Abstract
N-ethyl maleimide sensitive factor (NSF) belongs to the AAA family of ATPases and is involved in a number of cellular functions, including vesicle fusion and trafficking of membrane proteins. We present the three-dimensional structure of the hydrolysis mutant E329Q of NSF complexed with an ATP–ADP mixture at 11 Å resolution by electron cryomicroscopy and single-particle averaging of NSF·α-SNAP·SNARE complexes. The NSF domains D1 and D2 form hexameric rings that are arranged in a double-layered barrel. Our structure is more consistent with an antiparallel orientation of the two rings rather than a parallel one. The crystal structure of the D2 domain of NSF was docked into the EM density map and shows good agreement, including details at the secondary structural level. Six protrusions corresponding to the N domain of NSF (NSF-N) emerge from the sides of the D1 domain ring. The density corresponding to α-SNAP and SNAREs is located on the 6-fold axis of the structure, near the NSF-N domains. The density of the N domain is weak, suggesting conformational variability in this part of NSF.
Keywords: AAA ATPase/electron cryomicroscopy/FREALIGN/NSF/vesicle fusion
Introduction
In eukaryotic cells, trafficking of vesicles is essential for diverse cellular functions such as exocytosis, endocytosis, transcytosis and subcellular compartmentalization (Rothman, 1994). NSF belongs to the AAA (ATPases Associated with a variety of cellular Activities) family of ATPases and is an essential component of the protein machinery that regulates vesicle fusion with target membranes. NSF acts together with α-SNAP (Soluble NSF Attachment Protein) to disassemble SNARE (SNAP REceptor) complexes. SNAREs, in a concerted action with additional proteins, facilitate docking and fusion of vesicles, and they are recycled and reactivated through disassembly by NSF (Brunger, 2001b; Whiteheart et al., 2001). NSF also interacts with other membrane proteins, such as glutamate [GluR2 (Nishimune et al., 1998), AMPA (Braithwaite et al., 2002)], GABA (Kittler et al., 2001) and β2 adrenergic receptors (Cong et al., 2001), indicating a possible additional role in trafficking of membrane proteins. In its functional state, NSF is a homohexamer (Fleming et al., 1998). Each of the NSF protomers contains three domains. The N-terminal domain of NSF (NSF-N) is essential for the binding of α-SNAP and is followed by ATPase domains D1 and D2. Binding and hydrolysis of ATP by the NSF-D1 domain induces conformational changes in NSF leading to disassembly of the SNARE complex. NSF-D2 has a lower ATPase activity than NSF-D1. ATP probably acts as a structural component for NSF-D2, since its binding is required for the hexamerization of NSF (Yu et al., 1998; Whiteheart et al., 2001).
Electron microscopy (EM) of NSF and NSF·α-SNAP·SNARE (20S) complexes has provided a basic understanding of the overall structure of NSF and 20S complexes at low resolution, as well as demonstrating possible conformational changes of the NSF hexamer during ATP hydrolysis (Hanson et al., 1997; Hohl et al., 1998). Of the three NSF domains, crystal structures of NSF-N (May et al., 1999; Yu et al., 1999) and hexameric NSF-D2 (Lenzen et al., 1998; Yu et al., 1998) have been obtained, but there are no three-dimensional (3D) structures available for the D1 domain or the complete NSF molecule, and consequently the quarternary arrangement of the NSF domains is uncertain. Here we present the 3D structure of the hydrolysis mutant E329Q of NSF complexed with α-SNAP, SNAREs and a mixture of ATP and ADP at 11 Å resolution, determined by electron cryomicroscopy (cryo-EM) and single-particle averaging. The hydrolysis mutant was used to slow ATP turnover and help stabilize a single conformation of NSF. The NSF domains have been assigned by imaging of truncated NSF constructs in negative stain and by docking of the NSF-D2 crystal structure into the 3D density map.
Results
As a prelude to cryo-EM, the E329Q mutant of NSF (NSFm) (Whiteheart et al., 1994) and the complex of NSFm, α-SNAP and SNAREs (20Sm) were studied by negative stain EM. The initial studies using negatively stained protein served to determine the location of NSF domains within the 20Sm complex, which assisted in the interpretation of the density map obtained by cryo-EM at higher resolution.
NSFm in negative stain
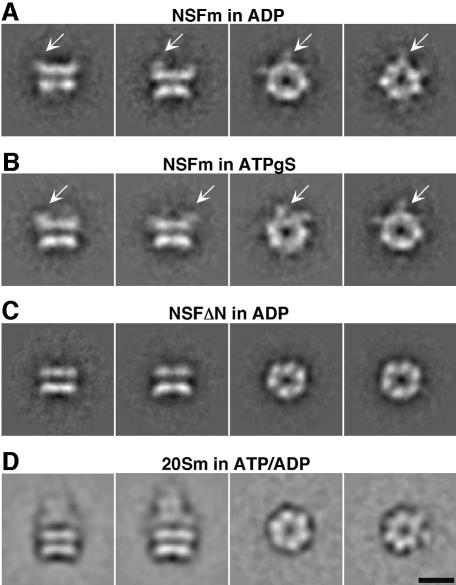
NSFm was purified, applied to carbon-coated copper grids and negatively stained with methylamine tungstate or uranyl acetate as described in Materials and methods. Similar to recombinantly expressed wild-type NSF (Hanson et al., 1997; Hohl et al., 1998), single particles of NSFm adopt a preferred orientation with a donut-like appearance and 6-fold symmetry on carbon support films. This orientation will henceforth be referred to as the 6-fold view. Other views that are only infrequently detected in uranyl acetate, but more regularly observed in methylamine tungstate, show the typical barrel-shaped double-layered structure observed for NSF (Hohl et al., 1998). These views will be referred to as side views. Small arms, corresponding to the globular density detected in 6-fold views of NSFm, extend at various angles upwards from only one of the two layers (Figure 1A and B). Deletion of the N-terminal domain of NSF removes these densities (Figure 1C), establishing that the double-layered barrel contains the two ATPase domains NSF-D1 and NSF-D2, while the N-terminal NSF-N domain is flexible and extends sideways and upwards from the barrel.
