Abstract
H2A.Z has been shown to regulate transcription in yeast, and that function resides in its C-terminal region as the reciprocal portion of H2A cannot substitute for the latter. We show that fusion of a transcriptional activating region to the C-terminal region of H2A, which is substituted for that of H2A.Z, can allow the chimera to fulfil the special role of H2A.Z in positive gene regulation, as well as complement growth deficiencies of htz1Δ cells. We further show that the ‘transcription’ function of H2A.Z is linked to its ability to preferentially localize to certain intergenic DNA regions. Our results suggest that H2A.Z modulates functional interactions with transcription regulatory components, and thus increases its localization to promoters where it helps poise chromatin for gene activation.
Keywords: H2A.Z/PUR5/transcription/yeast
Introduction
Chromatin is known to present a formidable obstacle to many cellular processes including gene transcription. Thus it is generally believed that nucleosomes must be remodelled at certain regions of a given gene in order to trigger efficient gene induction. Current views suggest that there are three classes of chromatin remodelling mechanism. The first involves the action of proteins that hydrolyze ATP as a source of energy. Such complexes include, for example, the well-studied Swi/Snf complex (Vignali et al., 2000; Havas et al., 2001; Becker and Hörz, 2002). The second mechanism involves protein factors that chemically modify histone proteins, such as the SAGA histone acetyltransferase complex (Roth et al., 2001). Both of the remodelling complexes mentioned above have been shown to be recruited to promoter regions, namely by the action of gene-specific transcriptional activators (Utley et al., 1998; Neely et al., 1999; Yudkovski et al., 1999; Kuo et al., 2000; Brown et al., 2001). The third mechanism involves the incorporation of histone variants that can presumably create ‘specialized’ regions of chromatin allowing it to become more permissive to the process of transcription. An example of this can be illustrated by histone variant H3.3 in Drosophila. H3.3 differs from regular H3 by only four amino acids, and it gradually replaces H3 in differentiated cells in a replication-independent fashion upon exiting the cell cycle (Ahmad and Henikoff, 2002). The replacement of H3 by H3.3 forces the remodelling of nucleosomes, and thus the quick removal of post-transcriptional modifications to histones. The outcome of this scenario gives rise to rapid induction of genes that were previously repressed by the chemical modification of histones (Ahmad and Henikoff, 2002). Another variant histone that could potentially create such specialized chromatin is H2A.Z.
Variant histone H2A.Z has been identified in such diverse eukaryotic organisms as humans (Hatch and Bonner, 1988), mice (Faast at al., 2001), sea urchins (Ernst et al., 1987), chickens (Harvey et al., 1983), Xenopus laevis (Iouzalen et al., 1996), Drosophila (van Daal et al., 1988), Tetrahymena (White et al., 1988) and yeasts (Carr et al., 1994; Jackson et al., 1996), and it has been found to be essential in many of these organisms including mice (Faast et al., 2001). In Tetrahymena, H2A.Z has been shown to be associated with transcriptionally active chromatin (Stargell et al., 1993). Experiments carried out in Saccharomyces cerevisiae have validated the importance of the H2A.Z variant histone in both positive and negative gene transcription (Dhillon and Kamakaka, 2000; Santisteban et al., 2000; Adam et al., 2001; Meneghini et al., 2003). Importantly, H2A.Z has been shown to be essential for the recruitment of both RNA polymerase II (pol II) and the TATA-binding protein (TBP) to the GAL1-10 promoters (Adam et al., 2001), and it also performs redundant functions with both the Swi/Snf and SAGA chromatin remodelling complexes (Santisteban et al., 2000). A recent report has suggested that the function of H2A.Z in yeast is also redundant with H2B ubiquitination in the formation of active chromatin (Hwang et al., 2003). An insightful study in Drosophila has revealed that the essential function of H2A.Z, compared with regular H2A, resides in its C-terminal region (Clarkson et al., 1999). These authors have shown that substitution of H2A.Z amino acids for those of H2A in a short α-helix at the C-terminus was essential for fly viability. We have previously shown that substitution of the H2A.Z C-terminal region by that of H2A in yeast could not fulfil the role of H2A.Z in GAL1-10 gene induction in a yeast strain bearing a deletion of HTZ1 (htz1Δ), the H2A.Z-encoding gene (Adam et al., 2001). On the other hand, fusion of the H2A.Z C-terminal region to H2A could complement a htz1Δ mutation for GAL1-10 gene induction. These results suggest that the essential role of H2A.Z in GAL gene transcription resides in the C-terminal region of the variant histone and that this function of H2A.Z cannot be provided by regular H2A. Importantly, structure determination of an H2A.Z-containing nucleosome particle (Suto et al., 2000) has revealed an altered surface that includes a metal ion. The authors of that study suggest that this surface could lead to changes in higher-order structure and/or could be involved in mediating interactions between H2A.Z and other nuclear proteins. In addition, the H2A.Z C-terminal region, through its docking domain, is essential in creating a large interaction surface with (H3–H4)2 tetramers. A recent report has shown that H2A.Z-containing nucleosome particles facilitate intranucleosomal interactions while simultaneously inhibiting intermolecular nucleosome associations (Fan et al., 2002). In contrast to H2A, H2A.Z forms a non-uniform binding pattern on Drosophila polytene chromosomes (Leach et al., 2000), and it preferentially binds promoter DNA of some, but not all genes in S.cerevisiae (Santisteban et al., 2000; Meneghini et al., 2003). Meneghini and colleagues (Meneghini et al., 2003) have also shown that many H2A.Z-activated genes cluster near telomeres in yeast, and that variant histone protects euchromatin from telomeric silencing as reduced expression of those genes in htz1Δ cells is reversed by deletion of SIR2. These important results suggest that H2A.Z possesses an additional function that allows it to bind selectively to certain genomic regions, the significance of which is still unknown.
In order to study the mechanism by which H2A.Z can modulate positive gene transcription, we set out to study further the functional role of the C-terminal region of the variant histone. In this study, we show that transcriptional activation domains can provide the special function of the H2A.Z C-terminal region in gene induction, and they can also complement various growth deficiencies of htz1Δ cells. We also demonstrate that the H2A.Z C-terminal region allows H2A.Z to bind preferentially the promoter regions of the PUR5 and PHO5 genes. Finally, we show that both the ‘locator’ and ‘transcription’ functions of the H2A.Z C-terminal region are linked.
