Abstract
Phosphorylation of the Neurospora circadian clock protein FREQUENCY (FRQ) regulates its degradation and the proper function of the clock. The mechanism by which FRQ undergoes degradation has not been established. Here we show that FRQ is likely ubiquitylated in vivo, and its proper degradation requires FWD1, an F-box/WD-40 repeat-containing protein. In the fwd1 disruption strains, FRQ degradation is severely impaired, resulting in the accumulation of hyperphosphorylated FRQ. Furthermore, the circadian rhythms of gene expression and the circadian conidiation rhythms are abolished in these fwd1 mutants. Finally, FRQ and FWD1 interact physically in vivo, suggesting that FWD1 is the substrate-recruiting subunit of an SCF-type ubiquitin ligase responsible for FRQ ubiquitylation and degradation. Together with the recent finding that Slimb (the Drosophila homolog of FWD1) is involved in the degradation of the Period protein in flies, our results indicate that FWD1 regulates the degradation of FRQ in Neurospora and is an evolutionarily conserved component of the eukaryotic circadian clock.
Keywords: circadian clock/FREQUENCY/FWD1/Neurospora/proteasome
Introduction
Circadian clocks are key cellular mechanisms that regulate a wide variety of physiological and molecular activities in eukaryotic and certain prokaryotic organisms. Similar to Drosophila and mammals, the circadian oscillator in Neurospora consists of autoregulatory negative feedback loops that include three essential components: FREQUENCY (FRQ), WHITE COLLAR-1 (WC-1) and WC-2 proteins (Loros and Dunlap, 2001; Young and Kay, 2001). In constant darkness, WC-1 and WC-2, the two PAS domain-containing transcription factors, form a heterodimeric complex through their PAS domains and bind to the frq promoter to activate its transcription (Crosthwaite et al., 1997; Talora et al., 1999; Cheng et al., 2001b, 2002, 2003; Froehlich et al., 2002, 2003). In addition to their essential role in the circadian oscillator, WC-1 and WC-2 are essential for virtually all light responses in Neurospora, including light resetting of the clock. Moreover, WC-1 has been shown recently to be the blue light photoreceptor (Froehlich et al., 2002; He et al., 2002). On the other hand, the FRQ homodimer represses its own transcription through its physical interactions with the WC-1–WC-2 complex (Aronson et al., 1994a; Cheng et al., 2001a; Denault et al., 2001; Merrow et al., 2001; Froehlich et al., 2003). Thus, FRQ acts as the negative element in this circadian feedback loop. Due to this critical role of FRQ in clock function, in addition to the aforementioned transcriptional regulation, its protein level is also regulated by protein degradation mechanisms. However, little is known about the degradation mechanisms of FRQ.
In several eukaryotic circadian organisms, phosphorylation of the circadian clock proteins has been shown to control their stability (Dunlap, 1999; Liu et al., 2000; Young and Kay, 2001). In Neurospora, like the circadian negative elements in other eukaryotic systems, FRQ is phosphorylated immediately following its synthesis (Edery et al., 1994; Garceau et al., 1997; Kloss et al., 1998; Liu et al., 2000; Lee et al., 2001). It then becomes extensively phosphorylated prior to its degradation. We and others have shown that phosphorylation of FRQ plays key roles in the clock function in Neurospora. Molecular, genetic and biochemical studies have identified three kinases that phosphorylate FRQ (Gorl et al., 2001; Yang et al., 2001, 2002). These kinases include casein kinase I (CKI), CKII and a calcium/calmodulin-dependent kinase (CAMK-1). Several lines of evidence strongly suggest that phosphorylation of FRQ regulates its stability. First, a kinase inhibitor that blocks FRQ phosphorylation in vivo reduces the degradation rate of FRQ and lengthens the period of the clock (Liu et al., 2000). Secondly, mutations of the FRQ phosphorylation sites lead to the stabilization of the FRQ protein and the lengthening of the periods of the clock (Liu et al., 2000; Gorl et al., 2001; Yang et al., 2003). Finally, disruption of the Neurospora CKII catalytic subunit or one of its regulatory subunits resulted in higher FRQ protein levels and slower FRQ degradation rate (Yang et al., 2002, 2003).
In addition to its role in mediating FRQ degradation, phosphorylation of FRQ by CKII has also been implicated in regulating the FRQ–WC interaction and the closing of the negative feedback loop (Yang et al., 2002). In the strain where the only catalytic subunit of CKII is disrupted, the levels of FRQ protein and frq RNA are high and arrhythmic, and the circadian control of gene expression is abolished (Yang et al., 2002). Consistent with this, disruption of one of the CKII regulatory subunits led to long period and low amplitude rhythms (Yang et al., 2003). Interestingly, CKII is also shown to be an important clock component in Arabidopsis and Drosophila (Sugano et al., 1998; Lin et al., 2002; Akten et al., 2003). Furthermore, a form of CKI in Drosophila and mammals is critical for the proper degradation of Period (Per) proteins (Kloss et al., 1998; Price et al., 1998; Suri et al., 2000; Akashi et al., 2002).
Aside from the role of phosphorylation of FRQ, little is known about the mechanism that mediates its degradation. In Drosophila and mammals, the degradation of Per proteins is mediated by the ubiquitin–proteasome pathway (Akashi et al., 2002; Yagita et al., 2002). In Drosophila, Slimb, an F-box/WD40 repeat-containing protein, recently has been suggested to be a component of the SCF (Skp1/Cullin/F-box protein) ubiquitin ligase (E3) that mediates the ubiquitylation of the phosphorylated Per protein (Grima et al., 2002; Ko et al., 2002). In slimb mutant flies, Per is hyperphosphorylated and loses its rhythmic fluctuation in protein levels and phosphorylation patterns. We show herein that FWD1 (F-box and WD40 repeat-containing protein 1), the Neurospora homolog of Drosophila Slimb, is critical for both FRQ degradation and clock function. The direct physical interaction between FRQ and FWD1 also suggests that FWD1 is part of the ubiquitin ligase complex responsible for FRQ ubiquitylation.
Results
Ophiobolin A promotes protein ubiquitylation and the formation of high molecular weight conjugates of FRQ in vivo
To understand the degradation mechanism of phosphorylated FRQ, we tested whether FRQ is ubiquitylated and then degraded by the proteasome in vivo. However, all the commonly used proteasome inhibitors, including LLN, MG115, MG132 and lactacystin, failed to enter the Neurospora cells, as no growth phenotypes were observed even after prolonged drug treatments (our unpublished data). The inability of these drugs to penetrate the Neurospora cell wall and/or cell membrane is not unusual since they also fail to enter the wild-type yeast cells. For Saccharomyces cerevisiae, the effects of these drugs on proteasome can only be examined in mutant strains with enhanced permeability (Lee and Goldberg, 1996). No such mutant Neurospora strains are currently available.
