Abstract
One manifestation of RNA silencing, known as post-transcriptional gene silencing (PTGS) in plants and RNA interference (RNAi) in animals, is a nucleotide sequence-specific RNA turnover mechanism with the outstanding property of propagating throughout the organism, most likely via movement of nucleic acids. Here, the cell-to-cell movement of RNA silencing in plants is investigated. We show that a short-distance movement process, once initiated from a small group of cells, can spread over a limited and nearly constant number of cells, independent of the presence of homologous transcripts. There is also a long-range cell-to-cell movement process that occurs as a relay amplification, which requires the combined activity of SDE1, a putative RNA-dependent RNA polymerase, and SDE3, a putative RNA helicase. Extensive and limited cell-to-cell movements of silencing are triggered by the same molecules, occur within the same tissues and likely recruit the same plasmodesmata channels. We propose that they are in fact manifestations of the same process, and that extensive cell-to-cell movement of RNA silencing results from re-iterated short-distance signalling events. The likely nature of the nucleic acids involved is presented.
Keywords: movement/RNA silencing/siRNA/suppressor
Introduction
In eukaryotes, RNA silencing is the suppression of gene expression through nucleotide sequence-specific interactions involving RNA. This phenomenon is experimentally activated by double-stranded (ds)RNA (Fire et al., 1998), which is cleaved into 21–25 nt long dsRNA—the short interfering (si)RNA—by an RNase III-like enzyme named Dicer (Bernstein et al., 2001). The siRNA is then incorporated into a multi-subunit complex, the RNA-induced silencing complex (RISC), so ensuring that it specifically degrades any RNA sharing sequence similarity with the inducing dsRNA (Hammond et al., 2000).
Natural roles of RNA silencing include transposon taming and antiviral defence. As a counter-defensive strategy, viruses have evolved proteins that suppress various steps of the RNA silencing mechanism (reviewed in Voinnet et al., 2001). RNA silencing is also involved in development, as illustrated by the discovery, in both plants and animals, of endogenous 21–24 nt long micro (mi)RNAs. These miRNAs arise from Dicer-mediated processing of precursor RNAs with extensive secondary structure and show partial or complete homology with subsets of developmental mRNAs, which are targeted at the level of stability or translation (reviewed in Bartel and Bartel, 2003).
In some organisms RNA silencing is amplified. Thus, in plants and Caenorhabditis elegans, a phenomenon known as transitivity increases the initial pool of siRNAs by producing new siRNAs corresponding to sequences located outside the primary targeted regions of a transcript (Sijen et al., 2001; Vaistij et al., 2002). In C.elegans and Arabidopsis, transitivity requires the action of putative endogenous RNA-dependent RNA polymerases (RdRps) (Dalmay et al., 2000; Sijen et al., 2001). However, transitivity in C.elegans proceeds mainly from 3′→5′ parts of target transcripts, whereas it is bi-directional in plants (Voinnet et al., 1998; Vaistij et al., 2002). Additionally, the occurrence of transitivity seems to be transcript-dependent in plants: some mRNAs engage in this process, whereas others do not (Vaistij et al., 2002). The reason for this disparity remains unclear, but transcript accessibility/subcellular localization may be involved. Another apparently plant-specific aspect of RNA silencing is the involvement of two functionally distinct siRNAs that likely arise from separate Dicer activities (Hamilton et al., 2002; Tang et al., 2003). A 21 nt siRNAs is sufficient for RISC-mediated cleavage of target transcripts, while a 25 nt siRNA correlates positively with DNA methylation and transmission of silencing over long distances (Hamilton et al., 2002).
Non-cell-autonomous RNA silencing has been best documented in plants and C.elegans (Palauqui et al., 1997; Voinnet et al., 1998; Winston et al., 2002), where localized activation of silencing causes sequence-specific degradation of transcripts in tissues located away from the initiation zone. This specificity of action suggests that the signals involved have nucleic acid components. In plants, indirect evidence indicates that RNA silencing moves over long distances through the phloem and, upon unloading, spreads from cell to cell through plasmodesmata in recipient tissues (Voinnet et al., 1998). However, the underlying mechanism and molecules implicated remain largely elusive. Recently, a putative transmembrane protein necessary for systemic RNA silencing has been identified in C.elegans, but its function is as yet uncharacterized (Winston et al., 2002).
We provide here a comprehensive analysis of the cell-to-cell movement of RNA silencing in plants, combining the use of diverse silencing triggers, viral-encoded suppressor proteins and silencing deficient mutants of Arabidopsis. This study revealed a short-distance movement process that is initiated from a small group of cells and spreads over a nearly constant number of 10–15 cells in the absence of trigger amplification. Production of 21 nt siRNAs at the site of silencing initiation correlates with the onset of short-range movement, whereas 25 nt siRNAs appear to be dispensable. There is also a long-range cell-to-cell communication mechanism that requires the activity of SDE1, a putative RdRp, and SDE3, a putative RNA helicase. This extensive movement is associated with de novo synthesis of secondary siRNAs produced by transitivity, which belong exclusively to the 21 nt size class. Limited and extensive cell-to-cell movement of silencing are triggered by the same molecules, occur within the same tissues and likely recruit the same plasmodesmata channels. Based on those results, we propose that both types of movement are in fact manifestations of the same process. A model is presented in which extensive cell-to-cell spread of RNA silencing results from re-iterated short-distance signalling events involving movement of 21 nt siRNAs.