Fig. 1. NSFm and 20Sm complexes in negative stain. (A) NSFm in 1 mM ADP stained with 2% methylamine tungstate. Left: side views show the typical double-layered barrel described for wild-type NSF (Hohl et al., 1998). Right: 6-fold views are donut-shaped particles from which density extends sideways (arrows). Arm-like densities (arrows) corresponding to the NSF-N domain extend at various angles from one end of the double-layered NSFm barrel. (B) NSFm in 1 mM ATPγS stained with 2% methylamine tungstate. Single particles of NSFm in ATPγS are indistinguishable from single particles of NSFm in ADP. Arrows point to the flexible NSF-N domains. Left: side views. Right: 6-fold views (C) NSF-N deletion mutant (NSFΔN) in 1 mM ADP stained with 2% methylamine tungstate. Although the overall structure of NSFΔN is similar to that of NSFm in either ADP or ATPγS, no densities extending sideways or upwards from NSF can be observed, indicating that the protrusions seen in NSFm correspond to the NSF-N domain, while the double-layered barrel contains the NSF domains D1 and D2. Left: side-views. Right: 6-fold views (D) 20Sm complexes in 0.1 mM ATP/0.01 mM ADP stained with 1% uranyl acetate show all features described for wild-type 20S complexes (Hohl et al., 1998), i.e. the barrel-shaped NSFm particle that carries a cone-shaped complex at one end. Scale bar, 100 Å.
20Sm complexes in negative stain
Similar to NSFm in negative stain, 20Sm complexes consisting of NSFm, a truncated SNARE complex fused to MBP and α-SNAP adopt a preferred orientation on the carbon support film. Six-fold views (Figure 1D) were prominent and side views were only infrequently observed, regardless of the negative stain used (uranyl acetate or methylamine tungstate). These 6-fold views appear indistinguishable from 6-fold views of NSFm (compare Figure 1A, B and D). However, class averages of side views (height ∼190 Å, width ∼130 Å) could be obtained and show the features previously described for complexes of wild-type 20S complexes (Hohl et al., 1998), i.e. a double layered NSF barrel (height ∼105 Å, width ∼130 Å) and a cone-shaped complex ∼80 Å long, corresponding to α-SNAP and SNAREs (Figure 1D). The cone-shaped extension in the images shown by Hohl et al. (1998) is more pronounced than in our images since they used full-length SNARE proteins, including the transmembrane domains which were bound to detergent micelles.
20Sm by electron cryomicroscopy
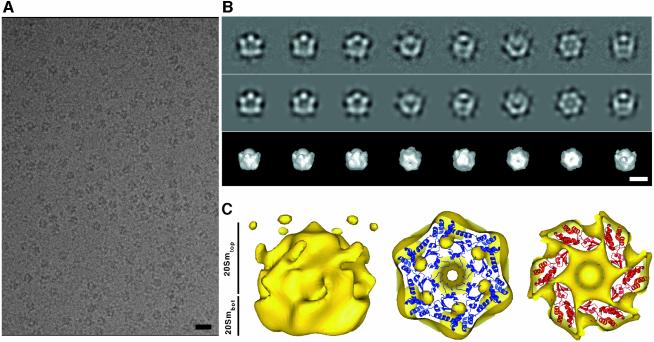
20Sm was frozen in vitreous ice and examined at liquid nitrogen temperature under low dose conditions (∼10 e–/A2). Unlike NSFm (data not shown), 20Sm complexes produced high contrast in ice (Figure 2A), enabling the selection of a total of 31 592 images of single particles from digitized micrographs. An initial 3D density map of 20Sm with imposed 6-fold symmetry was calculated from a subset of 10 002 particle images using the IMAGIC image processing package (van Heel et al., 1996), as described in Materials and methods. Class averages obtained in the final round of classification, using IMAGIC, correspond well to matching projections of the 3D structure (Figure 2B). This initial 3D density map of 20Sm was subsequently further refined to 20 Å resolution and corrected for the contrast transfer function (CTF) of the electron microscope using FREALIGN (Grigorieff, 1998) (Figure 2C).
Fig. 2. Initial 3D structure of 20Sm complexes in ice. (A) 20Sm particles produce high contrast in ice. Scale bar, 200 Å. (B) Class averages (first row) of selected views of 20Sm complexes demonstrating good correspondence with matching projections (second row) from the 3D density map calculated by angular reconstitution as implemented in IMAGIC. The third row shows 3D surface views with orientations corresponding to the projections in the second row. Scale bar, 100 Å. (C) Preliminary 3D density map of 20Sm. Left: NSFm forms a double-layered barrel. Protrusions, similar to those observed in NSFm in negative stain, extend sideways from one end of the barrel. Near the protrusions, a cap-like density that coincides with the location of α-SNAP and SNARE in 20Sm complexes in negative stain is clearly visible around the 6-fold symmetry axis. The protrusions fuse with disconnected density next to the cap, when the molecular weight of the structure is increased, indicating flexibility in this region. Right: the reasonably good fit of the NSF-D2 crystal structure (PDB entry code, 1NSF.pdb) into both layers of the NSFm barrel demonstrates that the major density in 20Sm contains NSF-D1 and NSF-D2. For these fits, we assumed an antiparallel arrangement of the D1 and D2 domains (see text).
As shown in Figure 3C for the entire data set of 31 592 particles (see below), Euler angles of 20Sm particles sample the entire 3D space, with a slight preference for 6-fold and side views, allowing a reliable 3D reconstruction of 20Sm. To interpret the initial 20Sm density map, the crystal structure of NSF-D2 (Yu et al., 1998) (PDB entry code: 1NSF.pdb) was manually docked into the density map of 20Sm. The resulting model produced good agreement for both layers of the barrel-like density of 20Sm, demonstrating that the reconstruction procedure described above produced a reliable density map in the region of the two NSF ATPase rings (Figure 2C).
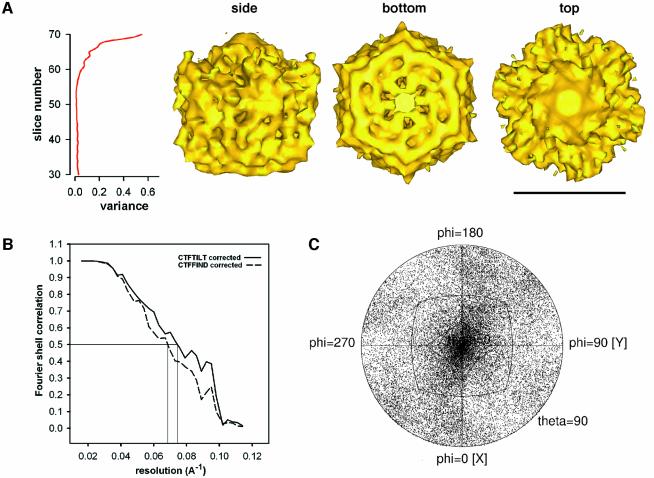
Fig. 3. 20Sm in ice at 11 Å resolution: Fourier shell correlation (FSC) and distribution of Euler angles. (A) Right: side view and the two end views of the structure (scale bar, 100 Å). Left: analysis of the variance in the 20Sm density map. The variance increases towards the end of the structure that is close to the NSF-N domains of 20Sm, indicating considerable flexibility in this region. (B) FSC indicates a resolution of 13.5 Å at a correlation value of 0.5 for the 20Sm structure with tilt-corrected defocus values. (C) 20Sm particles in ice sample the entire 3D space with a slight preference for end and side views.