In the process of characterizing CAMK-1 (Yang et al., 2001), we tested the effects of several known calmodulin inhibitors. When wild-type Neurospora cells were treated with one of the calmodulin inhibitors, ophiobolin A (Leung et al., 1984), we noticed that the FRQ protein became hyperphosphorylated and a high molecular weight FRQ-specific smear was observed in these samples, reminiscent of the polyubiquitylated proteins (see Supplementary data available at The EMBO Journal Online). In addition, ophiobolin A treatment led to a significant increase in the ubiquitylated proteins as shown by the same blot probed with a monoclonal antibody against ubiquitin (see Supplementary data). Interestingly, the high molecular weight species of FRQ were not observed after treatment of cells with other calmodulin inhibitors (e.g. W-7), suggesting that they were not caused by the inhibition of calmodulin and might be due to an unknown effect of ophiobolin A. Furthermore, we showed that ophiobolin A treatment enhanced the ubiquitylation of cyclin B examined in Xenopus egg extracts (Tang et al., 2001; data not shown). Taken together, these data suggest that ophiobolin A has a general stimulatory effect on the ubiquitylation of eukaryotic proteins, and FRQ might be ubiquitylated in Neurospora cells.
Disruption of fwd1, the Neurospora homolog of Drosophila Slimb
If FRQ is ubiquitylated in Neurospora, its degradation should be mediated by the proteasome. Proteins that regulate the ubiquitylation of FRQ may be important components of the Neurospora circadian clock system. We took a candidate approach to identify the ubiquitin ligase for FRQ ubiquitylation. Because the Drosophila Slimb protein recently has been implicated in the degradation of Per, we searched in the Neurospora genome database for genes with similarity to Slimb and the mammalian β-TRCP proteins. Two such hypothetical proteins were identified in Neurospora, both of which contain an F-box and WD40 repeats. They share high similarity to Slimb and β-TRCP proteins and were named FWD1 (EAA26744) and FWD2 (EAA29133). FWD1 shares higher sequence similarity with the Slimb and β-TRCP proteins than does FWD2.
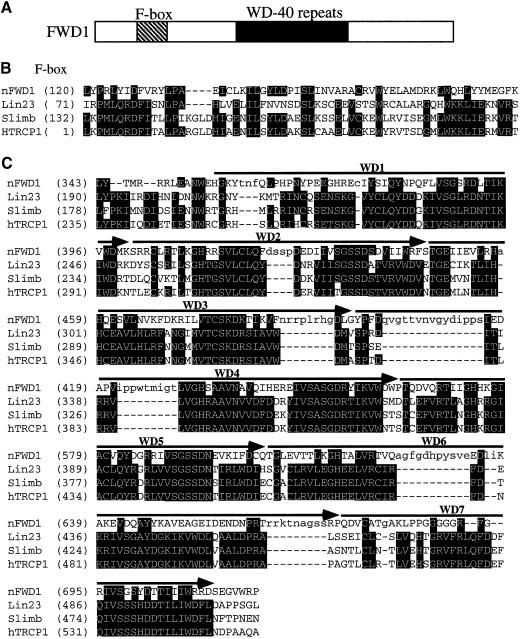
Sequence analysis of the cDNA revealed that the fwd1 gene contains one continuous exon and encodes a protein of 1010 amino acids. Although FWD1 is considerably larger than the Slimb homologs in other organisms, this difference is mostly due to the presence of the extra ∼300 residues at the C-terminus of the FWD1 protein (Figure 1A). When the FWD1 protein sequence was used in a Blast search against protein databases, it was found to be most similar to various animal Slimb homologs, including the Caenorhabditis elegans Lin-23, the human and mouse β-TRCP1 and 2, and the Drosophila Slimb (in the order of e values). The amino acid sequence alignment of the F-box and WD-40 regions of FWD1 and several animal Slimb homologs is shown in Figure 1B and C. The F-box regions of these proteins exhibit modest but significant sequence similarities. In contrast, the sequence similarities in their WD40 repeat regions are considerably higher (∼40% identity). Like the Slimb homologs, FWD1 appears to contain seven copies of WD40 repeats, which may form a seven-bladed β-propeller structure and bind to its phosphorylated substrates. The sequence similarity between FWD1 and the animal Slimb homologs suggests that FWD1 is a Slimb homolog in Neurospora.
Fig. 1. (A) Schematic depiction of the domain structure of FWD1. (B and C) Amino acid sequence alignment of the F-box domains (B) and the WD-40 repeat regions (C) from the Neurospora FWD1, C.elegans Lin23, Drosophila Slimb and human β-TRCP1. The alignments were based on the structural-based alignments (Orlicky et al., 2003). The seven WD40 repeats are labeled above.
F-box/WD40 repeat-containing proteins are often found to be the substrate-recruiting subunits of the SCF ubiquitin ligase complexes that mediate the ubiquitylation of phosphorylated substrates (Orlicky et al., 2003). To mediate substrate ubiquitylation, the F-box of these proteins interacts with Skp1, which in turn binds to the Cullin/RING modules responsible for the recruitment of the ubiquitin-conjugating enzyme (E2). The WD40 repeat region of these proteins confers substrate specificity through binding directly and selectively to the phosphorylated substrates. In several cases, the removal of the F-box domain in these proteins resulted in mutants that inhibited the functions of the full-length proteins in a dominant-negative manner. These mutants presumably retain their ability to bind to the substrates, but they cannot establish interactions with the rest of the SCF complex (Spencer et al., 1999).
To investigate the function of FWD1 in Neurospora, the fwd1 gene was disrupted in Neurospora by repeat-induced point mutation (RIP) (Cambareri et al., 1989). To mutate fwd1, a 3.6 kb genomic DNA containing the entire fwd1 open reading frame (ORF) and its 3′-untranslated region (UTR) was introduced into a wild-type strain at the his-3 locus. A transformant carrying an extra copy of fwd1 was then crossed with a wild-type strain, and individual sexual spores were picked and germinated. During the sexual cycle, RIP introduces random, but exclusively G–C to A–T, point mutations to the duplicated gene. After the spores were germinated, we noticed that some of the progenies exhibited slower growth rate and produced fewer aerial hyphae and conidia compared with those of the wild-type. To verify that the fwd1 gene was disrupted in these strains, the endogenous fwd1 genes from some of the strains were cloned and sequenced. DNA sequencing revealed that they all contained many G–C to A–T point mutations in the fwd1 gene, including premature stop codons in the FWD1 ORF. A fwd1RIP progeny (330-1), which contains a single copy of the fwd1 gene and grows on histidine-free medium, was used in most of the molecular analysis described herein. In this strain, there are multiple premature stop codons in the FWD1 ORF, including two in the F-box (residues 163 and 173). Thus, this strain can only produce a short truncated and non-functional FWD1, and it can be considered as a fwd1-null strain.