Results
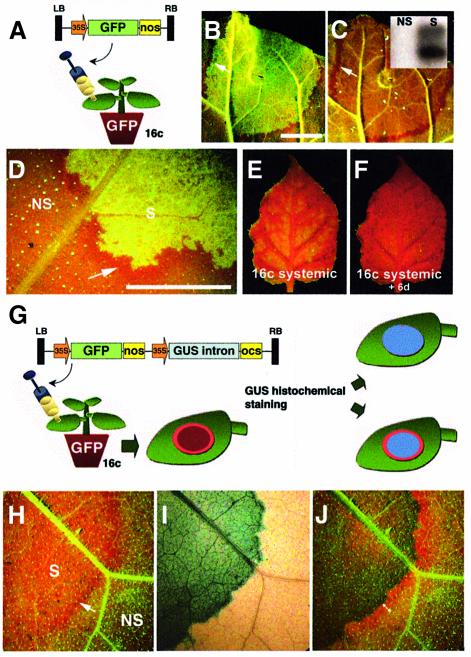
We used Nicotiana benthamiana line 16c, carrying a highly expressed green fluorescent protein (GFP) transgene. These plants are green under ultra violet (UV) light, whereas non-transgenic plants are red due to chlorophyll fluorescence. In line 16c, silencing of GFP is induced by leaf infiltration of Agrobacterium cultures (agro-infiltration) in which the T-DNA carries a copy of the GFP transgene (Figure 1A; Voinnet et al., 1998). The infiltrated tissue is initially strongly green fluorescent due to superimposed expression of the ectopic and stably integrated GFP transgenes (Figure 1B). However, at 10 days post-infiltration (d.p.i.), it has turned red, due to silencing of both transgenes (Figure 1C). This tissue contains low levels of GFP mRNA and accumulates GFP siRNAs (Figure 1C, inlay; data not shown). This local silencing precedes ‘systemic silencing’, which is manifested at 7–9 d.p.i. in non-infiltrated leaves, upon long-distance transport of the silenced state. Silencing first occurs around the veins (Figure 1E) and progressively invades the whole lamina (Figure 1F) through extensive cell-to-cell spread of a signal (Voinnet et al., 1998).
Fig. 1. Short-range movement of GFP silencing. (A–D) Analysis of Agrobacterium-infiltrated tissues in line 16c. (A) Principle of the experiment. LB and RB, left and right borders of the pBin19 T-DNA, respectively; 35S, CaMV 35S promoter; Nos, nopaline synthase terminator. (B) Upon expression of transgenic and ectopic GFPs, a thin border of red tissue is visible at 5 d.p.i. (arrow). (C) At 11 d.p.i., the silenced patch accumulates both GFP siRNAs (inlay). The red border is now clearly visible (arrow). (D) Microscopic inspection of the red border (arrow) at 5 d.p.i. Bar, 1 cm. (E and F) Systemic silencing in a developing leaf of line 16c, imaged at a 6 day interval. (G–J) The red border results from movement of a silencing signal. (G) Principle of the experiment. OCS, octopine synthase terminator. (H) The red border (arrow) triggered by the construct in (G) at 10 d.p.i. (I) GUS histochemical staining of the leaf in (H). (J) Overlay of the images in (H) and (I). S, silenced; NS, non-silenced.
Short-range movement of RNA silencing targeted against a GFP transgene
We consistently observe a thin border of GFP-silenced cells at the margin of agro-infiltrated zones in leaves of line 16c (Figure 1B and C, arrow). This border is most evident at ∼10 d.p.i. (Figure 1C, arrow). Its size does not expand at further time points and it closely mirrors the edge of the infiltrated zone (Figure 1D, arrow). To further investigate this phenomenon, these experiments were repeated with a construct containing the GFP transgene (as an inducer of GFP silencing) together with a reporter gene encoding the β-glucuronidase (GUS) (Figure 1G). The presence of an intron in the GUS open reading frame (ORF) prevents its expression from the Agrobacterium cells. Thus, histochemical staining would unambiguously indicate the zone of T-DNA transfer and thereby reveal the precise outline of the agro-infiltrated patch. The pictures in Figure 1H–J illustrate one such experiment at 10 d.p.i. GFP silencing occurred within the infiltrated area and the red border was visible (Figure 1H, arrow). Subsequent histochemical staining and ethanol clearing (Figure 1I) showed that GFP silencing expanded beyond the GUS-stained tissues in a region that coincided with the red border (Figure 1J, arrow). Moreover, the width of the border was remarkably constant: an average of 13 cells (±2 cells) were consistently affected. In addition, the outmost cell layer of the infiltrated area (5–10 cells) was sufficient for this process to occur (see Supplementary data 1, available at The EMBO Journal Online). We conclude, therefore, that the red border results from limited cell-to-cell movement of a signal triggered in a small number of cells, a phenomenon that had so far escaped our attention.