Results
Transcriptional activating regions can confer the special function of H2A.Z in positive gene transcription and growth on hydroxyurea
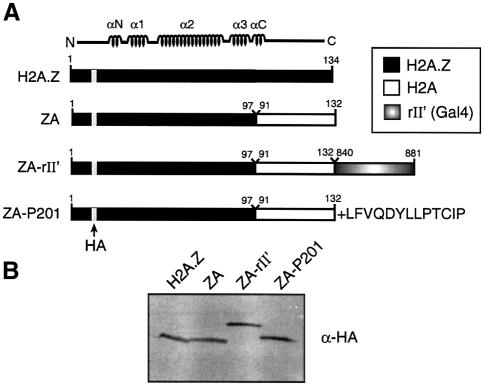
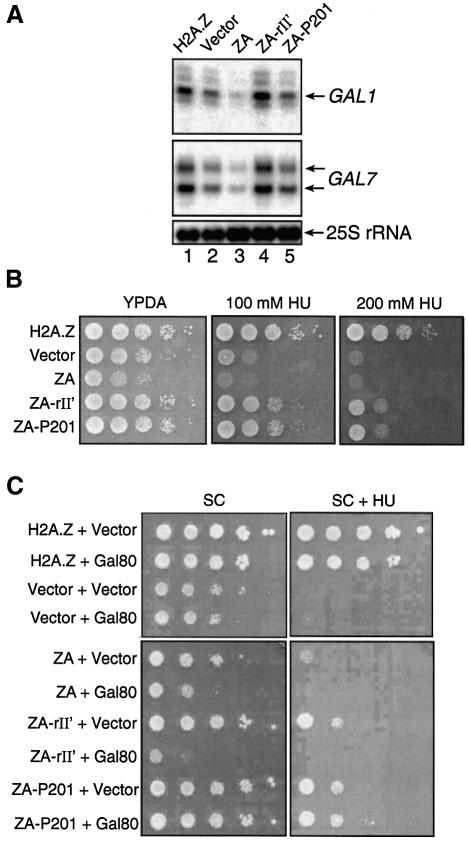
Recent reports have shown that H2A.Z is important for gene expression in yeast, with possible roles in chromatin remodelling and transcriptional elongation (Santisteban et al., 2000; Adam et al., 2001; Krogan et el., 2003). Importantly, H2A.Z is required for appropriate recruitment of RNA pol II at the yeast GAL1-10 genes under inducing conditions, and the C-terminal region of H2A.Z also interacts with RNA pol II components in vitro (Adam et al., 2001). It is interesting to note that both of the above mentioned functions can also be elicited by a DNA-tethered transcriptional activating region (e.g. Ptashne and Gann, 1997). In addition, it has been suggested (Suto et al., 2000) that within an H2A.Z-containing nuclesome particle, the latter variant histone has an acidic patch in its C-terminal region that extends to H2B. Based on all these facts, we hypothesized that one special function of H2A.Z in gene transcription, as compared with regular H2A, would be to contact components of the transcriptional machinery and/or chromatin remodelling components directly. According to this prediction, the H2A.Z C-terminal region should harbour a function reminiscent of a transcriptional activating region when incorporated into a nucleosomal particle. However, investigation of the specific function of the H2A.Z C-terminal region in gene transcription is difficult to assess because of its importance in nucleosome stability, namely in the docking domain (Suto et al., 2000). Accordingly, we have found that deletion of the H2A.Z C-terminal region prevented stable expression of the latter in yeast (M.Larochelle and L.Gaudreau, unpublished data). Hence, in order to test our model directly, we first made use of the ZA fusion protein fused to the Gal4 acidic transcriptional activating region (ZA-rII′) (see Figure 1A). ZA bears the N-terminal region as well as the core domain of H2A.Z, fused to the C-terminal region of H2A (Figure 1A). Such a fusion protein is expected to be able to assemble into nucleosome particles but would lack the special function of H2A.Z in positive gene transcription (Adam et al., 2001). We reasoned that if ZA-rII′ could complement the phenotypes of htz1Δ cells and rescue transcription defects, then the function of H2A.Z in positive gene transcription would overlap with that of a transcriptional activating region. We also wanted to test a non-acidic activating region that functions in yeast. Hence we made a chimera consisting of ZA fused to a synthetic activating region, P201 (Lu et al., 2000) (Figure 1A). Western blotting experiments reveal that all fusion proteins used in our experiments were expressed at comparable levels (Figure 1B). Deletion of HTZ1 negatively affects GAL1 and GAL7 gene induction as previously reported (Figure 2A, lane 2) (Adam et al., 2001). In addition, expression of the ZA fusion also negatively affects GAL gene induction (lane 3) to a further extent than the empty vector alone in htz1Δ cells. We have observed in certain experiments that expression of ZA downregulates GAL transcription to a greater extent than expressing no H2A.Z derivative (e.g. empty vector in the htz1Δ strain). Importantly, expression of ZA-rII′ could efficiently restore full activation of the GAL1 and GAL7 genes in htz1Δ cells (lane 4). ZA-P201 could reverse the negative effect of ZA on transcription but could not fully restore GAL transcription (lane 5).
Fig. 1. Structure and expression of variant histone protein chimeras used in this study. (A) Schematic representation of the chimeras. The location of the HA tag is shown as a gray bar. (B) Immunoblot analysis of H2A.Z chimeras. Histone protein levels were determined by immunoblotting with an anti-HA antibody.
Fig. 2. Fusion of a transcriptional activating region to ZA is sufficient to restore GAL1 and GAL7 transcription, as well as the HU-sensitivity phenotype of htz1Δ yeast cells. (A) Ability of H2A.Z derivatives to induce GAL1 and GAL7 expression in htz1Δ cells. Primer extension analyses of GAL1 and GAL7 expression upon galactose induction. The 25S rRNA is shown at the bottom of the figure as a loading control. (B) Ability of variant H2A.Z derivatives to complement htz1Δ- mediated HU growth deficiency. Ten-fold serial dilutions of yeast cell suspensions were dropped on YPDA plates supplemented or not with either 100 or 200 mM HU. Plates were incubated at 30°C for 2–5 days. (C) Overexpression of Gal80 in Δhtz1 cells expressing ZA-rII′. Δhtz1 cells containing the indicated chimeras were transformed with either an empty vector or a Gal80 expression vector under the control of the ACT1 promoter on a 2µ plasmid. Serial dilutions of cells were spotted on plates without or with HU and grown for 3–6 days at 30°C.
In addition to their inability to grow on certain carbon sources, slow growth and thermosensitivity (ts) phenotype, we have also previously observed that htz1Δ cells are highly sensitive to hydroxyurea (HU) (J.Brodeur and L.Gaudreau, unpublished data). HU is known to arrest cells in S phase, and previous reports have shown that a complete replication block can occur when yeast cells are treated with 200 mM HU (Allen et al., 1994). Hence, we also tested if our ZA constructs fused to activating regions could rescue HU sensitivity of htz1Δ yeast cells. Figure 2B first shows that htz1Δ yeast cells bearing either an empty vector or expressing the ZA fusion have a marked growth deficiency on rich media containing HU. As expected, expression of wild-type (WT) H2A.Z rescues that deficiency, as does the expression of both ZA-rII′ and ZA-P201. Again, these results support the possibility that the H2A.Z C-terminal region harbours a function that resembles that of a transcriptional activating region since both types of activating region allow ZA to complement htz1Δ-associated defects.