FRQ is hyperphosphorylated and more stable in the fwd1RIP strain
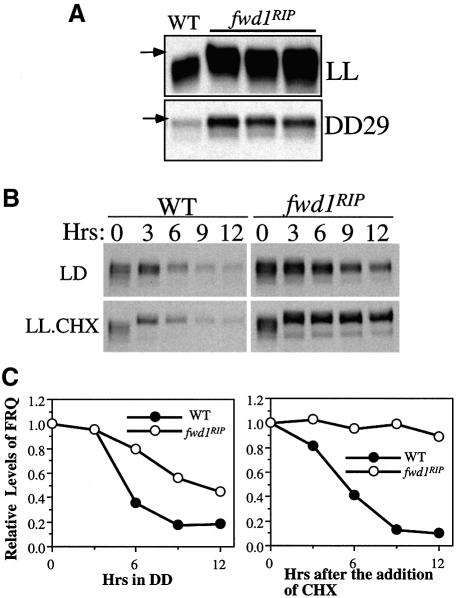
It has been shown previously that phosphorylation of FRQ is required for its degradation (Liu et al., 2000; Gorl et al., 2001; Yang et al., 2002). We have shown in this study that FRQ is ubiquitylated and presumably degraded by the proteasome. Thus, phosphorylation of FRQ may be a prerequisite for its ubiquitylation. If FWD1 is part of the ubiquitin ligase that mediates the ubiquitylation of FRQ, we would predict that, in the fwd1RIP strain, the degradation of FRQ should be blocked, resulting in higher FRQ protein levels. In addition, FRQ should become hyperphosphorylated in the fwd1 mutants. We thus examined the FRQ expression profile in the fwd1RIP strains. Western blot analyses were performed to examine the FRQ expression profile for Neurospora cultures grown in constant light (LL) or in constant darkness (DD29). As shown in Figure 2A, the levels of FRQ in all fwd1RIP progenies were significantly higher than that of the wild-type strain under both conditions. Furthermore, FRQ was hyperphosphorylated in the mutant strains, and some hyperphosphorylated FRQ species (indicated by the arrows) were not observed in the wild-type samples. The levels of FRQ in DD were lower than in LL in the fwd1RIP strains, suggesting that the expression of FRQ is still regulated by the light/dark transition. In contrast to the significant changes in the FRQ expression profile, the levels and phosphorylation patterns of WC-1 and WC-2 were not affected in the fwd1RIP strains (data not shown), indicating that the WC proteins were not substrates for FWD1.
Fig. 2. In the fwd1RIP strains, FRQ is hyperphosphorylated, and the FRQ protein levels are higher due to increased protein stability. (A) Western blot analysis showing that, in the fwd1RIP strains, FRQ was mostly hyperphosphorylated and its levels were higher than those of the wild-type strains. Cultures harvested in LL or DD29 were used. The arrows indicate the hyperphosphorylated FRQ species observed only in the fwd1RIP strains. (B and C) Western blot analyses showing that FRQ was more stable in the fwd1RIP strain after LD transition or cycloheximide (CHX) treatment (10 µg/ml). Cultures were first grown in LL for 1 day prior to the addition of CHX or were transferred into constant darkness and harvested at the indicated time. Densitometric analysis of the western blot results of (B) is shown in (C).
To examine whether the elevated levels of the FRQ protein were due to a decrease in the rate of degradation in the fwd1RIP strain, we measured the degradation rates of FRQ following a light to dark (LD) transition or after the addition of the protein synthesis inhibitor, cycloheximide (CHX). Both methods are reliable for comparing FRQ stability (Liu et al., 1997, 2000; Gorl et al., 2001; Yang et al., 2002). As shown in Figure 2B and C, the degradation of FRQ in the fwd1RIP strain was significantly slowed down under both conditions. The blockage of FRQ degradation was especially profound with the CHX treatment (in LL). In this case, ∼90% of FRQ was still not degraded after 12 h in the fwd1RIP strain, compared with only <10% of FRQ remaining in the wild-type strain. Some amounts of FRQ were degraded after the LD transition (Figure 2B and C), indicating that FRQ can also be degraded through a fwd1-independent mechanism under this condition. However, the degradation of FRQ was, nonetheless, significantly slowed down in the fwd1RIP strain after an LD transition. These results indicate that FWD1 is required for the proper degradation of FRQ.
Loss of circadian rhythms at the molecular level in the fwd1RIP strain
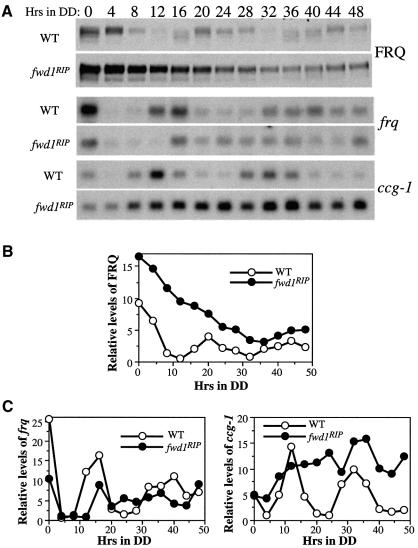
Because the degradation of FRQ was significantly impaired in the fwd1RIP strain, we expected that its circadian clock function was also severely compromised. To confirm this, we performed western and northern blot analyses to examine whether the levels of FRQ, frq and clock-controlled gene-1 (ccg-1) retained their circadian oscillation in constant darkness in the fwd1RIP strain. Figure 3 shows the results of the representative rhythmic experiments. Similar to previously described results (Garceau et al., 1997), robust circadian rhythms of the steady-state levels and phosphorylation patterns of the FRQ protein were observed in the wild-type strain in DD (Figure 3A and B). In contrast, FRQ remained hyperphosphorylated in DD and the circadian oscillations of the protein levels and phosphorylation patterns of FRQ were lost in the fwd1RIP strain. After the LD transition, the FRQ protein level in the mutant cells decreased gradually and reached a basal level that was higher than the peak FRQ level in the wild-type strain. Similar results were obtained in more than four independent experiments.
Fig. 3. Loss of circadian rhythms of gene expression in the fwd1RIP strain. (A) Cultures were harvested at the indicated time in constant darkness. The representative results of the western blot analysis of FRQ and northern blot analyses of frq and ccg-1 mRNA in both the wild-type and the fwd1RIP strains are shown. Densitometric analyses of the western blot results in (A) are shown in (B) and (C).
The decrease of FRQ level in the fwd1RIP strain after the LD transition may be partially explained by the rapid degradation of frq RNA immediately following the transition. As shown in Figure 3A and C, the frq RNA quickly disappeared after the LD transfer in the fwd1RIP strain and remained at a low level for ∼12 h. Afterwards, frq transcription in the mutant appeared to be reactivated and the level of the frq RNA randomly fluctuated between the peak and trough levels of the wild-type strain. The constant high levels of the FRQ protein and the intermediate levels of the frq RNA in the fwd1RIP strain suggest that the hyperphosphorylated FRQ protein might not be efficient in repressing its own transcription, thus failing to close the circadian feedback loop. This result also indicates that frq transcription can still respond to light in the fwd1RIP strain.
In contrast to the robust circadian rhythm of ccg-1 in the wild-type strain, the level of the ccg-1 message initially increased in DD in the fwd1RIP strain, and it then remained at a level comparable with its peak level in the wild-type strain throughout the rest of the time course (Figure 3A and C). The high level of ccg-1 RNA in the fwd1RIP strain was similar to that in a strain lacking the catalytic subunit of CKII (Yang et al., 2002), again suggesting that the hyperphoshorylated FRQ in the fwd1RIP strain, like the hypophosphorylated FRQ in the cka mutant strain, might not be fully functional. Taken together, these molecular data indicate that the circadian clock is dysfunctional in the fwd1RIP strain.