The effect of viral suppressors on short-range movement of GFP silencing
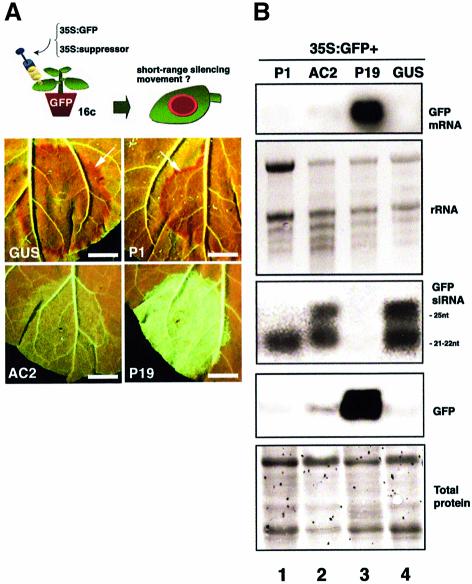
To further investigate this limited movement, we exploited virus-encoded suppressors of RNA silencing that were co-delivered with the GFP construct via Agrobacterium infiltration (Figure 2A, diagram). A bacterium strain with a GUS transgene provided a negative control for silencing suppression (Hamilton et al., 2002). Plants of line 16c were infiltrated with each mix of cultures and subsequently assayed for (i) intracellular silencing of GFP within the agro-inoculated patch, and (ii) limited movement of silencing at the edge of the patch, as assessed by occurrence of the red border (Figure 2A, diagram). We present the results with the P1, AC2 and P19 proteins (Voinnet et al., 1999; Hamilton et al., 2002).
Fig. 2. The effect of viral-encoded silencing suppressors on short distance movement of RNA silencing. (A) The effect of the P1, AC2 and P19 proteins on the onset of localized signalling (arrows) at 7 d.p.i. (B) Molecular analysis of GFP, GFP mRNA and GFP siRNA levels in the co-infiltrated tissues depicted in (A). rRNA, ethidium bromide staining of ribosomal RNA provides a control for RNA loading. Coomassie staining of total protein provides a control for protein loading. Bar, 5 mm.
Intracellular silencing and its short-range movement developed as quickly and as extensively in the P1- as in the GUS-treated plants. Thus, at 7 d.p.i., with both treatments, the GFP and its mRNA were bellow detection in the co-infiltrated tissues (Figure 2B, tracks 1 and 4). In addition, a layer of red-fluorescent tissue was visible at the edge of both types of patches (Figure 2A, top panels, arrows), and it affected a similar number (13 ± 2) of epidermal cells, as assessed by the approach described in Supplementary data 1. The 21 nt GFP siRNA accumulated to similar levels in the P1- and GUS-treated samples (Figure 2B, lanes 1 and 4) but the 25 nt siRNA was undetectable in P1-treated tissues (Figure 2B, lane 1), whereas it was abundant in the controls (Figure 2B, lane 4). The P19-treated tissues were intensely green fluorescent at 7 d.p.i. (Figure 2A). They contained elevated GFP and GFP mRNA levels and were devoid of both GFP siRNA species (Figure 2B, lane 3). Limited movement of silencing was also abolished in the P19-treated tissues (Figure 2A). Patches co-expressing AC2 were slightly green fluorescent at 7 d.p.i. and, accordingly, contained low levels of GFP and GFP mRNA (Figure 2B, lane 2). Both GFP siRNA species accumulated (although to a slightly lower extent than in the GUS-treated controls), indicating intracellular RNA silencing. However, short-range silencing movement around the infiltrated patches could not be detected (Figure 2A, left-hand bottom panel). These results were reproduced in three independent experiments.
From these results, we rule out the possibility that occurrence of the red border is a non-specific stress response due, for example, to Agrobacterium, because it was readily affected by silencing suppressors. Secondly, short-range silencing movement was the same in the P1-treated samples as in the control samples, despite the lack of 25 nt GFP siRNAs in the P1-infiltrated tissues. Therefore, occurrence of the 21 nt siRNAs species is sufficient for limited movement of silencing to take place.
Short-range movement of GFP silencing does not require transcription of a homologous transgene in recipient tissues
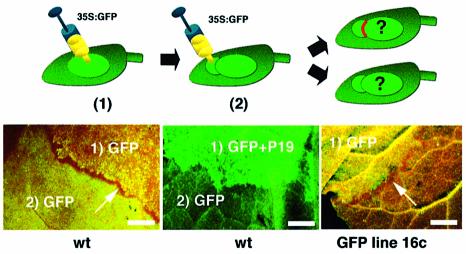
To test whether limited movement of GFP silencing required the presence of GFP mRNA in surrounding cells, we performed sequential infiltrations of the GFP Agrobacterium strain in leaves of wild-type (wt) N.benthamiana, such that the second infiltrated patch overlapped with the first one (Figure 3, diagram). The two infiltrations were carried out at a 5 day interval. If a localized GFP silencing signal had moved from the first infiltrated patch to adjacent, non-transgenic cells, GFP expression from the second overlapping patch would be prevented at the edge of the first one. In the absence of localized signalling from the first patch, however, expression of GFP from the second patch would coincide with the edge of the first patch. The image in Figure 3 (left-hand panel) shows that localized signalling indeed occurred in non-transgenic leaves and that its extent was the same as in leaves of line 16c (right-hand panel). Moreover, as in line 16c (Figure 2, right-hand bottom panel) this process was abolished if the first patch had received a P19 co-treatment (middle panel). We conclude that limited movement of silencing is not influenced by prior transcription of a homologous transgene in recipient tissues.