GAL gene transcription is strongly induced by the Gal4 activator, which in turn can be negatively regulated by Gal80, a specific inhibitor that binds the Gal4 rII′ activating region in the absence of galactose (Ma and Ptashne, 1987a; Bash and Lohr, 2001). In order to verify whether the action of the Gal4 activating region was indeed responsible for the observed effects with ZA-rII′, we overexpressed Gal80 to ensure efficient titration of ZA-rII′. Thus we made use of the HU growth deficiency phenotype of htz1Δ cells to test the specificity of rII′ in our assays. Figure 2C shows that ZA-rII′ can reverse the slow growth and HU-sensitivity phenotypes of htz1Δ cells, and that overexpression of Gal80 can reverse the complementation effect of the rII′ activating region. Gal80 has no effect on yeast cells expressing WT H2A.Z and ZA-P201, an anticipated result since Gal80 is not expected to bind either of these two proteins. Similar results were also obtained with the ts phenotype of htz1Δ cells (data not shown).
Replacement of the H2A.Z C-terminal region by the corresponding H2A region strongly inhibits PUR5 induction
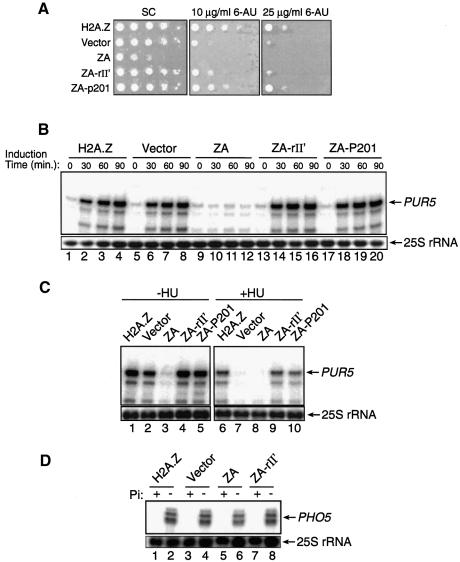
We have previously observed that htz1Δ cells are somewhat sensitive to 6-azauracil (6-AU) (M.Adam and L.Gaudreau, unpublished data), a result suggesting that H2A.Z can be important for transcriptional elongation since 6-AU is known to deplete GTP and UTP cellular pools (Shaw and Reines, 2000). This depletion makes RNA pol II more susceptible to pausing during elongation, thus making the role of elongation factors essential under those conditions. We thus decided to investigate further the behaviour of our ZA fusions with growth assays in the presence of 6-AU. Figure 3A shows that htz1Δ cells are significantly more sensitive to 6-AU when compared with either cells expressing WT H2A.Z or the ZA-rII′ and ZA-P201 fusions. The figure further shows that expression of the ZA fusion can completely prevent yeast growth even at a concentration of 10 µg/ml 6-AU, and is consistent with the results obtained with ZA in our GAL gene assays (e.g. Figure 2A). Since we observed a growth defect on 6-AU plates, especially with the ZA fusion, we decided to investigate the transcriptional activity of the PUR5 gene with our different ZA fusion proteins. PUR5 is a gene that encodes an IMP dehydrogenase that is activated by the presence of 6-AU (Exinger and Lacroute, 1992; Shaw and Reines, 2000; Escobar-Henriques and Daignan-Fornier, 2001). In the presence of 6-AU, yeast cells harbouring a defect in PUR5 expression are usually defective in transcriptional elongation, but cells sensitive to 6-AU when PUR5 is fully expressed are not (Shaw and Reines, 2000). Thus, to test if H2A.Z is important for PUR5 induction, yeast cells were grown in synthetic complete (SC) medium lacking uracil and 6-AU was added to a final concentration of 75 µg/ml to induce PUR5 expression fully. Figure 3B shows primer extension analyses of PUR5 transcription. Upon addition of 6-AU, the PUR5 gene can be strongly induced when WT H2A.Z is present (Figure 3B, lanes 1–4). The PUR5 gene can still be efficiently induced even in the absence of any H2A.Z derivative (Figure 3B, lanes 5–8), whereas the expression of ZA completely prevented PUR5 induction (Figure 3B, lanes 9–12). Finally, expression of either ZA-II′ (Figure 3B, lanes 13–16) or ZA-P201 (Figure 3B, lanes 17–20) rescued PUR5 induction as compared with ZA. These results suggest that the 6-AU sensitivity of htz1Δ cells is not solely due to a PUR5 transcriptional defect, but perhaps to other genes inhibited by 6-AU such as those encoding for an orotidylate decarboxylase (Shaw and Reines, 2000).
Fig. 3. Effect of H2A.Z derivatives on PUR5 and PHO5 transcription. (A) Growth of cells in the absence or presence of 6-AU. Cells were grown and then spotted onto plates containing no 6-AU, 10 µg/ml 6-AU or 25 µg/ml 6-AU. (B) Primer extension analysis of the PUR5 gene in the presence of 6-AU. Induction time in the presence of 6-AU is shown in minutes. The 25S rRNA is shown at the bottom of the figure as a loading control. (C) Transcription of the PUR5 gene in the presence of HU. Primer extension analysis of the PUR5 gene after yeast cells were grown and incubated or not for 3.5 h with 200 mM HU. The 25S rRNA is shown at the bottom of the figure as a loading control. (D) Transcription of the PHO5 gene. Primer extension analyses of the PHO5 locus after yeast cells were grown in the presence (+) or absence (–) of phosphate (Pi). The 25S rRNA is shown at the bottom of the figure as a loading control.
We next wanted to determine if preventing DNA replication would affect PUR5 induction in htz1Δ cells. We reasoned that if H2A.Z has a redundant function with other cellular factors possibly involved in transcription, then generally crippling DNA synthesis-dependent chromatin remodelling might make PUR5 transcription dependent on H2A.Z. In order to block DNA replication, we made use of HU as in Figure 2B. Thus Figure 3C shows similar PUR5 primer extension analyses to those in Figure 3B but all are under activated conditions. In the absence of HU, PUR5 induction is only slightly decreased in htz1Δ cells (compare lanes 1 and 2), whereas expression of ZA prevents PUR5 induction as previously observed (compare lanes 1 and 3). Lanes 4 and 5 show transcription levels at PUR5 when ZA-rII′ and ZA-P201 are expressed. When HU is added to yeast cultures to arrest DNA synthesis, PUR5 induction is dramatically affected in htz1Δ cells (compare lanes 6 and 7) as well as when ZA is expressed (lane 8). Expression of ZA-rII′ and ZA-P201 is able to recover PUR5 transcription levels fully (lanes 9 and 10). These results suggest that H2A.Z function is essential for PUR5 induction in the absence of DNA replication.