20Sm at 11 Å resolution
Inclusion of all 31 592 single particles and further refinement using FREALIGN produced a density map at 11 Å resolution (see Materials and methods). Similar to the initial 20Sm density map described above, the 11 Å density map of 20Sm is characterized by a double-layered barrel that measures ∼120 Å along the symmetry axis and has a diameter of ∼130 Å. At one end of 20Sm, six protrusions emerge (top of the side view in Figure 3A) and a protruding feature located on the 6-fold axis of the density map is visible. The location of the six protrusions coincides with the location of the NSF-N domains, as evidenced by the low-resolution images of the NSF N-terminal deletion mutant determined in negative stain (Figure 1C). The density on the 6-fold axis overlaps with density attributed to α-SNAP and SNAREs in Figure 1D. The NSF-N domains, α-SNAP and SNAREs are not clearly resolved in the density map. Since the SNARE complex does not possess 6-fold symmetry, one expects to lose structural detail in this part of the complex upon imposition of 6-fold symmetry. The unresolved density may also be due to a variable number of SNAREs present in the 20S complexes. However, the loss of structural detail in the region of the NSF-N domains is most likely due to flexibility in this part of the molecule. The lower quality of the density map in the region of NSF-N is also indicated by an increased density variance (Figure 3A).
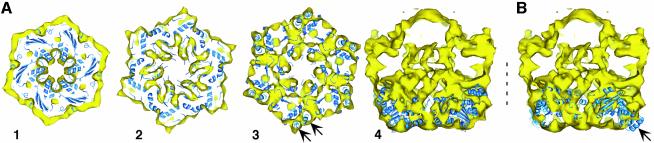
Docking of the NSF-D2 crystal structure
The crystal structure of NSF-D2 complexed with ATP (Yu et al., 1998) (PDB entry code: 1NSF.pdb) was docked into the 11 Å EM density map both manually and using SITUS 2.0 (Wriggers et al., 1999). Consistent with visual inspection and manual docking, the volumetric docking algorithm of SITUS 2.0 placed the crystal structure of NSF-D2 into the layer of the EM density map that is opposite to the one with the six protrusions (Figure 4). When fitted into 20Smbot, the crystal structure produced the highest correlation coefficient (0.86) with the C-terminus and the helical subdomain facing towards the interface between the two ATPase domain rings (Figure 4A, panels 1–4). The unusually high quality of the fit is illustrated, for example, by the good agreement of two density features at the perimeter of the D2 ring with two α-helices of the helical subdomain of NSF-D2 (Figure 4A, panel 3, arrows). Placement of the D2 ring in the opposite orientation, i.e. with the helical subdomains pointing away from the other NSF domains, produced a lower correlation coefficient (0.79) and there were clear discrepancies between the crystal structure and the 20Sm density map (Figure 4B, arrow). We interpret residual discrepancies between the fitted crystal structure of NSF-D2 in the orientation shown in Figure 4A (panels 1–4) and the EM map as an indication of small conformational differences between the crystallized NSF-D2 structure and the EM density map.
Fig. 4. Docking of the NSF-D2 crystal structure into 20Smbot. (A) Docking of the NSF-D2 crystal structure (PDB entry code, 1NSF.pdb) by SITUS 2.0 (Wriggers et al., 1999) reveals good agreement with the layer distal to the protrusions of 20Sm (20Smbot, bottom of panel 4). Panels 1–3: slices through 20Smbot perpendicular to the symmetry axis starting from the bottom of the structure. Arrows point to two α-helices of the C-terminal helical NSF-D2 subdomain that fit remarkably well into two separated densities in 20Smbot. The separation between these two α-helices suggests a resolution of the 20Sm structure of 11 Å. Panel 4: slice parallel to the symmetry axis through 20Sm. The best fit of NSF-D2 places it into 20Smbot, such that the N-terminus of NSF-D2 faces away from the interface with 20Smtop and the C-terminal helical NSF-D2 subdomain faces toward it. (B) Slice corresponding to panel 4 through 20Sm with NSF-D2 rotated 180° around an axis perpendicular to the symmetry axis and docked into 20Smbot. Considerable mismatch between NSF-D2 and the density map of 20Sm can be observed (arrow).
Comparison of 20Smbot and 20Smtop by correlation analysis
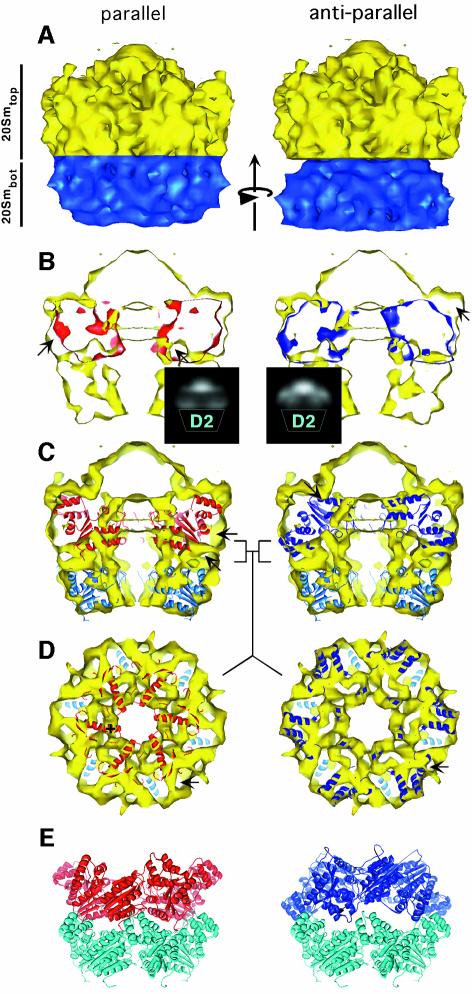
No detailed structural information on NSF-D1 is available as it has not been crystallized so far. AAA ATPase domains are generally believed to be structurally homologous, especially for the α/β subdomain (Ogura and Wilkinson, 2001). However, larger differences have been observed for the helical subdomain (DeLaBarre and Brunger, 2003). Since NSF-D1 and NSF-D2 are structurally related, one should expect some structural similarity between the two density regions in the EM density map designated as 20Smbot and 20Smtop (see above and Figure 6A). To obtain further insight into the similarity between these two density regions, and to determine the orientation of the NSF-D1 domain with respect to NSF-D2, the 20Smbot density was rotationally and translationally fitted into the 20Smtop density by maximizing a correlation coefficient. This approach represents a model-free analysis of the arrangement of the two ATPase domains within NSF. Two different orientations were tested: the parallel orientation, in which 20Smbot is shifted into the 20Smtop density, and the antiparallel orientation, in which 20Smbot is rotated by 180° around an axis perpendicular to the 6-fold symmetry axis and shifted into the 20Smtop density, as depicted in Figure 6. For each orientation of 20Smbot, a rotational search around the 6-fold symmetry axis was carried out, and the rotation angle with the best correlation coefficient was chosen for further analysis.