Fig. 3. Short-range silencing movement in wt plants. The two sequential infiltrations with 35S-GFP are performed at a 5 day interval (numbered 1 and 2, respectively). In the middle panel, the first infiltration was performed with the P19 protein, as in Figure 2A. Arrows, short-range movement of silencing. Bar, 5 mm.
A phloem-restricted virus-induced gene silencing system based on potato virus X
All the above experiments were based on GFP transgenic plants and involved delivery of the silencing trigger via leaf infiltration of Agrobacterium. Thus, it could be argued that short-range silencing movement was merely a silencing-based defence response to T-DNA molecules and/or a transgene-specific process. This prompted the development of an alternative system to trigger non-cell-autonomous RNA silencing in N.benthamiana, potentially targeting endogenous mRNAs. Potexviruses such as potato virus X (PVX) silence host mRNAs if they share sequence homology with the corresponding nuclear genes, a process named virus-induced gene silencing (VIGS; Baulcombe, 1999). Thus, we sought to elaborate a tissue-specific VIGS system.
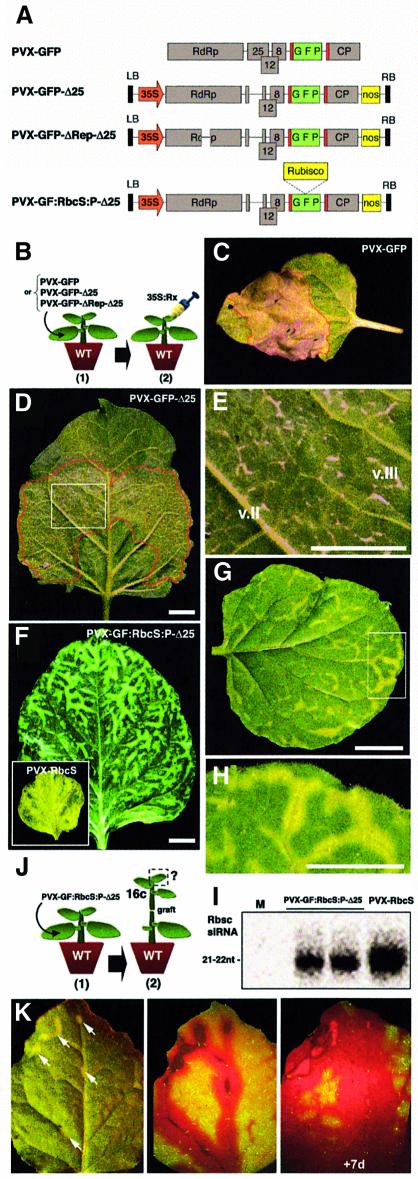
Strawberry mild yellow edge virus (SMYEV), unlike other potexviruses, is restricted to the phloem because its genome lacks the ORF for a 24–26 kDa protein, which is strictly required for cell-to-cell spread of potexviruses (Jelkmann et al., 1992). Thus, we anticipated that disruption of the 25 kDa ORF in the PVX genome would result in phloem restriction of the virus. Since the PVX 25 kDa protein is also a silencing suppressor (Voinnet et al., 2000), its removal would prevent any interference with movement of silencing potentially induced by PVX. A full-length PVX clone expressing GFP (PVX–GFP; Figure 4A) was thus deleted for the 25 kDa ORF. In contrast to previous PVX–GFP mutants (Voinnet et al., 2000), the resulting virus (PVX–GFP-Δ25; Figure 4A: Supplementary data 2) produced the coat protein (CP), essential for systemic spread of PVX (Santa Cruz et al., 1998).
Fig. 4. Vein-limited recombinant PVX triggers short-range movement of RbcS silencing. (A) Viral constructs. The PVX genome has four ORFs. The RdRp is required for replication. The 25, 12 and 8 kDa and the coat (CP) proteins ensure cell-to-cell and systemic movement. Expression of GFP is from a duplicated CP promoter (red bar). (B) The viruses in (A) were inoculated in one leaf (1). At 15 d.p.i., systemic leaves were infiltrated with the Rx Agrobacterium strain (2). (C) Extensive necrosis (red outline) elicited by the Rx-CP interaction in a PVX–GFP-infected leaf. (D) Same as in (C), but on a PVX–GFP-Δ25-infected leaf. (E) Enlarged view of the inlay in (D). Necrosis is mainly restricted to the class III veins (v.III). (F) RbcS silencing due to PVX-GF:RbcS:P-Δ25 on a systemic, mature leaf. (G) Same as (F) but on a developing leaf. (H) Enlarged view of the inlay in (G). (I) Accumulation of RbcS siRNAs in PVX-RbcS- and PVX-GF:RbcS:P-Δ25-infected plants. M, mock. (J) Wt plants were first inoculated with PVX-GF:RbcS:P-Δ25 (1). The inoculated leaf was removed and, at 15 d.p.i., plants were used as rootstocks in grafts involving shoots of line 16c as scions (2). (K) Silencing of RbcS and GFP in a leaf of a 16c scion, 10 days post-grafting. Left: RbcS silencing around a few class III veins (arrows). Middle: UV imaging of the same leaf. GFP silencing is much more extensive. Right: 1 week later, GFP silencing affects the whole lamina, whereas RbcS silencing remains the same. Chlorosis due to RbcS silencing alters the red fluorescence of chlorophyll under UV, providing an internal control in these images.