We next attempted to test the effect of ZA on PHO5 transcription in order to determine if the dominant negative effect of the latter molecule was manifested at all genes that we tested. PHO5 expression is repressed in the presence of inorganic phosphate but is strongly induced when starved of the latter (Svaren and Hörz, 1997). Thus, Figure 3D shows primer extension analyses of the PHO5 locus from yeast cells expressing various H2A.Z derivatives under repressed and induced conditions. As previously reported (Santisteban et al., 2000), PHO5 induction is not significantly affected in htz1Δ cells when compared with WT cells or cells expressing ZA-rII′ (compare lanes 2, 4 and 8). Moreover, expression of ZA also had little or no effect on PHO5 induction (lane 6). In addition, the results demonstrate that expression of ZA-rII′ does not derepress PHO5 transcription (compare lanes 1, 3 and 7). Similar results were obtained with the PHO8 gene (data not shown). These results first suggest that fusion of an activating region to ZA does not generally derepress gene transcription (see also Figure 3B for PUR5) and, secondly, that the dominant negative effect of ZA is somewhat gene specific.
H2A.Z derivatives are able to bind the PUR5 gene
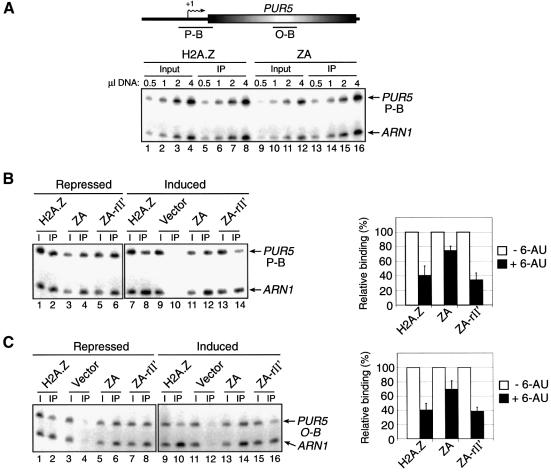
Previous reports have shown that H2A.Z binding is gradually lost upon gene induction (Santisteban et al., 2000; Adam et al., 2001) implying that the variant histone is remodelled prior to mRNA synthesis. In order to ensure that our ZA, ZA-rII′ and ZA-P201 fusion proteins can associate with chromatin, we tested their ability to bind the PUR5 gene using chromatin immunoprecipitation (ChIP) experiments, followed by quantitative PCR analysis. For our ChIP assays, we made use of the HA tag at the N-terminal end of the H2A.Z derivatives for specific immunoprecipitation. As an internal control for our ChIP experiments, we made use of the ARN1 gene as we found that its expression was not affected by H2A.Z nor induced in our yeast culture conditions. The ARN1 region analyzed by PCR lies within the promoter region and is constant throughout our experiments. The lower part of Figure 4A shows a PCR titration of input and immunoprecipitated material at the PUR5 promoter. The results show that both H2A.Z and ZA can efficiently bind the PUR5 promoter region, and also that our PCR amplifications are within linear range. Such titrations have been carried out for all the ChIP experiments presented in this paper (data not shown). Figure 4B (left panel) shows PCR analyses from a ChIP experiment designed to assess the presence of H2A.Z, ZA and ZA-rII′ over a PUR5 promoter region under repressed and induced conditions. In this experiment, ARN1 serves as an internal control to normalize signals for each lane. The right panel of Figure 4B shows a graphic representation of the binding efficiency at a region of the PUR5 promoter of the H2A.Z derivatives. The results indicate that all three derivatives can efficiently bind that promoter region under repressed conditions and, upon induction, H2A.Z and ZA-rII′ binding is significantly reduced compared with ZA binding. Figure 4C shows a similar experiment but with PCR analyses carried out in the coding region of PUR5. We have also been able to locate ZA-P201 at the PUR5 locus as well as all the H2A.Z derivatives at the GAL1 locus (data not shown). Taken together, these results show that all our H2A.Z derivatives are able to bind the PUR5 gene and are thus likely to incorporate into nucleosomes.
Fig. 4. Binding of H2A.Z derivatives at the PUR5 gene. ChIP experiments were performed using anti-HA antibodies to monitor the binding of H2A.Z, ZA and ZA-rII′ to PUR5 under repressed (–6-AU) or induced (+6-AU) conditions. All PCRs contain ARN1 primers used here as an internal control to normalize signals for each lane. (A) Top: representation of the PUR5 locus. Transcription start site (arrows with +1), open reading frame (open shaded rectangle) and regions amplified by PCR (black bars) are represented. Bottom: PCR titration of input (lanes 1–4 and 9–12) and immunoprecipitated material (lanes 5–8 and 13–16) for H2A.Z (lanes 1–8) and ZA (lanes 9–16) at the PUR5 promoter. (B) ChIP analysis of the binding of H2A.Z derivatives at the PUR5 promoter. Binding of the derivatives under repressed (–6-AU, lanes 1–6) and induced (+6-AU, lanes 7–14) conditions (left panel). The right part of the figure depicts a quantification of that experiment. Binding of each H2A.Z derivative under repressed conditions is normalized to 100%. (C) Same experiment as in (B) with the exception that the PUR5 region analyzed was in the open reading frame.
The localization patterns of H2A.Z and ZA are different at the PUR5 gene
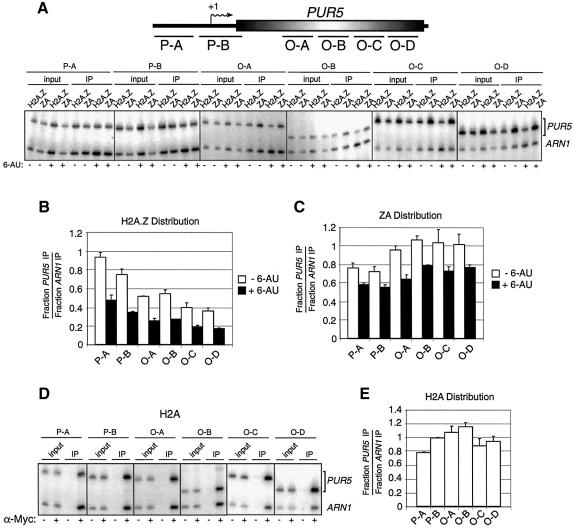
Santisteban et al. (2000) have previously demonstrated that H2A.Z is preferentially associated with intergenic regions as compared with open reading frames at the PHO5 and GAL1 loci. To test if the H2A.Z C-terminal region is important for proper localization of the variant histone on DNA, we used ChIP experiments to compare the binding of H2A.Z and ZA at the PUR5 gene under repressed and induced conditions. The upper part of Figure 5A depicts the regions of PUR5 that were analyzed by PCR. Figure 5A shows the PCR analyses of the different PUR5 regions. Figure 5B shows a graphic representation of the WT H2A.Z distribution over the PUR5 gene under either repressed or induced conditions. The results suggest that H2A.Z is located more abundantly in the promoter region and significantly less in the coding region. However, we observe a totally different distribution pattern with ZA (Figure 5C) in that it is found to be less abundant in the promoter region than in the coding region. In addition, it appears that H2A.Z binding throughout the PUR5 locus is more strongly diminished upon induction of PUR5 as compared with ZA.