Fig. 6. Parallel versus antiparallel arrangement of NSF-D1 and NSF-D2. (A) 20Sm structure with 20Smtop in yellow and 20Smbot in blue. The arrow points in the direction of the rotational and translational fit carried out to compare 20Smtop with 20Smbot, and to determine the orientation of NSF-D1 with respect to NSF-D2. (B) Slice through 20Sm parallel to the symmetry axis. In the parallel arrangement of 20Smbot and 20Smtop, considerable density mismatch can be observed. The arrows point to density that does not overlap. Protrusions corresponding to the NSF-N domains are partially occupied. The inset shows a 2D projection of the difference map between 20Smbot and 20Smtop. Apart from the peaks in the region of the N domains, there are also strong peaks at the D1–D2 interface. In the antiparallel arrangement 20Smbot (blue) fits well into 20Smtop without major discrepancies between these two densities. For this arrangement, protrusions corresponding to the NSF-N domain remain unoccupied and are marked by an arrow. The difference map in the inset shows strong peaks in the region of the N domains, but not at the D1–D2 interface. (C) Fit of the NSF-D2 crystal structure into both 20Smbot and 20Smtop. The discrepancy seen between the parallel fit of the 20Smbot density into the 20Smtop density can also be seen between the NSF-D2 crystal structure and 20Smtop density. The arrows indicate density occupied in the antiparallel fit that is not occupied in the parallel fit. (D) Slice through 20Sm (yellow) at the interface between 20Smbot and 20Smtop, perpendicular to the symmetry axis. The position of the slice is marked by square brackets ] [ in (C). The NSF-D2 crystal structure fitted into the 20Smbot density is shown in light blue, and the NSF-D2 crystal structure fitted into the 20Smtop density is shown in dark blue or red for the antiparallel or parallel arrangement, respectively. The interface between 20Smbot and 20Smtop is well structured. For the antiparallel arrangement, almost the entire density is occupied and no significant discrepancies can be observed between the NSF-D2 crystal structure in 20Smbot and the NSF-D2 crystal structure in 20Smtop. The arrow points to a well-refined tube-like density that is likely to contain the N-terminus of NSF-D2 and/or the linker between NSF-D1 and NSF-D2. In the parallel arrangement, the NSF-D2 crystal structure does not fit well into 20Smtop at the interface between 20Smbot and 20Smtop. An α-helix of the NSF-D2 crystal structure that does not have a corresponding density in 20Sm is marked by a + sign. The arrows point to unoccupied density. (E) NSF-D2 crystal structures arranged in the parallel and antiparallel orientations.
In the parallel orientation of 20Smbot and 20Smtop, the correlation coefficient is 0.47, and the outer surface of 20Smbot agrees well with the outer surface of 20Smtop over the entire superimposed area, including the transition zone to the protrusions on the side of 20Smtop (Figures 5, panels A1–A5 and 6B, left panel). However, internal surfaces show considerable density mismatches, especially at the interface between 20Smbot and 20Smtop (Figure 5, panels A1 and A2, arrows) and in the area close to the protrusions on 20Smtop (Figure 5, panels A4 and A5, arrows). To illustrate the differences between the two densities further, the aligned 20Smbot density was subtracted from 20Smtop to produce a difference map. A two-dimensional (2D) projection of this difference map is shown as an inset in Figure 6B, left panel. Apart from large difference peaks near the putative locations of the N domains, there are also strong peaks at the interface between 20Smbot and 20Smtop.
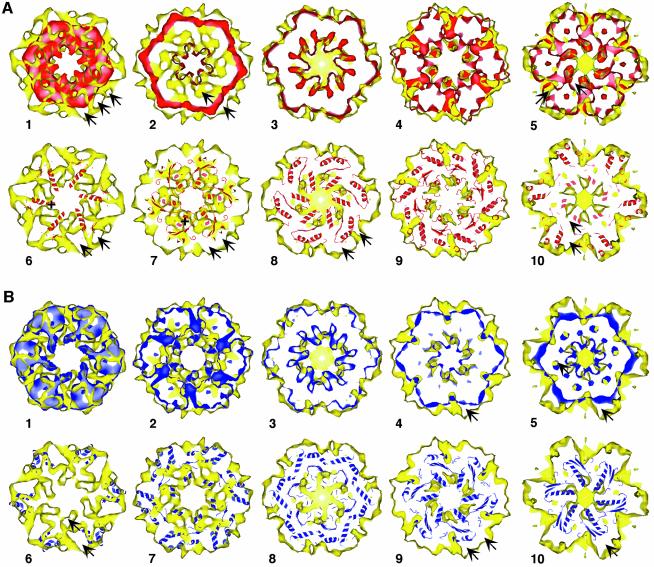
Fig. 5. Comparison of 20Smbot and 20Smtop. (A) Comparison of 20Smbot with 20Smtop arranged in a parallel orientation (see also Figure 6). 20Smbot (red) was rotationally and translationally fitted into 20Smtop (yellow) as described in the Results and shown in Figure 6. Panels A1–A5: slices through 20Smtop perpendicular to the symmetry axis, starting at the interface between 20Smbot and 20Smtop. Arrows point to densities that do not overlap in 20Smtop and 20Smbot. Panels A6–A10: the transformation of the best parallel fit of 20Smbot into 20Smtop was used to dock NSF-D2 into 20Smtop. The slices through 20Smtop correspond to panels A1–A5. It is evident from panels A6 and A7 that the NSF-D2 crystal structure does not agree well with 20Smtop near the interface with 20Smbot. In the transition zone toward the protrusions on 20Smtop (panels A8 and A9), NSF-D2 shows better agreement with 20Smtop. For this fit, the C-terminal α-helical NSF-D2 subdomain extends upward into the protrusions at one end of 20Smtop (panel A10). The arrows point to density that is not occupied by 20Smbot and the NSF-D2 crystal structure; the + signs in panels 6 and 7 show the NSF-D2 α-helices that do not fit into the 20Smtop density. (B) Comparison of 20Smbot with 20Smtop arranged in the antiparallel orientation. 20Smbot (blue) was rotationally and translationally fitted into 20Smtop (yellow) as described in the Results. Panels B1–B5: slices through 20Smtop perpendicular to the symmetry axis, starting at the interface between 20Smbot and 20Smtop. Arrows point to non-overlapping density. Panels B6–B10: the transformation of the best antiparallel fit of 20Smbot into 20Smtop was used to dock NSF-D2 into 20Smtop. The slices through 20Smtop correspond to panels B1–B5. NSF-D2 agrees well with the 20Sm density map throughout the entire volume. All major densities overlap. The arrows point to non-overlapping densities.