Wt N.benthamiana plants were inoculated with PVX–GFP-Δ25, alongside the replication-deficient PVX–GFP-ΔRep-Δ25 (Figure 4A; Supplementary data 2) and the movement-competent PVX–GFP (Figure 4A). At 15 d.p.i., PVX–GFP-inoculated plants showed a systemic, mild chlorosis, whereas both PVX–GFP-Δ25- and PVX–GFP-ΔRep-Δ25-inoculated plants remained asymptomatic (data not shown). To develop a sensitive and visual assay for replication and systemic movement of PVX–GFP-Δ25, we used the Rx gene from potato, which confers resistance to PVX. Transient expression of Rx in N.benthamiana triggers a CP-dependent hypersensitive response (HR), causing rapid cell death (Bendahmane et al., 1999). Occurrence of an HR upon Rx treatment would thus indicate the presence of replicating PVX, due to CP synthesis (Figure 4B). Systemic PVX–GFP-infected leaves infiltrated with the Rx strain of Agrobacterium indeed developed extensive necrosis that covered most tissues of the infiltrated area (Figure 4C).
Two days post-infiltration of the Rx construct, many flecks of HR were observed in systemic leaves of PVX–GFP-Δ25-inoculated plants (Figure 4D and E). However, in contrast to the extensive HR in PVX–GFP-infected leaves, these flecks coincided precisely with the class III (and occasionally class II) veins, from which they never expanded (Figure 4E). HR never occured in systemic leaves of PVX–GFP-ΔRep-Δ25-inoculated plants (data not shown). Thus, we conclude that PVX–GFP-Δ25 had moved systemically but was limited to the class III veins, where PVX normally unloads (Roberts et al., 1997). This vein-restricted infection persisted for >1.5 months.
Short-range movement of silencing targeted against an endogenous mRNA
We then used PVX–GFP-Δ25 to trigger silencing against an endogenous mRNA. Since this virus appears to be phloem limited, a VIGS phenotype developing away from the class III veins would indicate silencing movement. We chose the Rubisco small subunit (RbcS) mRNA as target, because VIGS of RbcS causes a distinctive yellow chlorosis (Ratcliff et al., 2001). A fragment of the N.benthamiana RbcS cDNA was mobilized into the GFP insert of PVX–GFP-Δ25, leading to PVX-GF:RbcS:P-Δ25 (Figure 4A; Supplementary data 2). As control, we used PVX-RbcS (Jones et al., 1999). Fifteen days after inoculation of PVX-GF:RbcS:P-Δ25 to wt N.benthamiana, systemic leaves exhibited silencing of Rbsc. However, unlike the widespread VIGS caused by PVX-Rbsc (Figure 4F, inlay), silencing of RbcS from PVX-GF:RbcS:P-Δ25 was restricted to 10–15 cells around the class III (and occasionally class II) veins (Figure 4F–H). Northern analysis of those silenced tissues showed accumulation of RbcS-specific siRNAs that were mostly of the 21 nt class (Figure 4I).
We conclude that movement of RbcS silencing occurred from the class III veins but did not expand further in the lamina, unlike GFP silencing in similar tissues of line 16c (Figure 1E and F). However, a strict comparison was difficult because the two systems involved silencing triggers that differed in nature and mode of delivery.
Vein-restricted replication of PVX-GF:RbcS:P-Δ25 triggers limited movement of RbcS silencing and extensive movement of GFP silencing in leaves of line 16c
To compare strictly the cell-to-cell movement of GFP and RbcS silencing, we exploited the fact that PVX-GF:RbcS:P-Δ25 carries a GFP insert as well as an RbcS insert (Figure 4A). We performed grafting experiments involving nontransgenic PVX-GF:RbcS:P-Δ25-infected plants as rootstocks and shoots of line 16c as scions (Figure 4J). Two weeks later, most scions (eight out of 10) showed vein-centred silencing of RbcS (Figure 4K, left panel) that was caused by graft-transmission of the vein-restricted PVX-GF:RbcS:P-Δ25, as assessed by Rx treatments (data not shown). In those leaves, GFP silencing triggered by PVX-GF:RbcS:P-Δ25 initially coincided with RbcS silencing (Figure 4K, middle panel). However, it progressively invaded the entire lamina (Figure 4K, right panel), while RbcS silencing remained the same. Thus, both limited (affecting RbcS) and extensive (affecting GFP) movement of silencing were activated by a common trigger and were manifested within the same tissue, a key circumstance that could not be achieved in the previous experiments.
Why was GFP silencing movement more extensive than RbscS silencing movement? We ruled out an effect of target mRNA level because both accumulated to a similar level in the 16c scions (data not shown). We also ruled out an effect of PVX-GF:RbcS:P-Δ25 replication, because the RbcS and GFP inserts were produced as transcriptional fusion from the same subgenomic RNA (Figure 4A). A possible explanation was in the differential capacity of the target mRNAs to sustain relay amplification of the signal outside the veins. In this regard, it was significant that the GFP transgene mRNA supports transitive RNA silencing, whereas the RbcS endogenous mRNA does not (Vaistij et al., 2002).