Fig. 5. Localization of H2A.Z, ZA and H2A at the PUR5 locus. (A) Top: representation of the PUR5 locus. Regions amplified by PCR in the promoter region (P-A and P-B) or in the open reading frame (O-A to O-D) are represented. Bottom: ChIP analysis of the binding of H2A.Z and ZA at the PUR5 locus. All PCRs contain ARN1 primers used here as an internal control to normalize signals for each lane. (B and C) Distribution of H2A.Z and ZA at the PUR5 locus. Quantification of (B) H2A.Z or (C) ZA under repressed (–6-AU, open bar) and induced (+6-AU, black bar) conditions. (D) ChIP analysis of the binding of Myc-H2A at the PUR5 locus. Cells were grown under repressed condition and chromatin was immunoprecipitated with (+) or without (–) an anti-Myc antibody. (E) Distribution of H2A over the PUR5 locus.
We next wanted to compare the distribution of H2A over the PUR5 gene and assess whether it would be similar to that of H2A.Z or ZA. For this purpose, we made use of a yeast strain in which one H2A-encoding gene has been Myc tagged (Adam et al., 2001). Thus, Figure 5D shows PCR analyses of a ChIP experiment using anti-Myc antibodies to assess the distribution of H2A over the PUR5 locus under repressed conditions. Figure 5E shows a quantification of the H2A distribution over the PUR5 locus. The results clearly indicate that the latter’s distribution closely resembles that of ZA. Taken together, these results suggest that H2A.Z is more abundant in the PUR5 promoter region than in the coding region, whereas the distribution of ZA and H2A is spread out more uniformly and is even slightly enriched in the coding region compared with the promoter region.
The localization patterns of H2A.Z and ZA are different at the PHO5 gene
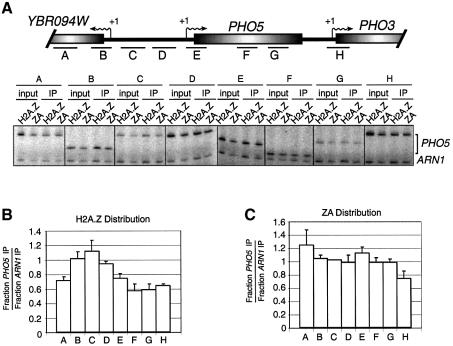
Since it has been shown (Santisteban et al., 2000) that H2A.Z preferentially localizes to the PHO5 intergenic region and that the distribution of H2A is rather uniform, we decided to investigate the distribution of our ZA fusion at that genomic region. As can be seen in Figure 6B, the distribution of H2A.Z over that locus is very similar to the previous report (Santisteban et al., 2000), i.e. H2A.Z is preferentially localized at the PHO5 promoter region compared with the coding region. However, as shown in Figure 6C, the distribution of ZA is more uniform and certainly not preferentially located to the intergenic region, a result similar to the previously published distribution of H2A at that locus (Santisteban et al., 2000). We have also obtained a similar distribution for H2A at PHO5 (data not shown) to that reported previously (Santisteban et al., 2000). These results further support our observations at the PUR5 gene and suggest that the H2A.Z C-terminal region is also responsible for the preferential binding to the PHO5 intergenic region.
Fig. 6. Distribution of H2A.Z and ZA at the PHO5 genomic region. (A) Top: representation of a 4.3 kb genomic region spanning the PHO5 gene. The transcription start sites (arrows with +1), open reading frames (open shaded rectangle) and regions amplified by PCR (black bars) are represented. Bottom: ChIP analysis of H2A.Z and ZA binding over the PHO5 genomic region. Cells were grown under PHO5 repression conditions. All PCRs contain ARN1 primers used here as an internal control to normalize signals for each lane. (B and C) Distribution of (B) H2A.Z and (C) ZA over the PHO5 genomic region.
The ZA-rII′ fusion has a distribution similar to H2A.Z over the PUR5 and PHO5 genes
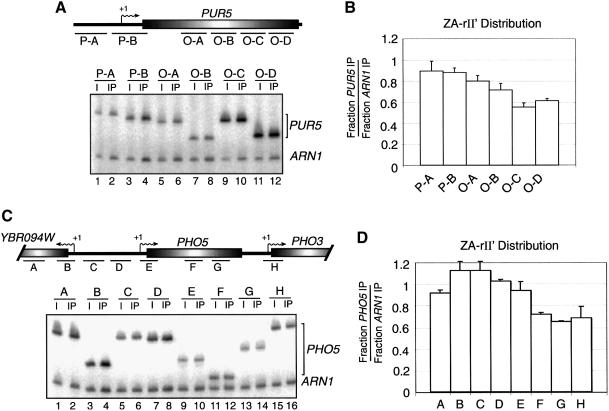
In order to test if the ‘locator’ function of the H2A.Z C-terminal region can be functionally separable from its function in transcription, we first carried out a ChIP experiment to determine the distribution of ZA-rII′ over the PUR5 gene under repressed conditions. We reasoned that if the locator function of the H2A.Z C-terminal region can be dissociated from its function in transcription, then the ZA-rII′ distribution should be similar to that of ZA and H2A. Figure 7A depicts PCR analyses of the ZA-rII′ ChIP over the PUR5 locus. Figure 7B shows a graphical representation of the ZA-rII′ distribution over that locus. Figure 7C and D shows a similar experiment, but at the PHO5 genomic region. At both genomic regions investigated, the results show that the distribution of the ZA-rII′ chimera is similar to that of WT H2A.Z since it is more abundant over the promoter regions than the coding regions of both PUR5 and PHO5.
Fig. 7. Distribution of the ZA-rII′ fusion over the PUR5 and PHO5 genomic regions. (A) Binding of ZA-rII′ at PUR5 gene under repressed conditions. (B) Distribution of ZA-rII′ at the PUR5 gene. (C) Same experiment as in (A) but the PHO5 genomic region was analyzed. (D) Distribution of ZA-rII′ at the PHO5 genomic region. All PCRs contain ARN1 primers used as an internal control to normalize signals for each lane.