Loss of circadian conidiation rhythms in the fwd1RIP strains
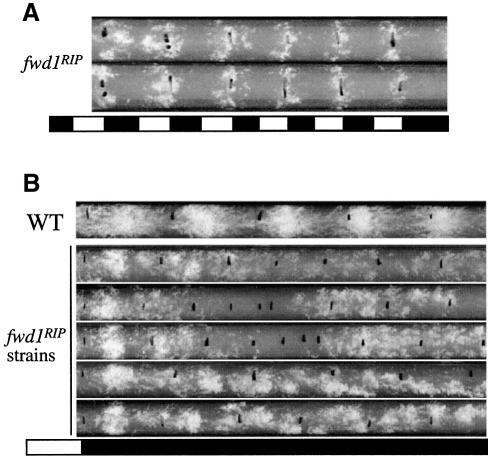
The lack of molecular rhythms suggests that the overt physiological rhythms should also be defective in the fwd1RIP strains. To confirm this prediction, race tube assays were used to examine their circadian conidiation rhythms. Although the fwd1RIP strains exhibited slower growth rates and produced fewer aerial hyphae and conidia, their growth and conidiation process could still be monitored by the race tube assay. Because the expression of both the frq RNA and the FRQ protein is still under the light/dark control in the fwd1RIP strains, the conidiation should be rhythmic in these strains under LD cycles. As expected, robust conidiation rhythms were observed under LD cycles in the fwd1RIP strains (Figure 4A).
Fig. 4. Loss of circadian conidiation rhythms in the fwd1RIP strains. (A and B) Race tube analysis results in light/dark cycles (A) or in constant dark after a LD transition (B). Severe independent fwd1RIP strains were examined, and the race tubes shown are representative samples from more than six replicate tubes for each strain.
The phenotypes of the fwd1RIP strains in the race tube assay in DD vary to some extent from strain to strain and from race tube to race tube. Representative race tube results from several independent fwd1RIP strains are shown in Figure 4B. During the first day in DD, one major conidiation band could be observed for all fwd1RIP strains. However, the phase of this conidiation band was 2–4 h more advanced than that of the wild-type strain. The appearance of this band in the first day was probably the result of the LD transition, as frq transcription was still light responsive. After the first day, however, no clear circadian conidiation rhythms could be observed in DD in all experiments. For some race tubes, arrhythmic conidiation was observed after the first day in DD (second race tube). In some race tubes (the two middle ones), the growth rate of the mutants significantly decreased after 2–3 days in DD. This slow growth phase lasted for several days, resulting in very few conidia. Afterwards, normal growth resumed and arrhythmic conidiation was observed. For some race tubes (the two bottom ones), unstable and very short period (∼12 h) conidiation rhythms were observed starting from the end of the first day in DD. The period and the phase of this short period banding rhythm varied significantly within the same race tube and from tube to tube. This race tube phenotype is reminiscent of the conidiation rhythms observed in frq null strains under certain conditions (Loros and Feldman, 1986; Aronson et al., 1994b), due to the existence of a residual frq-independent oscillator that controls the conidiation process. Together with the molecular data presented above, these results indicate that FWD1 is a crucial component of the Neurospora circadian clock.
FRQ physically interacts with FWD1 in vivo
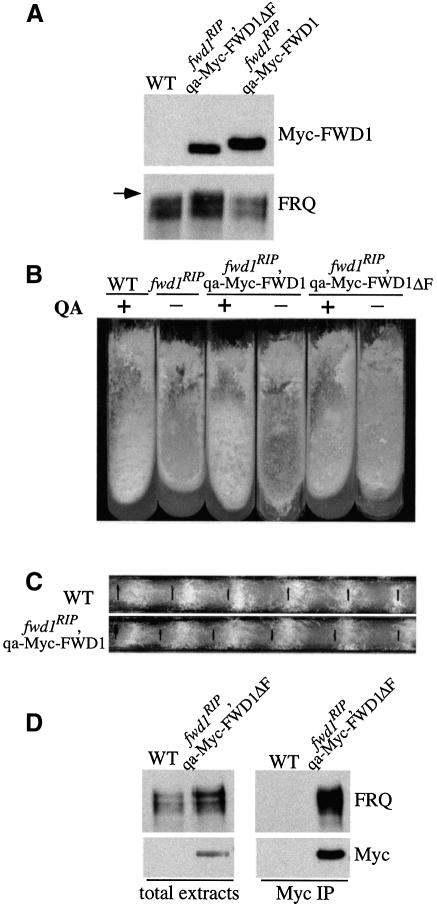
Our results described above suggest that FWD1 is a part of an SCF-type ubiquitin ligase complex that directly mediates the ubiquitylation of FRQ. If so, the WD-40 domain of FWD1 is expected to bind directly to the phosphorylated FRQ, which can then be ubiquitylated by other components in the FWD1-containing SCF complex. To detect a direct interaction between FRQ and FWD1, we made two constructs that contained the FWD1 ORF or the FWD1 ORF lacking the F-box (with residues 131–177 deleted, termed FWD1ΔF) under the control of the quinic acid (QA)-inducible promoter (Aronson et al., 1994a). In addition, five copies of the c-Myc epitope (Cheng et al., 2001a) were inserted at the N-terminus of the FWD1 ORF to facilitate the detection of the expression of Myc-FWD1 or Myc-FWD1ΔF using a c-Myc monoclonal antibody (9E10). Both constructs were transformed into the wild-type strain or the fwd1RIP strain, and the expression of the Myc-FWD1 or the Myc-FWD1ΔF in these two strains was confirmed by western blot analysis using the c-Myc antibody.
As shown in Figure 5A, Myc-FWD1 or Myc-FWD1ΔF was expressed in the fwd1RIP strains in the presence of QA. Furthermore, expression of Myc-FWD1 in the fwd1RIP strain led to a decrease of the FRQ protein level and the disappearance of the hyperphosphorylated forms of FRQ. When the fwd1RIP,qa-Myc-FWD1 strain was grown in minimal slants containing QA, the growth rate and the production of conidia and aerial hyphae (note the conidia and aerial production in the middle of the slants) were very similar to that of the wild-type strain (Figure 5B, comparing slants 1 with 3 and 4). In QA-containing race tubes, the circadian conidiation rhythms of the fwd1RIP,qa-Myc-FWD1 strains were also similar to those of the wild-type strains (Figure 5C). These data indicate that Myc-FWD1 can complement the function of the endogenous FWD1 protein in the fwd1RIP strain.