Extensive as opposed to limited movement of RNA silencing is conditioned by SDE1 and SDE3 in GFP transgenic Arabidopsis
Transitivity in Arabidopsis depends on SDE1/SGS2, a putative RNA-dependent RNA polymerase required for transgene silencing (Dalmay et al., 2000; Vaistij et al., 2002). Based on genetic analyses, SDE1/SGS2 was proposed to synthesize a dsRNA trigger of silencing, using transgene ssRNA as template (Dalmay et al., 2000). If transitivity was key to the development of extensive but not limited silencing movement, we anticipated that extensive silencing movement would not occur in an sde1 mutant whereas localized movement would remain unaffected. The availability of the sde1 null mutant and GFP transgenic Arabidopsis line GFP142 (both in ecotype C24; Dalmay et al., 2000) provided the opportunity to test directly this hypothesis.
First, we developed a transgenic system mimicking the vein-centred signalling of silencing in N.benthamiana. We used the promoter of the Arabidopsis SUC2 sucrose symporter (atSUC2), which is exclusively active in the phloem of Arabidopsis ecotype C24 (Imlau et al., 1999) to trigger phloem-specific GFP silencing. We created an inverted repeat construct corresponding to the 5′ part (‘GF’) of the GFP transgene expressed in Arabidopsis line GFP142. This pan-handled construct, predicted to produce a dsRNA and referred to as GF-FG (Figure 5A), was mobilized downstream of the atSUC2 promoter, leading to atSUC2-GF-FG (Figure 5A). This study involved Arabidopsis line GFP142 and line GFP142/sde1, which also carries an sde1 null mutation (Figure 5A). Both lines appear uniformly green under UV.
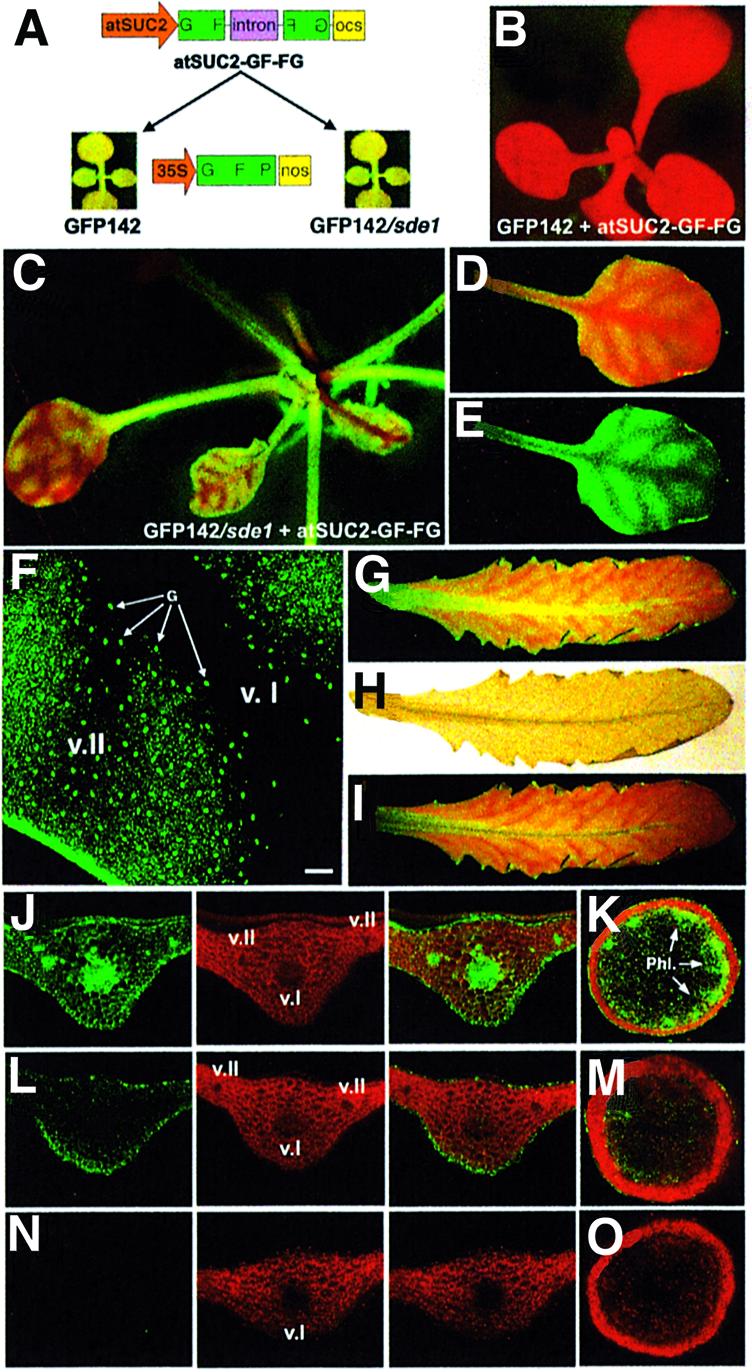
Fig. 5. Extensive, but not limited, movement of silencing is conditioned by SDE1 in transgenic Arabidopsis. (A) Experimental set up. (B) A GFP142 plant transformed with atSUC2-GF-FG. (C) Typical phenotype of GFP142/sde1 plants transformed with atSUC2-GF-FG. (D and E) A leaf of a seedling from the plant in (C) imaged under UV without (D) or with (E) a band-pass filter. (F) Confocal imaging of vein-centred, silenced tissues as seen in (E). G, guard cells. Bar, 300 µm. (G–I) Silencing in a mature leaf of the plant in (C). (H) Acetic acid clearing of the leaf in (G). (I) Overlay of (G) and (H): silencing expands outside the vein network. (J) Transversal section of a mature leaf from a GFP142 plant. Right/middle split channels: GFP and chlorophyll fluorescence, respectively. Left channel: overlay. (K) Transversal section of the stem of a GFP142 plant. Phl, phloem. (L and M) Same as (J) and (K) but in a GFP142/sde1 plant transformed with atSUC2-GF-FG. (N and O) same as (J) and (K) but in a GFP142 plant transformed with atSUC2-GF-FG. v.I, class I vein; v.II, class II vein.