Discussion
In this work, we have shown that addition of two different transcriptional activating regions to ZA, one taken from a natural activator (rII′) and one synthetic (P201), can allow the latter to complement efficiently most transcription and growth deficiencies of htz1Δ cells that we have tested so far. In addition, at least in the case of ZA-rII′, the activating region alone specifically confers to the C-terminal region of H2A the missing function found in H2A.Z since this activity can be specifically inhibited by Gal80. Thus we suggest that the H2A.Z C-terminal region is different from the reciprocal H2A region in that it harbours an ‘activating region-like surface’, but only when it is incorporated into a nucleosome particle. In fact, the crystal structure of a H2A.Z-containing nucleosome reveals an uninterrupted acidic patch at the surface of the nucleosome that starts at the H2A.Z C-terminus and extends to H2B, thus creating a potential surface that could mediate novel protein–protein interactions (Suto et al., 2000). In contrast, an H2A-containing nucleosome does not bear such a surface. Thus it is likely that this acidic patch might serve as a specialized activating region whose integrity would be dependent on the formation of a nucleosome. The primary mechanism by which transcriptional activating regions are believed to stimulate transcription, at least in yeast, is by recruitment of the transcriptional machinery to a target promoter (e.g. Ptashne and Gann, 1997; Barberis and Gaudreau, 1998; Keaveney and Struhl, 1998). The Gal4 activating region (rII′) has been shown to interact with numerous transcription factors including TBP and TFIIB (Wu et al., 1996), Srb4 (Koh et al., 1998), the SAGA histone acetyltransferase complex (Melcher and Johnston, 1995; Bhaumik and Green, 2001; Brown et al., 2001; Larschan and Winston, 2001) and the NuA4 acetyltransferase complex (Brown et al., 2001). The P201 synthetic activating peptide has been shown to interact with the Gal11 mediator component (Lu et al., 2002). It thus seems likely that activating regions can interact with a plethora of different factors involved in the process of gene transcription. The fact that both activating regions fused to ZA can complement htz1Δ defects to a different extent may reflect their intrinsic ability to recruit appropriate factors for a given gene. Thus we imagine that H2A.Z could recruit certain transcription factors that would, for example, help poise promoter chromatin for gene activation once an appropriate induction signal is triggered. Importantly, in the absence of such a gene induction signal, H2A.Z would not be expected to derepress genes since expression of ZA-rII′ does not derepress the PUR5, PHO5 or PHO8 genes prior to their activation.
We have also shown that the function of the H2A.Z C-terminal region may act in concert with other nuclear factors for PUR5 gene induction. Support for this claim is based on the fact that the ZA fusion strongly prevents PUR5 induction while the absence of any H2A.Z derivative does not significantly affect PUR5 induction. However, preventing DNA replication, a condition that makes transcription-dependent chromatin remodelling necessary, renders htz1Δ cells incapable of efficiently inducing PUR5 transcription. Similarly, PHO5 gene induction has been shown to be independent of H2A.Z as well as the Swi/Snf chromatin remodelling complex, but mutation of a Swi/Snf subunit renders H2A.Z essential for PHO5 induction (Santisteban et al., 2000). Thus it seems likely that the function of H2A.Z in transcription will become essential under a variety of conditions that may either compact chromatin, such as in mitosis, or even prevent DNA replication. Intriguingly, GAL1 gene induction, which is normally independent of Swi/Snf function (Gaudreau et al., 1997) becomes highly dependent on the latter complex when cells are arrested in mitosis (Krebs et al., 2000). Also, the function of H2A.Z could be essential where a number of transcription factors, including chromatin remodelling complexes, are crippled.
Our results also suggest that once H2A.Z performs its positive role in gene activation, it must then be remodelled for efficient gene induction to occur. In addition, our results suggest that, at least for the PUR5 gene, H2A.Z-bearing nucleosomes are more stable than H2A-bearing nucleosomes. This possibility is highlighted by the fact that ZA exerts a strong dominant negative effect at PUR5. In addition, a recent report has shown that H2A.Z-containing chromatin is more stably folded than H2A-containing chromatin (Fan et al., 2002), a result that may explain the requirement for more localized alterations caused by chromatin remodelling complexes (Santisteban et al., 2000). It is conceivable that H2A.Z-poised chromatin, by virtue of its C-terminal region, would be easier to remodel than ZA-containing chromatin given possible interactions with chromatin remodelling components. We propose that without its ‘activating region’ function, H2A.Z could become particularly repressive at certain genes. However, this dominant negative effect of ZA is not general since there is no significant effect at the PHO5 and PHO8 genes, and ZA has a more modest effect at GAL genes. We have also shown that H2A.Z binding is strongly reduced upon gene induction as compared with the ZA fusion, a result again suggesting that H2A.Z is more readily remodelled than ZA at PUR5. These results also correlate with the transcriptional activity of yeast cells expressing these fusion proteins. At this point, we do not know if the role of H2A.Z at PUR5 is exerted at the level of transcription initiation or elongation, or at both.
In Tetrahymena, H2A.Z has been shown to be associated with active chromatin, a result that further implies the importance of the variant histone in gene transcription, but also the fact that it has a distinct distribution pattern (Stargell et al., 1993). Experiments in Drosophila (Leach et al., 2000) and yeast (Santisteban et al., 2000; Meneghini et al., 2003) have shown that H2A.Z has a non-uniform binding pattern. In addition to its important role in gene induction, we have shown that the H2A.Z C-terminal region was also necessary to preferentially locate this variant histone to the PUR5 and PHO5 promoter regions as the distribution of the ZA fusion more closely resembled that of H2A. Importantly, the distribution of the ZA-rII′ chimera had more similarity to that of H2A.Z than ZA, suggesting that this ‘locator’ function of the C-terminus is linked to its role in gene induction. Thus we propose a model in which H2A.Z would first poise chromatin at an inactive gene by interaction with a transcription-related factor, or chromatin remodelling component, that might have certain gene-specific determinants. The fact that H2A.Z interacts with such a factor(s) would also allow it to preferentially localize to promoter regions, and upon an appropriate gene induction signal H2A.Z would become remodelled by virtue of the same interaction(s). The H2A.Z-poised chromatin would allow a gene to be remodelled and activated quickly. It is also conceivable that the so-called transcription-related factors, or chromatin remodelling factors, might be recruited to promoter regions because local chromatin architecture might readily accommodate these factors at promoter regions, but not in other regions of the genome, thus providing the required specificity function. This last suggestion may be more plausible since H2A.Z does not preferentially localize to the promoter regions of all genes (Meneghini et al., 2003). Consistent with our model is the fact that acetylation levels of H3 (lysine 9) and H4 (lysine 12) are predominant over the PHO5 promoter region, as compared with its coding region, and surprisingly under gene repression conditions (Vogelauer et al., 2000). This acetylation pattern remarkably resembles the distribution of H2A.Z over the same locus. In addition, the H3–H4 acetylation state at the PHO5 promoter region does not increase upon gene activation (Vogelauer et al., 2000), and these authors suggest that this preacetylated state of chromatin at PHO5 might also poise the promoter for gene activation. It is tempting to speculate that different chromatin-modifying enzymes could also share a similar distribution over promoter regions of inactive genes. Of course, our model does not exclude the possibility that H2A.Z-containing chromatin itself would be more permissive to transcription, for example by mimicking an acetylated state of chromatin as suggested by the in vitro studies of Fan et al. (2002).