In the antiparallel orientation, the best correlation coefficient between 20Smbot and 20Smtop is 0.59, significantly better than that found for the parallel orientation. The internal surfaces of 20Smtop and 20Smbot are in good agreement throughout the entire overlapping area, as can be seen from the side view in Figure 6B, right panel and the perpendicular slices in Figure 5, panels B1–B5. Although all major densities of 20Smtop overlap with corresponding densities in 20Smbot, discrepancies in the overall appearance and location of these densities (Figure 5, panels B1–B5) are indicative of somewhat different conformations of the helical and α/β subdomains of NSF-D1 compared with those of NSF-D2, and unaccounted density corresponding to NSF-N. A density mismatch near the N domain would be consistent with a difference in the conformation of the N-terminus of NSF-D1 and that of the N-terminus of NSF-D2. The N-terminus of NSF-D1 is part of the linker to NSF-N and is likely to undergo considerable conformational changes along with NSF-N during ATP hydrolysis. As before, a difference map between 20Smbot and 20Smtop was calculated, and a projection is shown as an inset in Figure 6B, right panel. Unlike the parallel arrangement, the antiparallel arrangement of the two ATPase domains does not produce strong difference peaks at the domain interface. The main differences occur on the opposite side of the D1 domain, near the putative locations of the N domains.
Fitting of the NSF-D2 crystal structure into 20Smtop
The observed differences between the two ‘barrel’ densities, 20Smtop and 20Smbot, prevented straightforward fitting of NSF-D2 into 20Smtop by automated docking. In order to obtain a fit of NSF-D2 into 20Smtop, we used the transformations found for the best parallel and antiparallel alignments of 20Smbot to 20Smtop, as described above, to fit the NSF-D2 crystal structure, previously aligned to 20Smbot, into 20Smtop. The modeling of the 20Smtop density by the NSF-D2 crystal structure is clearly approximate since there may be differences between the NSF-D1 and NSF-D2 domains, especially in the helical domains.
For the parallel fit, considerable density at the interface between 20Smbot and 20Smtop remains unoccupied, and an α-helix of the α/β subdomain extends into empty space inside the pore of the barrel (Figures 5, panel A6 and 6C, left panel). Overall, there is little agreement between the NSF-D2 crystal structure and 20Smtop close to the interface between the two rings (Figure 5, panels A6 and A7). Near the protrusions that emerge from 20Smtop (Figure 5, panels A8–A10), the agreement is somewhat better. The wider part of the NSF-D2 hexamer fits into the protrusions that presumably correspond to the NSF-N domains. However, considerable density within and adjacent to the protrusions remains vacant (Figures 5, panel A10 and 6C, left panel).
For the antiparallel fit, the NSF-D2 crystal structure shows good agreement with 20Smtop in the well-defined densities at the interface between the two ATPase rings (Figure 5, panels B6 and B7) and other densities inside the ring (Figures 6C, right panel and 5, panel B10). The protrusions and adjacent density remain unoccupied (Figure 5, panels B8–B10). Discrepancies between NSF-D2 and the 20Smtop density are small (Figure 5, panels B6–B10). Interestingly, the α-helices of the D2 helical subdomain that correspond to the two pronounced densities at the perimeter of 20Smbot (Figure 4A, panel 3) also fit well into density features of 20Smtop at the interface between 20Smbot and 20Smtop (Figure 5, panel B6). In contrast, for the parallel fit, these two NSF-D2 α-helices extend into the protrusions that presumably correspond to the NSF-N domains, but most of the surrounding density remains unoccupied. Taken together with the correlation analysis between 20Smtop and 20Smbot presented in the previous section, the 20Sm density map suggests that NSF-D1 and NSF-D2 are arranged in an antiparallel fashion with the two helical subdomains close to each other (Figure 6E, right panel).
Discussion
Type II AAA ATPases such as NSF or p97/valosin containing protein (VCP) play important roles in the homeostasis of mammalian cells, and their molecular physiology has been extensively studied over the past decade (Brunger, 2001a; Ogura and Wilkinson, 2001; Whiteheart et al., 2001). NSF interacts with the adaptor α-SNAP and the SNARE complex to form the 20S complex. Crystal structures of the NSF domains [N (May et al., 1999; Yu et al., 1999) and D2 (Lenzen et al., 1998; Yu et al., 1998)], Sec17/α-SNAP (Fasshauer et al., 1998; Rice and Brunger, 1999) and the SNARE complex (Sutton et al., 1998) have provided valuable information for the development of molecular models of vesicle fusion (Brunger, 2001a). However, detailed knowledge of the entire structure of the NSF hexamer, as well as of higher-order complexes, and, eventually, information on their dynamic behavior will be required to fully understand the vesicle fusion process.
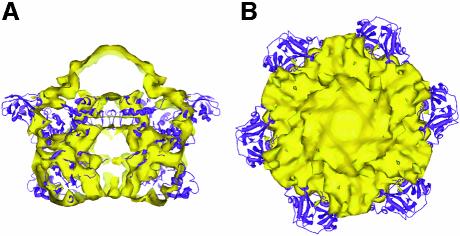
As a first step toward the characterization of full-length NSF and 20S complexes, we have determined the 3D structure of the E329Q mutant of NSF (Whiteheart et al., 1994) at 11 Å resolution by cryo-EM and single-particle averaging of images of 20Sm complexes. The 20Sm density map shows a double-layered barrel containing NSF-D1 and NSF-D2 domains, and six protrusions emerging from one of the rings (Figure 3). The protrusions appear near the putative locations of the NSF-N domains (Figures 1A and B, and 3). In the case of the homologous p97/VCP AAA ATPase, a crystal structure of the entire molecule has been obtained (DeLaBarre and Brunger, 2003). Figure 7 shows a superposition of the p97/VCP crystal structure on the 20Sm density map. A large difference between the 20Sm density map and the p97/VCP crystal structure occurs in the region of the N domain. Whereas p97/VCP shows an extended conformation of the N domain, the lack of density for 20Sm suggests considerable motion, as has also been observed for p97/VCP in complex with both ADP and AMP-PNP, a non-hydrolysable ATP analog (Rouiller et al., 2002). A more rigid conformation of the p97/VCP N domain was only observed for the ATP analog ADP·AlFx that mimics the transition for hydrolysis (Rouiller et al., 2002). One can speculate that the behavior of NSF might be similar to p97/VCP where the positions of the NSF-N domains become well defined only at the transition state.