Fig. 5. FRQ and FWD1 interact physically in vivo. (A) Western blot analyses of the expression of FRQ and the Myc-FWD1 or Myc-FWD1ΔF proteins in fwd1RIP strains. QA was added into all cultures grown in LL. Note that the FRQ expression profile in the fwd1RIP,qa-Myc-FWD1 strain was similar to that of the wild-type strain (WT), while in the fwd1RIP,qa-Myc-FWD1ΔF strain, the FRQ profile was similar to that of the fwd1RIP mutants. The arrow indicates the hyperphosphorylated FRQ species observed in the fwd1RIP,qa-Myc-FWD1ΔF strain. (B) Growth phenotype of different strains in slants showing the rescue of the fwd1RIP phenotypes by the expression of the Myc-FWD1 protein, but not Myc-FWD1ΔF. (C) Circadian conidiation rhythm in the fwd1RIP,qa-Myc-FWD1 strain in the presence of QA (10–3 M). (D) Immunoprecipitation assay showing that FRQ and Myc-FWD1ΔF form a complex in vivo. The wild-type strain (WT) was used as the negative control. Cultures were grown in LL in the presence of QA (10–2 M). The total lysates or the immunoprecipitates were analyzed by immunoblotting using FRQ or c-Myc antibodies. The result shown is a representative example of multiple independent experiments.
In contrast, Myc-FWD1ΔF failed to rescue any of the molecular, growth, developmental and circadian phenotypes of the fwd1RIP strain (Figure 5A and B, and data not shown). Thus, the F-box domain of FWD1, its putative Skp1 interaction domain, is essential for its proper function. However, because the substrate-binding domain of FWD1 (its WD-40 repeat region) is intact in the Myc-FWD1ΔF protein, the interactions with its substrates are expected to be maintained and may even be enhanced due to the lack of subsequent ubiquitylation and degradation of these substrates in the fwd1RIP,qa-Myc-FWD1ΔF strain. Indeed, FRQ was present in the c-Myc immunoprecipitates, indicating that FRQ and Myc-FWD1ΔF form a complex in vivo in this strain (Figure 5D). The direct association between FRQ and FWD1 strongly suggests that FWD1 is a part of the SCF complex that mediates the ubiquitylation of FRQ in vivo. Interestingly, both the hypophosphorylated and hyperphosphorylated FRQ co-immunoprecipitated with Myc-FWD1ΔF, suggesting that all of these FRQ species might be targeted by the SCF complex. However, because FRQ mostly exists as homodimers (Cheng et al., 2001a), the interactions between different FRQ forms may explain the pull-down of the hypophosphorylated FRQ species.
In contrast to the strong association between FRQ and FWD1ΔF protein found in the fwd1RIP,qa-Myc-FWD1ΔF strain, no significant interaction between FRQ and the Myc-tagged FWD1 proteins was observed in the wt,qa-Myc-FWD1 and wt,qa-Myc-FWD1ΔF strains (data not shown). This is not surprising because the interaction between FWD1 and phosphorylated FRQ is expected to be transient and the FWD1-bound FRQ might be rapidly ubiquitylated and degraded in the wild-type strain. This is also consistent with the fact that the hyperphosphorylated FRQ forms observed in the fwd1RIP strains were absent in the wild-type strain (Figure 2A). In the wt,qa-Myc-FWD1ΔF strains, the QA-induced expression levels of FWD1ΔF may not be high enough for it to inhibit the function of the endogenous FWD1 protein in a dominant-negative fashion. This possibility is supported by our observations that the wt,qa-Myc-FWD1ΔF strains did not exhibit abnormal growth or developmental phenotypes even when high concentrations of QA (0.01 M) were used (data not shown). In our experience using the qa-2 promoter, we found that it is not a strong promoter that can induce gene expression to very high levels (Cheng et al., 2001b, 2002).
Discussion
Regulated degradation of clock proteins is an important mechanism for controlling circadian clocks. In this study, we uncovered a novel role for the known calmodulin inhibitor, ophiobolin A. It promotes protein ubiquitylation globally in Neurospora and other eukaryotic systems. Using ophiobolin A, we demonstrated that the critical Neurospora clock protein, FRQ, was ubiquitylated in vivo, suggesting that FRQ is degraded through the ubiquitin–proteasome pathway. Because the commonly used proteasome inhibitors are not cell permeable for fungi, including Neurospora and yeast (Lee and Goldberg, 1996), ophiobolin A provides an alternative pharmacological approach to identify substrates of the ubiquitin–proteasome pathway in these organisms.
To understand the mechanism of FRQ degradation, we identified FWD1, an F-box/WD40 repeat-containing protein and a Neurospora homolog of Drosophila Slimb. Disruption of the fwd1 gene revealed its critical role in the Neurospora circadian clock. In the fwd1RIP strains, FRQ is hyperphosphorylated and its level is high due to a significant increase of the FRQ protein stability. Although frq expression can still respond to light and the conidiation process can still be driven by light/dark cycles, the circadian rhythms of the protein levels and the phosphorylation patterns of FRQ are abolished in constant darkness. The loss of normal circadian function in the fwd1RIP strain is indicated further by the loss of circadian rhythms of frq, ccg-1 and the conidiation process. Finally, we showed that FRQ and FWD1 interacted directly in vivo. Taken together, our results suggest that FWD1 is a part of an SCF-type ubiquitin ligase complex responsible for FRQ ubiquitylation and degradation.
FWD1 mediates the degradation of phosphorylated FRQ
Phosphorylation of FRQ has been shown previously to regulate the degradation of FRQ and the function of the clock (Liu et al., 2000; Gorl et al., 2001; Yang et al., 2002). On the other hand, F-box/WD40 repeat-containing proteins are often the substrate-recruiting components of the SCF-type ubiquitin ligases. They have been shown to bind to a wide variety of phosphorylated substrates, leading to their ubiquitylation and degradation (Orlicky et al., 2003). In this study, we showed that FWD1, an F-box/WD40 repeat-containing protein, is required for the proper degradation of FRQ and it physically interacts with phosphorylated FRQ. Therefore, it is very likely that phosphorylated FRQ is a substrate for an FWD1-containing SCF-type ubiquitin ligase complex. We failed to detect the interaction between FRQ and FWD1 in the wild-type strain. This is very probably due to the transient nature of this interaction. The FWD1-associated FRQ is expected to be rapidly ubiquitylated and degraded by the 26S proteasome in the wild-type strain. Consistent with this notion, a strong interaction between FRQ and FWD1 was indeed observed in a fwd1RIP strain containing the FWD1 protein lacking its F-box domain (Figure 5). Deletion of its F-box in FWD1 allows it to bind to the phosphorylated substrates, but abolished its ability to mediate their ubiquitylation, thus stabilizing the FWD1–FRQ interaction.