Materials and methods
Strains and plasmids
The htz1Δ and Myc-H2A strains used in this study are as described (Adam et al., 2001). All H2A.Z derivatives bear an HA tag inserted at the BglII site of the HTZ1 gene and were expressed from the ACT1 promoter. The ZA fusion was made by fusing amino acids 91–132 of H2A to the C-terminal part of amino acids 1–97 of H2A.Z (Adam et al., 2001). ZA-rII′ was generated by adding amino acids 840–881 of Gal4 to the C-terminal end of ZA. ZA-P201 was constructed by adding the amino acid sequence LFVQDYLLPTCIP (Lu et al., 2000) to the C-terminus of ZA. The Gal80 expression plasmid was constructed by cloning its open reading frame in a high copy number vector (YEplac181, LEU2) under the control of the ACT1 promoter.
Phenotypic assays
For the HU-sensitivity growth assays, yeast cells were spotted on rich media (YPDA) with or without 100 or 200 mM HU, except for the Gal80 experiment where cells were spotted onto plates containing SC media lacking uracil and leucine. For the 6-AU-sensitivity growth assays, yeast cells were grown overnight to logarithmic phase in SC media lacking uracil and containing 2% glucose. Cells were washed, serially diluted and spotted on plates containing SC media lacking uracil but containing 2% glucose or 2% glucose plus 10 or 25 µg/ml 6-AU. Plates were incubated for 3–7 days at 30°C or as indicated.
Gene induction conditions and primer extension assays
PUR5 expression was induced essentially as described (Lee et al., 2002). Cells were grown to an OD600 of 0.6 and 6-AU was added to a final concentration of 75 µg/ml for 60 min. For GAL1–7 transcription, cells were grown to an OD600 of 0.6 in SC media containing 2% raffinose and galactose was then added to a final concentration of 5% for 60 min. In order to monitor PUR5 expression upon HU treatment, yeast cells were grown as above and subsequently washed with water. Cells were then blocked in S phase by incubating cultures in SC media lacking uracil with the indicated concentration of HU for 3.5 h at 30°C. After incubation, cultures were washed again in water and resuspended in SC media lacking uracil with 2% glucose containing HU and 75 µg/ml 6-AU for PUR5 induction. All inductions were carried out for 60 min. For PHO5 transcription, cells were grown to an OD600 of 0.6–1.0, washed with sterile water and resuspended in SC media lacking phosphate. Cells were then further incubated for 6 h. For primer extensions, 25 µg of RNA was used and the assays were carried out as previously described (Ma and Ptashne, 1987b). For RNA loading controls, 15 µg of total RNA was loaded onto agarose–formaldehyde gels.
ChIP experiments
ChIP experiments were performed essentially as described previously (Adam et al, 2001) with some modifications. Briefly, cells were grown to an OD600 of 0.6 in SC medium lacking uracil and containing 2% glucose. Then, 6-AU was added or not to a final concentration of 75 µg/ml for 60 min to induce PUR5. For the PHO5 ChIP, cells were grown in SC medium lacking uracil and containing 2% glucose and 500 µl of whole-cell extract was incubated with anti-HA antibodies (12CA5; Roche) coupled to magnetic beads (Dynal). Two microliters of DNA were used for the PCRs. ChIP experiments performed with anti-Myc antibodies were as previously described (Adam et al., 2001). All the ChIP data were quantified by phosphorImaging (PhosphorImager, Molecular Dynamics). Reactions with antibodies in untagged strains were also carried for all experiments but are not shown here since they were negative. The sequence of the oligonucleotides used for all of the experiments is available upon request. All experiments were carried out at least twice and gave similar results.
Acknowledgments
Acknowledgements
We thank Joëlle Brodeur, Marco Di Fruscio, Alain Lavigueur, Benoît Leblanc, Karine Lemieux and Annie Moisan for critical reading of the manuscript. This work was supported by a grant from the Canadian Institutes of Health Research (CIHR) to L.G. L.G. is a research scholar of the CIHR/Cancer Research Society Inc. of Canada.
References
- Adam M., Robert,F., Larochelle,M. and Gaudreau,L. (2001) H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol., 21, 6270–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad K. and Henikoff,S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell, 9, 1191–1200. [DOI] [PubMed] [Google Scholar]
- Allen J.B., Zhou,Z., Siede,W., Fried,E.C. and Elledge,S.J. (1994) The SAD1/RAD3 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev., 8, 2416–2428. [DOI] [PubMed] [Google Scholar]
- Barberis A. and Gaudreau,L. (1998) Recruitment of the RNA polymerase II holoenzyme and its implications in gene regulation. Biol. Chem., 379, 1397–1405. [DOI] [PubMed] [Google Scholar]
- Bash R. and Lohr,D. (2001) Yeast chromatin structure and regulation of GAL gene expression. Prog. Nucleic Acid Res. Mol. Biol., 65, 197–259. [DOI] [PubMed] [Google Scholar]
- Becker P.B. and Hörz,W. (2002) ATP-dependent nucleosome remodeling. Annu. Rev. Biochem., 71, 247–273. [DOI] [PubMed] [Google Scholar]
- Bhaumik S.R. and Green,M.R. (2001) SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev., 15, 1935–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.E., Howe,L., Sousa,K., Alley,S.C., Carrozza,M.J., Tan,S. and Workman,J.L. (2001) Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science, 292, 2333–2337. [DOI] [PubMed] [Google Scholar]
- Carr A.M., Dorrington S.M., Hindley,J., Phear,G.A., Aves,S.J. and Nurse,P. (1994) Analysis of a histone H2A variant from fission yeast: evidence for a role in chromosome stability. Mol. Gen. Genet., 245, 628–635. [DOI] [PubMed] [Google Scholar]
- Clarkson M.J., Wells,J.R.E., Gibson,F., Saint,R. and Tremethick,D.J. (1999) Regions of variant histone His2AvD required for Drosophila development. Nature, 399, 694–697. [DOI] [PubMed] [Google Scholar]
- Dhillon N. and Kamakaka,R.T. (2000) A histone variant, Htz1p and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol. Cell, 6, 769–780. [DOI] [PubMed] [Google Scholar]
- Ernst S.G., Miller,H., Brenner,C.A., Nocente-McGrath,C., Francis,S. and McIsaac,R. (1987) Characterization of a cDNA clone coding for a sea urchin histone H2A variant related to the H2A.F/Z histone protein in vertebrates. Nucleic Acids Res., 15, 4629–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar-Henriques M. and Daignan-Fornier,B. (2001) Transcriptional regulation of the yeast GMP synthesis pathway by its end products. J. Biol. Chem., 276, 1523–1530. [DOI] [PubMed] [Google Scholar]
- Exinger F. and Lacroute,F. (1992) 6-azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr. Genet., 22, 9–11. [DOI] [PubMed] [Google Scholar]
- Faast R. et al. (2001) Histone variant H2A.Z is required for early mammalian development. Curr. Biol., 11, 1183–1187. [DOI] [PubMed] [Google Scholar]