Fig. 7. Comparison of p97/VCP with 20Sm. (A) Side view of 20Sm (yellow) superimposed on the p97/VCP crystal structure (DeLaBarre and Brunger, 2003) (purple). The superposition was carried out by aligning the D1 domain of the p97/VCP structure to the model of the NSF-D1 domain consisting of the NSF-D2 crystal structure fitted to the 20Smtop density in the antiparallel orientation (Figure 5E, right panel). (B) End view of the 20Sm structure. Note the mismatch between the p97/VCP D2 domain with the 20Sm density map (bottom of figure). The orientation of the p97/VCP D2 domain is opposite to that of NSF (compare with Figure 4B).
NSF undergoes marked conformational changes upon ATP hydrolysis. These changes could be observed at low resolution by comparing images of NSF in the ADP and ATP bound state (Hanson et al., 1997). Protrusions extending from the NSF core could only be observed when ATP was present. This led to the hypothesis that the NSF-N domains are located on the side of the NSF-D1/D2 core during ATP hydrolysis (Hanson et al., 1997; May et al., 1999; Yu et al., 1999). In our study, the flexibility of the NSF-N domain is evidenced by the varying position of the protrusions visible in class averages of NSFm in negative stain (Figure 1A and B). This flexibility is also evident in the cryo-EM density map of 20Sm described here (Figure 3A). However, unlike in the previous study, we do not find a dependence of the position of the NSF-N domain on the bound nucleotide (Figure 1). It is possible that the lack of nucleotide-dependent conformational changes in the N domain is the result of the E329Q mutation that slows ATP hydrolysis.
Type II AAA ATPases such as NSF contain two ATPase domains. Structural studies of various type I and type II AAA ATPases have clearly shown that these ATPase domains share a common fold, although the sequence similarity among these proteins can be less than 25% (Swaffield and Purugganan, 1997; Patel and Latterich, 1998; Brunger, 2001b; Ogura and Wilkinson, 2001). For some AAA ATPases, such as Cdc48p, the amino acid sequence of the ATPase domains D1 and D2 is highly conserved. In NSF, sequence similarity between NSF-D1 and NSF-D2 is limited to regions that include the Walker nucleotide binding motifs (Swaffield and Purugganan, 1997). Automated docking of the crystal structure of NSF-D2 into the 20Sm structure with SITUS 2.0 placed the NSF-D2 crystal structure into the layer (20Smbot) of the 20Sm barrel that is opposite to the protrusions that presumably correspond to the NSF-N domains. The high correlation of 0.86 and the good agreement of the NSF-D2 crystal structure with the 20Sm density map (Figure 4) support this result. Therefore the only part of the density map that could accommodate NSF-D1 is 20Smtop, located between the density containing NSF-D2 and the protrusions. Although NSF-D1 and NSF-D2 are believed to share the same overall fold, structural differences can be expected due to the low sequence similarity (Swaffield and Purugganan, 1997), different conformations of structural elements (DeLaBarre and Brunger, 2003) and the different function within the NSFm hexamer (Patel and Latterich, 1998): NSF-D2 is responsible for the hexamerization of NSF, while NSF-D1 is believed to trigger conformational changes of NSF upon ATP hydrolysis. The correlation coefficient between the 20Smbot and 20Smtop densities was considerably lower than that between 20Smbot and the NSF-D2 crystal structure, suggesting that there are differences between the two ATPase rings. The lower correlation may also indicate a loss of structural features in some parts of the D1 domain that are affected by the flexibility of the adjacent N domain.
A crucial question concerns the relative orientation of the NSF-D1 and NSF-D2 domains within the hexamer. The correlation between the 20Smbot and 20Smtop densities was lower when they faced the same way than when they faced in opposite directions, suggesting that the NSF ATPases are arranged in an antiparallel fashion. This model-free correlation analysis assumes that the conformation of the two ATPase domains is sufficiently similar to observe a meaningful correlation. Therefore a parallel arrangement of NSF-D1 and NSF-D2 would be consistent with our data only if we assume large conformational differences between the NSF-D1 and NSF-D2 domains that would invalidate our correlation analysis. However, large conformational differences between the two ATPase domains appear unlikely in light of the good agreement observed between the two densities in Figure 5B, panels 1–3. An antiparallel arrangement is also indicated by the difference maps in Figure 6B. The large difference peaks between the 20Smbot and 20Smtop densities at the D1–D2 interface that are observed with a parallel arrangement of the two ATPase domains cannot be explained by the NSF-N domains that are on the other side of the D1 hexamer. These peaks are not observed with an antiparallel arrangement, making this the more plausible interpretation of our density map.
For the homologous AAA ATPase p97/VCP, a parallel arrangement of the ATPase rings has been found in the crystal structure of the entire protein (DeLaBarre and Brunger, 2003) where the helical subdomain of the p97/VCP D2 domain points away from the interface between the two rings (Figure 7). Our data thus support a significant difference between NSF and p97/VCP where the D2 domain is found in opposing orientations. This difference in the D2 domain might be related to the low sequence identity of NSF-D2 with NSF-D1 (23%) and p97/VCP-D2 (21%). By comparison, the sequence identity is 38% between NSF-D1 and p97/VCP-D1, and between p97/VCP-D1 and p97/VCP-D2. It is unlikely that the resolution of our density map of 20Sm obscures a different orientation of the D2 domain, considering the detailed agreement found, for example, for two of the helices of the helical subdomain. Rather, we believe that the different orientation of the D2 domain points to a mechanistically very different role of this domain in NSF versus p97/VCP. Indeed, the D2 domain is catalytically inactive in NSF, whereas in p97/VCP it is the D1 domain that is inactive and the D2 domain that is active (Whiteheart et al., 1994; DeLaBarre and Brunger, 2003; Song et al., 2003). Thus in p97/VCP the nucleotide state of the D2 domain needs to be transmitted to the N-terminal domain, whereas in NSF the transmission likely occurs within the D1 domain itself.