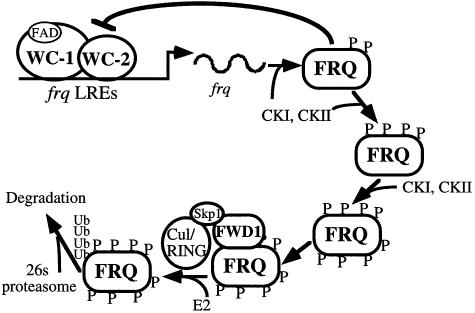
Like the Per proteins in Drosophila and mammals, FRQ is progressively phosphorylated and becomes extensively phosphorylated before its levels decrease (Edery et al., 1994; Garceau et al., 1997; Liu et al., 2000; Lee et al., 2001). What is the purpose of the progressive and extensive phosphorylation? Are the hyperphosphorylated FRQ species the only forms targeted for ubiquitylation by an FWD1-containing SCF complex? Mutagenesis studies showed that several of the identified and the potential phosphorylation sites of FRQ are important for FRQ degradation (Liu et al., 2000; Gorl et al., 2001; Yang et al., 2003). Interestingly, these sites are located at different regions of the FRQ protein, suggesting that the stability of FRQ is not solely determined by a single phosphorylation event. Instead, it might be controlled by phosphorylation at many sites in a dynamic and quantitative process. We showed in this study that the less phosphorylated forms of FRQ could still form complexes with FWD1, suggesting they may be targeted for ubiquitylation. However, this result could also be due to the FRQ–FRQ interactions (Cheng et al., 2001a). On the other hand, the hyperphosphorylated forms of FRQ should be better substrates for the SCF complex because they can only be detected in the fwd1RIP strains. Thus, it is likely that the SCF complex can recognize different phosphorylated motifs within FRQ. Extensive FRQ phosphorylation increases the number of potential FWD1-binding sites and its overall affinity toward FWD1, in a manner similar to that of the interaction between the F-box protein, Cdc4, and its phosphorylated substrates (Orlicky et al., 2003). Therefore, we propose that progressive phosphorylation of FRQ at multiple sites is a dynamic process that fine-tunes the stability of FRQ and thus the period of the clock (Figure 6). It may also help to generate a critical delay in the circadian negative feedback loop that allows the clock to cycle with a 24 h periodicity.
Fig. 6. A model for FWD1-mediated FRQ degradation by the ubiquitin–proteasome pathway. FRQ, WC-1 and WC-2 form the circadian negative feedback loop. FRQ is progressively phosphorylated by CKI, CKII and possibly other kinases. After FRQ is phosphorylated extensively, it interacts with FWD1 and is rapidly ubiquitylated and degraded.
Although the degradation of FRQ was significantly slower in the fwd1RIP strains after an LD transition, FRQ was still degraded at a modest rate, indicating that a fwd1-independent mechanism can mediate FRQ degradation under LD conditions. Similar observations have been reported for Per in Drosophila (Grima et al., 2002; Ko et al., 2002). The FRQ fluctuation in the fwd1RIP strains provides an explanation for the conidiation rhythms under LD cycles and the first conidiation band in DD (Figure 4). We do not yet know whether the fwd1-independent FRQ degradation mechanism is non-specific protein degradation or proteasome-mediated degradation. However, it is worth mentioning that ophiobolin A treatment can still induce the high molecular weight FRQ species in a fwd1RIP strain (data not shown), suggesting that additional F-box/WD40-containing proteins, such as FWD2, may mediate FRQ ubiquitylation in the absence of FWD1.
Conserved roles of the post-translational regulators of the eukaryotic circadian clocks
The cloning of frq 14 years ago (McClung et al., 1989) raised the question of whether the divergent eukaryotic circadian systems have evolved independently or are evolutionarily linked. The former possibility was supported by the apparent lack of bona fide functional and sequence homologs within the core oscillators among different circadian systems. The general architecture of the circadian feedback loops in Neurospora, including the negative feedback loop and the interconnected positive feedback loops, resembles those found in Drosophila and mammals (Dunlap et al., 1999; Lee et al., 2000; Cheng et al., 2001b; Young and Kay, 2001). However, FRQ and the WC proteins are not generally considered to be the sequence homologs of the Per, Clock and Bmal1 proteins in these systems, despite the fact that all of the positive elements in each system contain PAS domains. With the availability of the Neurospora and Arabidopsis genome sequences, it is clear that these organisms do not have sequence homologs of the animal core clock components. This had led to the belief that the fungal, plant and animal circadian clocks evolved independently from each other.
Since the identification of the Drosophila doubletime (dbt) gene that encodes a form of CKI and is the first known post-translational clock regulator (Kloss et al., 1998; Price et al., 1998), several lines of evidence accumulated in the last 2 years and presented in this study raise the possibility that the post-translational mechanisms are the evolutionary links among divergent eukaryotic circadian systems. First, the kinases that phosphorylate the negative elements appear to be conserved among fungi, plants and animals. As in Drosophila and mammals, a Neurospora CKI protein similar to DBT binds to and phosphorylates FRQ and possibly regulates the stability of FRQ (Kloss et al., 1998; Gorl et al., 2001; Lee et al., 2001). In addition, we showed that the Neurospora CKII functions as a critical clock component. It phosphorylates FRQ and regulates the stability of FRQ and its role in closing the feedback loop (Yang et al., 2002, 2003). CKII was implicated previously to play a role in the Arabidopsis circadian clock by phosphorylating CCA1 (Sugano et al., 1998), and it was also shown recently to be an important clock component in Drosophila (Lin et al., 2002; Akten et al., 2003). The conservation of CKII in different systems leads to the speculation that it may also be a clock component in the mammalian circadian clocks. It should be emphasized that, although both CKI and CKII are Ser/Thr protein kinases, they are significantly different in their structures and substrate specificities. Secondly, phosphorylation of the negative elements of the circadian clock leads to their degradation, another common theme of different eukaryotic clock systems (Kloss et al., 1998; Price et al., 1998; Liu et al., 2000; Akashi et al., 2002; Yagita et al., 2002). Finally, as demonstrated in this study, the mechanism of degradation of the phosphorylated negative clock elements and the components involved in the degradation machinery are conserved in different eukaryotic systems.
The ubiquitin–proteasome pathway is important for clock function in all eukaryotic circadian systems examined from fungi, plants, Drosophila to mammals (Naidoo et al., 1999; Akashi et al., 2002; Grima et al., 2002; Ko et al., 2002; Yagita et al., 2002; Kim et al., 2003). Although the ubiquitin ligases have not been identified in every system, this study showed that Neurospora and Drosophila use similar SCF-type ubiquitin ligase complexes to mediate the ubiquitylation and degradation of the negative components of their circadian feedback loops (Grima et al., 2002; Ko et al., 2002). This remarkable conservation suggests that the β-TRCP proteins may perform a similar role in the mammalian circadian clock.
Why do the Neurospora FRQ and the Drosophila Per proteins use similar ubiquitin ligases for their degradation even though their protein sequences are not related? The answer may lie in the fact that they are both phosphorylated by CKI and CKII. FRQ and Per may share similar short peptide motifs and/or structural features at the tertiary structure level that are recognized by these kinases. Phosphorylation of FRQ and Per by these common kinases generates similar phosphopeptide motifs that might mediate their binding to FWD1 and Slimb, respectively. In conclusion, the conservation of the post-translational regulators in divergent circadian systems suggests that the molecules mediating the modification and degradation of clock proteins may be the common foundation that allows the evolution of circadian clocks in eukaryotic systems.