All (10 out of 10) of the GFP142 plants transformed with atSUC2-GF-FG were uniformly red under UV light (Figure 5B). In contrast, most (seven out of 10) of the GFP142/sde1-transformed plants showed a vein-centred pattern of silencing in seedlings (Figure 5C–E) and mature leaves (Figure 5G). Overlays between GFP imaging and acetic acid clearing of mature leaves confirmed that this phenotype resulted from short-range movement of silencing from the phloem to adjacent cells (Figure 5G–I). This was also evident by comparing the vein-specific expression of the atSUC2 promoter (Imlau et al., 1999) with the extent of silencing observed here. As seen in the previous experiments, silencing movement affected a nearly constant number of cells (10–15 cells) and never expanded further (Figure 5D–I). Green fluorescence was markedly reduced in the vascular system of GFP142/sde1 plants transformed with atSUC2-GF-FG (Figure 5L, v.I and v.II, and M), but it was still detectable in epidermal, outer parenchyma and outer cortical cells. This was consistent with the silencing trigger being expressed in the phloem and a localized silencing signal moving in neighbouring cells. Notably, stomata guard cells were unaffected by silencing (Figure 5F, arrows). In contrast, GFP was high in all tissues of the parental GFP142 and GFP142/sde1 lines (Figure 5J and K; data not shown) and it was undetectable in tissues of GFP142 plants transformed with atSUC2-GF-FG (Figure 5N and O). These results indicate that extensive, but not limited, silencing movement was critically dependent upon SDE1.
Like SDE1, SDE3 is also required for transgene silencing in Arabidopsis (Dalmay et al., 2001). SDE3 has signatures of an RNA helicase and, like SDE1, has been genetically positioned upstream or at the step of dsRNA synthesis (Dalmay et al., 2001). In contrast to sde1 knockouts, however, null mutations in sde3 do not result in a complete loss of transgene silencing, presumably because of residual SDE1 activity (Dalmay et al., 2000). To test whether SDE3 also had an influence on extensive versus limited cell-to-cell movement of silencing, the atSUC2-GF-FG construct was introduced in line GFP142/sde3, which carries an sde3 null mutation (Figure 6A). As depicted in Figure 6B, the phenotype of most transformants (eight out of 10) was intermediate between that of the atSUC2-GF-FG-transformed GFP142/sde1 and GFP142 plants (Figures 5B and C). Thus, silencing was still vein-centred in young tissues (Figure 6B, leaf 1), but it expanded to a greater extent than in the GFP142/sde1-transformed plants (Figure 6B, leaves 2 and 3) and it had invaded nearly all the lamina in mature leaves (Figure 6B, leaf 4, C and D). Analysis of leaf transversal sections confirmed these observations (Supplementary data 3). These results indicate that extensive cell-to-cell movement of silencing was indeed dependent upon SDE3 but to a lesser extent than it was dependent upon SDE1.
Fig. 6. Effect of SDE3 on extensive signalling and northern analyses. (A) Experimental set up. (B) Typical phenotype of GFP142/sde3 plants transformed with atSUC2-GF-FG. GFP silencing is initially centred around the main vein (1) and expands in the lamina as the leaf age increase (2, 3, 4). (C and D) Mature leaves of the plant in (B). (E) Northern analysis of low and high molecular weight RNA extracted from the plants depicted in Figure 5 and this figure, 3 weeks post-germination. (F) Wt, sde1 and sde3 mutant Arabidopsis transformed with the atSUC2-SUL-LUS construct. (G) Wt Arabidopsis transformed with the atSUC2-RUB-BUR construct. The vein-centred chlorosis is similar in extent to that in (F).
Molecular analysis of the GFP142/sde1- and GFP142/sde3-transformed plants
Total RNA was extracted from GFP142 plants and from GFP142, GFP142/sde1, GFP/sde3 plants transformed with atSUC2-GF-FG. Northern analysis of high molecular weight RNA was in accordance with the phenotypes of these lines (Figure 6E). Thus, in line GFP142, the GFP mRNA was abundant (Figure 6E, lane 4). It was also abundant in lines GFP142/sde1 and GFP142/sde3 (data not shown). However, the GFP mRNA was below detection limit in the GFP142 plants transformed with atSUC2-GF-FG (Figure 6E, lane 3). In both the GFP/sde3 and GFP/sde1 plants transformed with atSUC2-GF-FG, there was a low level of the GFP mRNA, but it was higher in the GFP/sde1 transformants than in the GFP/sde3 transformants (Figure 6E, lane 1 compared with lane 2). These figures were reproduced in more than three independent analyses.