- Fan J.Y., Gordon,F., Luger,K., Hansen,J.C. and Tremethick,D.J. (2002) The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol., 9, 172–176. [DOI] [PubMed] [Google Scholar]
- Gaudreau L., Schmid,A., Blaschke,D., Ptashne,M. and Hörz,W. (1997) RNA polymerase II holoenzyme recruitment is sufficient to remodel chromatin at the yeast PHO5 promoter. Cell, 89, 55–62. [DOI] [PubMed] [Google Scholar]
- Harvey R.P., Whiting,J.A., Coles,L.S., Krieg,P.A. and Wells,J.R. (1983) H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc. Natl Acad. Sci. USA, 80, 2819–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch C.L. and Bonner,W.M. (1988) Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species. Nucleic Acids Res., 16, 1113–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havas K., Whitehouse,I. and Owen-Hughes,T. (2001) ATP-dependent chromatin remodeling activities. Cell. Mol. Life Sci., 58, 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang W.W., Venkatasubrahmanyam,S., Ianculescu,A.G., Tong,A., Boone,C. and Madhani,H.D. (2003) A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol. Cell, 11, 261–266. [DOI] [PubMed] [Google Scholar]
- Iouzalen N., Moreau,J. and Mechali,M. (1996) H2A.ZI, a new variant histone expressed during Xenopus early development exhibits several distinct features from the core histone H2A. Nucleic Acids Res., 24, 3947–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.D., Falciano,V.T. and Gorovsky,M.A. (1996) A likely histone H2A.F/Z variant in Saccharomyces cerevisiae. Trends Biochem. Sci., 21, 466–467. [DOI] [PubMed] [Google Scholar]
- Keaveney M. and Struhl,K. (1998) Activator-mediated recruitment of the RNA polymerase II machinery is the predominant mechanism for transcriptional activation in yeast. Mol. Cell, 1, 917–924. [DOI] [PubMed] [Google Scholar]
- Koh S.S., Ansari,A.Z., Ptashne,M. and Young,R.A. (1998) An activator target in the RNA polymerase II holoenzyme. Mol. Cell, 1, 895–904. [DOI] [PubMed] [Google Scholar]
- Krebs J.E., Fry,C.J., Samuels,M.L. and Peterson,C.L. (2000) Global role for chromatin remodelling enzymes in mitotic gene expression. Cell, 102, 587–598. [DOI] [PubMed] [Google Scholar]
- Krogan N.J. et al. (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol., 23, 4207–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo M-H., vom Baur,E., Struhl,K. and Allis,C.D. (2000) Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell, 6, 1309–1320. [DOI] [PubMed] [Google Scholar]
- Larschan E. and Winston,F. (2001) The S.cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev., 15, 1946–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach T.J., Mazzeo,M., Chotkowski,H.L., Madigan,J.P., Wotring,M.G. and Glaser,R.L. (2000) Histone H2A.Z is widely but nonrandomly distributed in chromosomes of Drosophila melanogaster. J. Biol. Chem., 275, 23267–23272. [DOI] [PubMed] [Google Scholar]
- Lee S.-K., Yu, S-L., Prakash,L. and Prakash,S. (2002) Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription: implications for cockayne syndrome. Cell, 109, 811–821. [DOI] [PubMed] [Google Scholar]
- Lu X., Ansari,A.Z. and Ptashne,M. (2000) An artificial transcriptional activating region with unusual properties. Proc. Natl Acad. Sci. USA, 97, 1988–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Ansari,A.Z., Lu,X., Ogirala,A. and Ptashne,M. (2002) A target essential for the activity of a nonacidic yeast transcriptional activator. Proc. Natl Acad. Sci. USA, 99, 8591–8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. and Ptashne,M. (1987a) The carboxy-terminal 30 amino acids of GAL4 are recognized by GAL80. Cell, 50, 137–142. [DOI] [PubMed] [Google Scholar]
- Ma J. and Ptashne,M. (1987b) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell, 48, 847–853. [DOI] [PubMed] [Google Scholar]
- Melcher K. and Johnston,S.A. (1995) GAL4 interacts with TATA-binding protein and coactivators. Mol. Cell. Biol., 15, 2839–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini M.D., Wu,M. and Madhani,H.D. (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell, 112, 725–736. [DOI] [PubMed] [Google Scholar]
- Neely K.E., Hassan,A.H., Wallberg,A.E., Steger,D.J., Cairns,B.R., Wright,A.P.H. and Workman,J.L. (1999) Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell, 4, 649–655. [DOI] [PubMed] [Google Scholar]
- Ptashne M. and Gann,A. (1997) Transcriptional activation by recruitment. Nature, 386, 569–577. [DOI] [PubMed] [Google Scholar]
- Roth S.Y., Denu,J.M. and Allis,C.D. (2001) Histone acetyltransferases. Annu. Rev. Biochem., 70, 81–120. [DOI] [PubMed] [Google Scholar]
- Santisteban M.S., Kalashnikova,T. and Smith,M.M. (2000) Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell, 103, 411–422. [DOI] [PubMed] [Google Scholar]
- Shaw R.J. and Reines,D. (2000) Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol., 20, 7427–7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell L., Bowen,J., Dadd,C.A., Dedon,P.C., Davis,M., Cook,R.G., Allis,C.D. and Gorovsky,M.A. (1993) Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev., 7, 2641–2651. [DOI] [PubMed] [Google Scholar]
- Suto R.K., Clarkson,M.J., Tremethick,D.J. and Luger,K. (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol., 12, 1121–1124. [DOI] [PubMed] [Google Scholar]
- Svaren J. and Hörz,W. (1997). Transcription factors vs nucleosomes: regulation of the PHO5 promoter in yeast. Trends Biochem. Sci., 22, 93–97. [DOI] [PubMed] [Google Scholar]
- Utley R.T., Ikeda,K., Grant,P.A., Côté,J., Steger,D.J., Eberharter,A., John,S. and Workman,J. (1998) Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature, 394, 498–502. [DOI] [PubMed] [Google Scholar]
- van Daal A., White,E.M., Gorovsky,M.A. and Elgin,S.C. (1988) Drosophila has a single copy of the gene encoding a highly conserved histone H2A variant of the H2A.F/Z type. Nucleic Acids Res., 16, 7487–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignali M., Hassan,A.H., Neely,K.E. and Workman,J.L. (2000) ATP-dependent chromatin-remodeling complexes. Mol. Cell. Biol., 20, 1899–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu,J., Suka,N. and Grunstein,M. (2000) Global histone acetylation and deacetylation in yeast. Nature, 408, 495–498. [DOI] [PubMed] [Google Scholar]
- White E.M., Shapiro,D.L., Allis,C.D. and Gorovsky,M.A. (1988) Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res., 16, 179–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Reece,R. and Ptashne,M. (1996) Quantification of putative activator-target affinities predicts transcriptional activating potentials. EMBO J., 15, 3951–3963. [PMC free article] [PubMed] [Google Scholar]
- Yudkovski N., Logie,C., Hahn, S and Peterson,C.L. (1999) Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev., 13, 2369–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]