Materials and methods
Strains and culture conditions
The bd, a and bd, his-3, A (wild-type clock) strains were used as the wild-type strains in this study. The bd, fwd1RIP and bd, fwd1RIP, his-3 strains were created in this study. The 301-6 (bd, his-3, A) strain and the bd, fwd1RIP, his-3 strain were the host strains for the his-3 targeting constructs. Liquid culture conditions were as described previously (Aronson et al., 1994a). Race tube assay medium contained 1× Vogel’s, 0.1% glucose, 0.17% arginine, 50 ng/ml biotin and 1.5% agar. For experiments in which the qa-Myc-FWD1 strains were used, 0.01 M QA pH 5.8 was added to liquid culture medium containing 1× Vogel’s, 0.1% glucose and 0.17% arginine (Cheng et al., 2001b). For ophiobolin A treatment, cultures were treated with 50 µM ophiobolin A (Sigma) for 3 h before harvesting. Monoclonal c-Myc and ubiquitin antibodies were purchased from Santa Cruz Biotechnology.
Disruption of fwd1 in Neurospora
The Neurospora fwd1 gene was disrupted by RIP. A 3.6 kb PCR fragment containing the entire FWD1 ORF and its 3′-UTR was cloned into pDE3dBH and introduced into the his-3 locus of a wild-type strain (301-6: bd, his-3, A) by electroporation (Margolin, 1999). Southern blot analysis was performed to identify transformants that carried the additional copy of fwd1 at the his-3 locus, and a positive transformant was crossed with a wild-type strain (bd, a). Sexual spores of the cross were picked individually and germinated on slants containing histidine. The endogenous fwd1 genes were amplified by PCR from the potential fwd1RIP progeny that grow on histidine-free medium, and DNA sequencing revealed that they all contained many G–C to A–T mutations in the endogenous fwd1 gene. A fwd1RIP strain (330-1) was then crossed with a bd, his-3 strain to obtain a bd, fwd1RIP, his-3 strain that was used as the host strain for Myc-FWD1-containing plasmids.
Plasmids
To make the his-3 targeting qa-FWD1 constructs, a PCR fragment containing the entire FWD1 ORF and its 3′-UTR was cloned into pqa.5Myc to create pqa-Myc-FWD1. The pqa.5Myc plasmid was derived from pDE3dBH-qa-2 (Cheng et al., 2001b) in which five copies of the c-Myc epitope tag were inserted downstream of the qa-2 promoter. The pqa-Myc-FWD1ΔF construct contained a deletion of residues 131–177 of the fwd1 ORF that was generated by a site-specific mutagenesis kit (Clontech Inc., Palo Alto, CA) using pqa-Myc-FWD1 as the template. The pqa-Myc-FWD1 and pqa-Myc-FWD1ΔF constructs were then transformed into the bd, his-3, A or bd, fwd1RIP, his-3 strain. Western blot analyses using a monoclonal c-Myc antibody (9E10, Santa Cruz Biotechnology) were used to identify transformants expressing the Myc-tagged FWD1 proteins.
Protein and RNA analyses
Protein extraction, quantification, western blot analysis and immunoprecipitation assay were performed as previously described (Garceau et al., 1997; Cheng et al., 2001a). Immunoprecipitates or equal amounts of total protein (40 µg) were loaded in each protein lane. After electrophoresis, proteins were transferred onto PVDF membrane, and western blot analysis was performed.
RNA extraction and northern blot analysis were performed as previously described (Aronson et al., 1994a; Crosthwaite et al., 1995). Equal amounts of total RNA (20 µg) were loaded onto agarose gels for electrophoresis; the gels were blotted and probed with RNA probes (Aronson et al., 1994a; Loros et al., 1989).
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Lixin Wang and Gabriel Pineda for excellent technical assistance, and three anonymous reviewers for suggestions. Supported by grants from the National Institutes of Health and Welch Foundation to Y.L. and H.Y. Y.L. and H.Y. are endowed scholars in Biomedical Research at UT Southwestern Medical Center.
References
- Akashi M., Tsuchiya,Y., Yoshino,T. and Nishida,E. (2002) Control of intracellular dynamics of mammalian period proteins by casein kinase Iε (CKIε) and CKIδ in cultured cells. Mol. Cell. Biol., 22, 1693–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akten B., Jauch,E., Genova,G.K., Kim,E.Y., Edery,I., Raabe,T. and Jackson,F.R. (2003) A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci., 6, 251–257. [DOI] [PubMed] [Google Scholar]
- Aronson B., Johnson,K., Loros,J.J. and Dunlap,J.C. (1994a) Negative feedback defining a circadian clock: autoregulation in the clock gene frequency. Science, 263, 1578–1584. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Johnson,K.A. and Dunlap,J.C. (1994b) The circadian clock locus frequency: a single ORF defines period length and temperature compensation. Proc. Natl Acad. Sci. USA, 91, 7683–7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambareri E.B., Jensen,B.C., Schabtach,E. and Selker,E.U. (1989) Repeat-induced G–C to A–T mutations in Neurospora. Science, 244, 1571–1575. [DOI] [PubMed] [Google Scholar]
- Cheng P., Yang,Y., Heintzen,C. and Liu,Y. (2001a) Coiled-coil domain mediated FRQ–FRQ interaction is essential for its circadian clock function in Neurospora. EMBO J., 20, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang,Y. and Liu,Y. (2001b) Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl Acad. Sci. USA, 98, 7408–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang,Y., Gardner,K.H. and Liu,Y. (2002) PAS domain-mediated WC-1/WC-2 interaction is essential for maintaining the steady state level of WC-1 and the function of both proteins in circadian clock and light responses of Neurospora. Mol. Cell. Biol., 22, 517–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P., Yang,Y., Wang,L., He,Q. and Liu,Y. (2003) WHITE COLLAR-1, a multifunctional Neurospora protein involved in the circadian feedback loops, light sensing and transcription repression of wc-2. J. Biol. Chem., 278, 3801–3808. [DOI] [PubMed] [Google Scholar]
- Crosthwaite S.K., Loros,J.J. and Dunlap,J.C. (1995) Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell, 81, 1003–1012. [DOI] [PubMed] [Google Scholar]
- Crosthwaite S.K., Dunlap,J.C. and Loros,J.J. (1997) Neurospora wc-1 and wc-2: transcription, photoresponses and the origins of circadian rhythmicity. Science, 276, 763–769. [DOI] [PubMed] [Google Scholar]
- Denault D.L., Loros,J.J. and Dunlap,J.C. (2001) WC-2 mediates WC-1–FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J., 20, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C. (1999) Molecular bases for circadian clocks. Cell, 96, 271–290. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C., Loros,J.J., Liu,Y. and Crosthwaite,S.K. (1999) Eukaryotic circadian systems: cycles in common. Genes Cells, 4, 1–10. [DOI] [PubMed] [Google Scholar]