Materials and methods
Protein purification
His6-tagged E329Q mutant (Chinese hamster, CHO) NSF (NSFm) was expressed in Escherichia coli and purified by immobilized metal affinity chromatography (IMAC). The His6 tag was removed by digestion with tobacco etch virus (TEV) protease followed by heparin–Sepharose chromatography. Hexameric NSFm was then size selected on a Superdex 200 (16/60) column. This NSFm preparation consistently yielded low-contrast images in ice, which could not be used for image processing. To further increase the purity of NSFm for electron microscopy and to ensure that the resulting NSFm preparation is indeed functional, NSFm was mixed in a 1:1 molar ratio with a fusion complex consisting of C-terminal maltose-binding protein (MBP) tagged SNAP25 (SNAP-25[1–83], SNAP-25[120–206]-MBP), syntaxin[188–262] and synaptobrevin[1-96]), followed by incubation overnight with a vast excess of bovine α-SNAP to promote assembly of NSFm·α-SNAP·SNARE complexes (20Sm). 20Sm was separated from free NSFm and unbound α-SNAP and MBP-SNARE by heparin–Sepharose and gel permeation chromatography (GPC) [Sephadex 200 (16/60) (Amersham Biosciences), equilibrated with NSF buffer (20 mM HEPES pH 7.4, 300 mM NaCl, 2 mM MgCl2, 5 mM β-mercaptoethanol, 0.01/0.1 mM ADP/ATP nucleotide or 0.1 mM ATPγS nucleotide)]. Nucleotide exchange was performed during GPC. The 20Sm complexes had a molecular weight of about 680 kDa as estimated by GPC and multiangle laser light scattering, which was in close agreement with the expected molecular weight of 610–680 kDa (the range of expected molecular weights is due to the unknown number of α-SNAPs per NSF hexamer which lies between one and three). Fractions containing 20Sm particles consisting of NSF, α-SNAP and SNARE in NSF buffer were combined and frozen in liquid nitrogen (see below).
Electron microscopy
Negative stain. Five microliters of NSFm (15 µg/ml) or 20Sm complexes (15 µg/ml) in NSF buffer were applied to glow-discharged carbon support film on copper grids. Excess protein was blotted off, and the grids were washed three times with NSF buffer, followed by three washes in negative stain (1% uranyl acetate or 2% methylamine tungstate pH 7.2). Grids were then examined on a Philips CM12 electron microscope equipped with an LaB6 filament under low-dose conditions. Images were taken at 60 000× magnification and 1.5 µm underfocus.
Electron cryomicroscopy. 20Sm samples were frozen in vitreous ice by applying 5 µl of protein solution (200 µg/ml 20Sm in NSF buffer) to Quantifoil holey carbon grids (SPI Supplies, West Chester, PA, USA), blotting off excess protein in a humidified chamber (∼80% relative humidity) and plunging into liquid ethane. Frozen grids were then transferred to liquid nitrogen and examined on a Philips CM12 or FEI Tecnai F20 FEG electron microscope under low-dose conditions and 2.0–4.0 µm underfocus.
Image processing. Micrographs were digitized on a SCAI scanner (Zeiss) with a pixel size of 7 µm. Groups of 3 × 3 pixels were averaged to give a pixel size of 21 µm on the micrograph or 3.5 Å on the specimen. Single-particle images were selected manually and image processing was performed as implemented in IMAGIC (van Heel et al., 1996). The first stages of the processing were the same for images of negatively stained protein and protein embedded in ice. Briefly, particle images were first bandpass filtered, normalized and then centered and rotationally aligned against an average of all non-aligned images. A new reference was calculated using the aligned images, and iterations of alignment and averaging were repeated another four times. Aligned images were then subjected to multivariate statistical analysis (MSA) and classified using their Eigenimage components. Class averages, showing different views of the particles, were calculated and the averages of the best classes were used as new references for multi-reference alignment (MRA). The cycle of MRA, MSA and classification was repeated until the classes were stable. For the cryodata, the relative orientations of the final class averages were determined by angular reconstitution and a 3D reconstruction with imposed 6-fold symmetry, calculated using the exact filter back-projection algorithm. Projections of 3D reconstructions were subsequently used in further cycles of MRA, MSA, classification and angular reconstitution until no further improvement of classes could be obtained. Particle orientations were then further refined and the 3D reconstruction corrected for the contrast transfer function of the electron microscope using FREALIGN (Grigorieff, 1998). The parameters used in FREALIGN were as follows: MODE 4 (randomized search and refinement with DANG = 100° and ITMAX = 100); PBC weighting factor, 4; symmetry, C6; amplitude contrast, 0.07; resolution cut-offs for the refinement, 22 Å (first three iterations) and 10 Å (next nine iterations). Defocus values for each image were determined using CTFFIND3 (Mindell and Grigorieff, 2003). FREALIGN applies a weighting scheme that reduces the weight of images contributing to the final 3D structure according to their phase residual or cross-correlation coefficient with respect to the reference structure. Twenty percent of a total of 31 592 particles with the highest phase residuals or lowest cross-correlation coefficients with respect to the reference structure were excluded from the reconstruction, thereby removing particles that did not agree well with the majority of the data. The resolution of the reconstruction was estimated by Fourier shell correlation (FSC), indicating a resolution of 15 Å after this initial refinement. At this stage, any local specimen tilt due to sample bending or cryo-wrinkling (Vonck, 2000) was determined using CTFTILT (Mindell and Grigorieff, 2003). The tilt information allowed us to use refined values for the local defocus for each particle. These new defocus values were used in two additional refinement iterations with FREALIGN (MODE 1; PBC weighting factor, 10; symmetry, C6; amplitude contrast, 0.07; resolution cut-off for the refinement, 10 Å). Although the resolution (as measured by the FSC) of the resulting 20Sm structure did not improve dramatically (Figure 3B), individual well-separated α-helices (Figure 4) could be resolved after a temperature factor (Havelka et al., 1995) of –1500 Å2 was applied. The distance between two well-separated α-helices suggests a resolution of this final 20Sm structure of about 11 Å, corresponding to an FSC of 0.39. The final reconstruction was filtered to 10.56 Å using a cosine edge filter. Docking of crystal structures into density maps was performed manually using the CHIMERA package (Computer Graphics Laboratory, University of California, San Francisco, CA) and by automated docking using SITUS 2.0 (Wriggers et al., 1999).
