berzofsky JA, Gattoni-Celli S, Shearer GM. Semi-allogeneic cell hybrids stimulate HIV-1 envelope-specific cytotoxic T lymphocytes. AIDS. 2000;14:1497–1506. doi: 10.1097/00002030-200007280-00005. [DOI] [PubMed] [Google Scholar]
- Myers RM, Maniatis T, Lerman LS. Detection and localization of single base changes by denaturing gradient gel electrophoresis. Meth Enzymol. 1987;155:501–527. doi: 10.1016/0076-6879(87)55033-9. [DOI] [PubMed] [Google Scholar]
- Selin LK, Nahill SR, Welsh RM. Cross-reactivities in memory cytotoxic T lymphocyte recognition of heterologous viruses. J Exp Med. 1994;179:1933–1943. doi: 10.1084/jem.179.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzl G, Tafuro S, Moss P, Openshaw PJ, Hussell T. Influenza virus lung infection protects from respiratory syncytial virus-induced immunopathology. J Exp Med. 2000;192:1317–1326. doi: 10.1084/jem.192.9.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HD, Fraire AE, Joris I, Brehm MA, Welsh RM, Selin LK. Memory CD8+ T cells in heterologous antiviral immunity and immunopathology in the lung. Nat Immunol. 2001;2:1067–1076. doi: 10.1038/ni727. [DOI] [PubMed] [Google Scholar]
- Selin LK, Varga SM, Wong IC, Welsh RM. Protective heterologous antiviral immunity and enhanced immunopathogenesis mediated by memory T cell populations. J Exp Med. 1998;188:1705–1715. doi: 10.1084/jem.188.9.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selin LK, Lin MY, Kraemer KA, Pardoll DM, Schneck JP, Varga SM, Santolucito PA, Pinto AK, Welsh RM. Attrition of T cell memory: selective loss of LCMV epitope-specific memory CD8 T cells following infections with heterologous viruses. Immunity. 1999;11:733–742. doi: 10.1016/s1074-7613(00)80147-8. [DOI] [PubMed] [Google Scholar]
- Loftus DJ, Chen Y, Covell DG, Engelhard VH, Appella E. Differential contact of disparate class I/peptide complexes as the basis for epitope cross-recognition by a single T cell receptor. J Immunol. 1997;158:3651–3658. [PubMed] [Google Scholar]
- Hemmer B, Gran B, Zhao Y, Marques A, Pascal J, Tzou A, Kondo T, Cortese I, Bielekova B, Straus SE, McFarland HF, Houghten R, Simon R, Pinilla C, Martin R. Identification of candidate T-cell epitopes and molecular mimics in chronic Lyme disease. Nat Med. 1999;5:1375–1382. doi: 10.1038/70946. [DOI] [PubMed] [Google Scholar]
- Oldstone MB. Molecular mimicry and autoimmune disease. Cell. 1987;50:819–820. doi: 10.1016/0092-8674(87)90507-1. [DOI] [PubMed] [Google Scholar]
- Loftus DJ, Castelli C, Clay TM, Squarcina P, Marincola FM, Nishimura MI, Parmiani G, Appella E, Rivoltini L. Identification of epitope mimics recognized by CTL reactive to the melanoma/melanocyte-derived peptide MART-1 (27–35) J Exp Med. 1996;184:647–657. doi: 10.1084/jem.184.2.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn KJ, Belz GT, Altman JD, Ahmed R, Woodland DL, Doherty PC. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- Greiner JW, Zeytin H, Anver MR, Schlom J. Vaccine-based therapy directed against carcinoembryonic antigen demonstrates antitumor activity on spontaneous intestinal tumors in the absence of autoimmunity. Cancer Res. 2002;62:6944–6951. [PubMed] [Google Scholar]
- Schlom J, Panicali D. Recombinant poxvirus vaccines. In: Rosenberg SA, editor. Biologic Therapy of Cancer: Principles and Practices. Philadelphia, JB Lippincott Co; 2000. pp. 686–694. [Google Scholar]
- Shimojo N, Maloy WL, Anderson RW, Biddison WE, Coligan JE. Specificity of peptide binding by the HLA-A2.1 molecule. J Immunol. 1989;143:2939–2947. [PubMed] [Google Scholar]
- Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. Cross-reactivity between hepatitis C virus and influenza A virus determinant-specific cytotoxic T cells. J Virol. 2001;75:11392–11400. doi: 10.1128/JVI.75.23.11392-11400.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalod M, Dupuis M, Deschemin J-C, Goujard C, Deveau C, Meyer L, Ngo N, Rouzioux C, Guillet J-G, Delfraissy J-F, Sinet M, Venet A. Weak anti-HIV CD8+ T-cell effector activity in HIV primary infection. J Clin Invest. 1999;104:1431–1439. doi: 10.1172/JCI7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJR, Altfeld MA, Rosenberg ES, Nguyen T, Tang Y, Eldridge RL, Addo MM, He S, Muckerjee JS, Phillips MN, Bunce M, Kalams SA, Sekaly RP, Walker BD, Brander C. Substantial differences in specificity of HIV-specific cytotoxic T cells in acute and chronic HIV infection. J Exp Med. 2001;193:181–193. doi: 10.1084/jem.193.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari G, Neal W, Jones A, Olender N, Ottinger J, Ha R, McElrath MJ, Goepfert P, Weinhold KJ. CD8 CTL responses in vaccines: emerging patterns of HLA restriction and epitope recognition. Immunol Lett. 2001;79:37–45. doi: 10.1016/S0165-2478(01)00264-4. [DOI] [PubMed] [Google Scholar]
- Douek DC, Betts MR, Brenchley JM, Hill BJ, Ambrozak DR, Ngai K-L, NJ Karandikar, Casazza JP, Koup RA. A novel approach to the analysis of specificity, clonality, and frequency of HIV-specific T cell responses reveals a potential mechanism for control of viral escape. J Immunol. 2002;168:3099–3104. doi: 10.4049/jimmunol.168.6.3099. [DOI] [PubMed] [Google Scholar]