- Edery I., Zweibel,L., Dembinska,M. and Rosbash,M. (1994) Temporal phosphorylation of the Drosophila period protein. Proc. Natl Acad. Sci. USA, 91, 2260–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froehlich A.C., Liu,Y., Loros,J.J. and Dunlap,J.C. (2002) White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science, 297, 815–819. [DOI] [PubMed] [Google Scholar]
- Froehlich A.C., Loros,J.J. and Dunlap,J.C. (2003) Rhythmic binding of a WHITE COLLAR-containing complex to the frequency promoter is inhibited by FREQUENCY. Proc. Natl Acad. Sci. USA, 100, 5914–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garceau N., Liu,Y., Loros,J.J. and Dunlap,J.C. (1997) Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell, 89, 469–476. [DOI] [PubMed] [Google Scholar]
- Gorl M., Merrow,M., Huttner,B., Johnson,J., Roenneberg,T. and Brunner,M. (2001) A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J., 20, 7074–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima B., Lamouroux,A., Chelot,E., Papin,C., Limbourg-Bouchon,B. and Rouyer,F. (2002) The F-box protein slimb controls the levels of clock proteins period and timeless. Nature, 420, 178–182. [DOI] [PubMed] [Google Scholar]
- He Q., Cheng,P., Yang,Y., Wang,L., Gardner,K.H. and Liu,Y. (2002) White collar-1, a DNA binding transcription factor and a light sensor. Science, 297, 840–843. [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Geng,R. and Somers,D.E. (2003) Circadian phase-specific degradation of the F-box protein ZTL is mediated by the proteasome Proc. Natl Acad. Sci. USA, 100, 4933–4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B., Price,J.L., Saez,L., Blau,J., Rothenfluh,A. and Young,M.W. (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell, 94, 97–107. [DOI] [PubMed] [Google Scholar]
- Ko H.W., Jiang,J. and Edery,I. (2002) Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature, 420, 673–678. [DOI] [PubMed] [Google Scholar]
- Lee C., Etchegaray,J.P., Cagampang,F.R., Loudon,A.S. and Reppert,S.M. (2001) Posttranslational mechanisms regulate the mammalian circadian clock. Cell, 107, 855–867. [DOI] [PubMed] [Google Scholar]
- Lee D.H. and Goldberg,A.L. (1996) Selective inhibitors of the proteasome-dependent and vacuolar pathways of protein degradation in Saccharomyces cerevisiae. J. Biol. Chem., 271, 27280–27284. [DOI] [PubMed] [Google Scholar]
- Lee K., Loros,J.J. and Dunlap,J.C. (2000) Interconnected feedback loops in the Neurospora circadian system. Science, 289, 107–110. [DOI] [PubMed] [Google Scholar]
- Leung P.C., Taylor,W.A., Wang,J.H. and Tipton,C.L. (1984) Ophiobolin A. A natural product inhibitor of calmodulin. J. Biol. Chem., 259, 2742–2747. [PubMed] [Google Scholar]
- Lin J.M., Kilman,V.L., Keegan,K., Paddock,B., Emery-Le,M., Rosbash,M. and Allada,R. (2002) A role for casein kinase 2α in the Drosophila circadian clock. Nature, 420, 816–820. [DOI] [PubMed] [Google Scholar]
- Liu Y., Garceau,N., Loros,J.J. and Dunlap,J.C. (1997) Thermally regulated translational control mediates an aspect of temperature compensation in the Neurospora circadian clock. Cell, 89, 477–486. [DOI] [PubMed] [Google Scholar]
- Liu Y., Loros,J. and Dunlap,J.C. (2000) Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl Acad. Sci. USA, 97, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros J.J. and Dunlap,J.C. (2001) Genetic and molecular analysis of circadian rhythms in Neurospora. Annu. Rev. Physiol., 63, 757–794. [DOI] [PubMed] [Google Scholar]
- Loros J.J. and Feldman,J.F. (1986) Loss of temperature compensation of circadian period length in the frq-9 mutant of Neurospora crassa. J. Biol. Rhythms, 1, 187–198. [DOI] [PubMed] [Google Scholar]
- Loros J.J., Denome,S.A. and Dunlap,J.C. (1989) Molecular cloning of genes under the control of the circadian clock in Neurospora. Science, 243, 385–388. [DOI] [PubMed] [Google Scholar]
- Margolin B.S., Freitag,M. and Selker,E.U (1999) Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet. Newslett., 44, 24–36. [Google Scholar]
- McClung C.R., Fox,B.A. and Dunlap,J.C. (1989) The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature, 339, 558–562. [DOI] [PubMed] [Google Scholar]
- Merrow M., Franchi,L., Dragovic,Z., Gorl,M., Johnson,J., Brunner,M., Macino,G. and Roenneberg,T. (2001) Circadian regulation of the light input pathway in Neurospora crassa. EMBO J., 20, 307–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naidoo N., Song,W., Hunter-Ensor,M. and Sehgal,A. (1999) A role for the proteasome in the light response of the timeless clock protein. Science, 285, 1737–1741. [DOI] [PubMed] [Google Scholar]
- Orlicky S., Tang,X., Willems,A., Tyers,M. and Sicheri,F. (2003) Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell, 112, 243–256. [DOI] [PubMed] [Google Scholar]
- Price J.L., Blau,J., Rothenfluh,A., Adodeely,M., Kloss,B. and Young,M.W. (1998) double-time is a new Drosophila clock gene that regulates PERIOD protein accumulation. Cell, 94, 83–95. [DOI] [PubMed] [Google Scholar]
- Spencer E., Jiang,J. and Chen,Z.J. (1999) Signal-induced ubiquitination of IκBα by the F-box protein Slimb/β-TrCP. Genes Dev., 13, 284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S., Andonis,C., Green,R., Wang,Z.Y. and Tobin,E.M. (1998) Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl Acad. Sci. USA, 95, 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V., Hall,J.C. and Rosbash,M. (2000) Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J. Neurosci., 20, 7547–7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talora C., Franchi,L., Linden,H., Ballario,P. and Macino,G. (1999) Role of a white collar-1–white collar-2 complex in blue-light signal transduction. EMBO J., 18, 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj,R., Li,B. and Yu,H. (2001) Mad2-independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell, 1, 162–164. [DOI] [PubMed] [Google Scholar]
- Yagita K., Tamanini,F., Yasuda,M., Hoeijmakers,J.H., van der Horst,G.T. and Okamura,H. (2002) Nucleocytoplasmic shuttling and mCRY-dependent inhibition of ubiquitylation of the mPER2 clock protein. EMBO J., 21, 1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Cheng,P., Zhi,G. and Liu,Y. (2001) Identification of a calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein FREQUENCY. J. Biol. Chem., 276, 41064–41072. [DOI] [PubMed] [Google Scholar]
- Yang Y., Cheng,P. and Liu,Y. (2002) Regulation of the Neurospora circadian clock by casein kinase II. Genes Dev., 16, 994–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Cheng,P., He,Q., Wang,L. and Liu,Y. (2003) Phosphorylation of FREQUENCY protein by casein kinase II is necessary for the function of the Neurospora circadian clock. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.W. and Kay,S.A. (2001) Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet., 2, 702–715. [DOI] [PubMed] [Google Scholar]