Acknowledgments
Acknowledgements
The authors would like to thank James Ernst for providing the truncated NSF construct, Michael Reese for help with the initial 20Sm purification, Byron DeLaBarre for insights into the project and help with multiangle light scattering analysis and Richard Yu for help with the NSF purification. Special thanks are due to Chen Xu and Yifan Cheng for their help with electron microscopy, Matthias Wolf, Mike Rigney, David Gutierrez, C. Miller, D. Oprian and M. Paulmichl for their support and Thomas Walz for providing access to his FEI F20 electron microscope. This work was supported by NIH grant 1-P01-GM62580 to N.G., NIH grant 1-RO1-MH63105-01 to A.T.B and a fellowship from the Max-Kade Foundation to J.F.
References
- Braithwaite S.P., Xia,H. and Malenka,R.C. (2002) Differential roles for NSF and GRIP/ABP in AMPA receptor cycling. Proc. Natl Acad. Sci. USA, 99, 7096–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. (2001a) Structural insights into the molecular mechanism of calcium-dependent vesicle–membrane fusion. Curr. Opin. Struct. Biol., 11, 163–173. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. (2001b) Structure of proteins involved in synaptic vesicle fusion in neurons. Annu. Rev. Biophys. Biomol. Struct., 30, 157–171. [DOI] [PubMed] [Google Scholar]
- Cong M., Perry,S.J., Hu,L.A., Hanson,P.I., Claing,A. and Lefkowitz,R.J. (2001) Binding of the beta2 adrenergic receptor to N-ethylmaleimide-sensitive factor regulates receptor recycling. J. Biol. Chem., 276, 45145–45152. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B. and Brunger,A.T. (2003) Complete structure of p97/Valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Sutton,R.B., Brunger,A.T. and Jahn,R. (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA, 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming K.G. et al. (1998) A revised model for the oligomeric state of the N-ethylmaleimide-sensitive fusion protein, NSF. J. Biol. Chem., 273, 15675–15681. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. (1998) Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J. Mol. Biol., 277, 1033–1046. [DOI] [PubMed] [Google Scholar]
- Hanson P.I., Roth,R., Morisaki,H., Jahn,R. and Heuser,J.E. (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell, 90, 523–535. [DOI] [PubMed] [Google Scholar]
- Havelka W.A., Henderson,R. and Oesterhelt,D. (1995) Three-dimensional structure of halorhodopsin at 7 A resolution. J. Mol. Biol., 247, 726–738. [DOI] [PubMed] [Google Scholar]
- Hohl T.M., Parlati,F., Wimmer,C., Rothman,J.E., Sollner,T.H. and Engelhardt,H. (1998) Arrangement of subunits in 20 S particles consisting of NSF, SNAPs and SNARE complexes. Mol. Cell, 2, 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler J.T., Rostaing,P., Schiavo,G., Fritschy,J.M., Olsen,R., Triller,A. and Moss,S.J. (2001) The subcellular distribution of GABARAP and its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABA(A) receptors. Mol. Cell. Neurosci., 18, 13–25. [DOI] [PubMed] [Google Scholar]
- Lenzen C.U., Steinmann,D., Whiteheart,S.W., Weis,W.I. (1998) Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein. Cell, 94, 525–536. [DOI] [PubMed] [Google Scholar]
- May A.P., Misura,K.M., Whiteheart,S.W. and Weis,W.I. (1999) Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein. Nat. Cell Biol., 1, 175–82. [DOI] [PubMed] [Google Scholar]
- Mindell J.A. and Grigorieff,N. (2003) Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol., 142, 334–347. [DOI] [PubMed] [Google Scholar]
- Nishimune A., Isaac,J.T., Molnar,E., Noel,J., Nash,S.R., Tagaya,M., Collingridge,G.L., Nakanishi,S. and Henley,J.M. (1998) NSF binding to GluR2 regulates synaptic transmission. Neuron, 21, 87–97. [DOI] [PubMed] [Google Scholar]
- Ogura T. and Wilkinson,A.J. (2001) AAA+ superfamily ATPases: common structure–diverse function. Genes Cells, 6, 575–597. [DOI] [PubMed] [Google Scholar]
- Patel S. and Latterich,M. (1998) The AAA team: related ATPases with diverse functions. Trends Cell Biol., 8, 65–71. [PubMed] [Google Scholar]
- Rice L.M. and Brunger,A.T. (1999) Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly. Mol. Cell, 4, 85–95. [DOI] [PubMed] [Google Scholar]
- Rothman J.E. (1994) Mechanisms of intracellular protein transport. Nature, 372, 55–63. [DOI] [PubMed] [Google Scholar]
- Rouiller I., DeLaBarre,B., May,A.P., Weis,W.I., Brunger,A.T., Milligan,R.A. and Wilson-Kubalek,E.M. (2002) Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat. Struct. Biol., 9, 950–957. [DOI] [PubMed] [Google Scholar]
- Song C., Wang,Q. and Li,C.C. (2003) ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity and D1 contributes to the heat-induced activity. J. Biol. Chem., 278, 3648–3655. [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer,D., Jahn,R. and Brunger,A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature, 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Swaffield J.C. and Purugganan,M.D. (1997) The evolution of the conserved ATPase domain (CAD): reconstructing the history of an ancient protein module. J. Mol. Evol., 45, 549–563. [DOI] [PubMed] [Google Scholar]
- van Heel M., Harauz,G., Orlova,E.V., Schmidt,R. and Schatz,M. (1996) A new generation of the IMAGIC image processing system. J. Struct. Biol., 116, 17–24. [DOI] [PubMed] [Google Scholar]
- Vonck J. (2000) Parameters affecting specimen flatness of two-dimensional crystals for electron crystallography. Ultramicroscopy, 85, 123–129. [DOI] [PubMed] [Google Scholar]
- Whiteheart S.W., Rossnagel,K., Buhrow,S.A., Brunner,M., Jaenicke,R. and Rothman,J.E. (1994) N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol., 126, 945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart S.W., Schraw,T. and Matveeva,E.A. (2001) N-ethylmaleimide sensitive factor (NSF) structure and function. Int. Rev. Cytol., 207, 71–112. [DOI] [PubMed] [Google Scholar]
- Wriggers W., Milligan,R.A. and McCammon,J.A. (1999) SITUS: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol., 125, 185–195. [DOI] [PubMed] [Google Scholar]
- Yu R.C., Hanson,P.I., Jahn,R. and Brunger,A.T. (1998) Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nat. Struct. Biol., 5, 803–811. [DOI] [PubMed] [Google Scholar]
- Yu R.C., Jahn,R. and Brunger,A.T. (1999) NSF N-terminal domain crystal structure: models of NSF function. Mol. Cell, 4, 97–107. [DOI] [PubMed] [Google Scholar]