The low molecular weight fractions were then assayed for GFP-specific siRNAs and for transitivity. Transitivity in plants is manifested by generation of secondary siRNAs corresponding to sequences located both in the 3′ and 5′ regions of the primary target (Voinnet et al., 1998; Vaistij et al., 2002). For instance, silencing initiated by siRNAs corresponding to the 5′ part of the GFP transcript (‘GF’) leads to production of secondary siRNAs with sequences of the non-overlapping 3′ part (‘P’). Here, the silencing trigger was limited to the 5′ part (‘GF’) of the GFP transgene (Figures 5A and 6A). Thus, detection of ‘P’ siRNAs would indicate transitive RNA silencing, whereas a probe corresponding to the ‘GF’ part of the GFP transcript would hybridize to primary siRNAs.
As expected, no siRNAs were detected in the GFP142, GFP142/sde1 and GFP142/sde3 parental lines, with either the ‘GF’ or ‘P’ probes (Figure 6E, lane 4; data not shown). In all of the atSUC2-GF-FG-transformed lines, however, there were high levels of ‘GF’ primary siRNAs of both size classes that accumulated to a similar extent (Figure 6E, lanes 1–3). ‘P’ siRNAs were readily detected in the GFP142 plants transformed with atSUC2-GF-FG, indicating transitivity (Figure 6E, lane 3). ‘P’ siRNAs also accumulated in the GFP142/sde3-transformed plants, but to a lesser extent, implying that transitivity had occurred less efficiently (Figure 6E, lane 2). However, ‘P’ siRNAs were not detected in the GFP142/sde1 plants transformed with atSUC2-GF-FG, indicating a lack of transitivity in the absence of SDE1 (Figure 6E, lane 1). This was in agreement with previous reports (Vaistij et al., 2002). Notably, and as opposed to ‘GF’ primary siRNAs, the secondary ‘P’ siRNAs were almost exclusively of the 21 nt class (Figure 6E).
First, these results indicated that SDE3 participates in transitivity, but to a lesser extent than SDE1, in agreement with the incomplete silencing deficient phenotype of the sde3 knockout (Dalmay et al., 2001). Secondly, there was no correlation between the level of ‘GF’ primary siRNAs and the extent of silencing movement. In particular, the ‘GF’ siRNAs were as abundant in the GFP142/sde1-transformed plants as they were in the GFP142-transformed plants (Figure 6E, lanes 1 and 3), although these plants exhibited drastically dissimilar movement phenotypes (Figure 5B and C). Thirdly, there was a strict correlation between the efficiency of transitive RNA silencing and the extent of silencing movement. Thus, in GFP142 plants transformed with atSUC2-GF-FG, there were high levels of ‘P’ siRNAs and extensive silencing movement. In the GFP142/sde3-transformed plants, moderate levels of ‘P’ siRNAs correlated with an intermediate movement phenotype. Finally, in the GFP142/sde1 plants transformed with atSUC2-GF-FG, the complete lack of ‘P’ siRNAs correlated with the lack of extensive movement, allowing short-range movement of silencing to be monitored. Therefore, extensive silencing movement was linked to production of secondary siRNAs and, in contrast, limited movement was not.
Cell-to-cell movement of RNA silencing targeted against endogenous mRNAs is limited to 10–15 cells in wt Arabidopsis and is SDE1- and SDE3-independent
The experiments described in Figure 4 suggested that it was the inability of the RbcS mRNA to sustain transitivity that resulted in limited silencing movement. If this was true, silencing of RbcS triggered by the atSUC2 promoter in Arabidopsis should only exhibit limited movement, independently of mutations affecting SDE1 and SDE3. To test this inference and assess how generally it applied to endogenous, as opposed to transgenic mRNAs, we designed constructs atSUC2-RUB-BUR and atSUC2-SUL-LUS (Figure 6F and G) to silence the Arabidopsis RbcS and sulphur mRNAs, which should result in chlorosis in both cases (Ratcliff et al., 2001). Wt, sde1 and sde2 Arabidopsis (ecotype C24) were transformed with the corresponding constructs. As shown in Figure 6F and G, 100% of the transformants (25 out of 25 for atSUC2-SUL-LUS and 10 out of 10 for atSUC2-RUB-BUR, respectively) exhibited chlorosis that affected a similar number of cells (10–15) outside of the veins and did not expand further. Moreover, this phenotype was unaltered by the presence or absence of SDE1 or SDE3 (Figure 6F; data not shown).
Discussion
Limited and extensive cell-to-cell movement of RNA silencing in plants
The various manifestations of limited silencing movement were related because they shared similar properties, despite the diversity of their triggers. In particular, in both N.benthamiana and Arabidopsis, a near constant number of 10–15 cells was affected, suggesting progressive dilution of the signal from its source and/or a highly controlled mechanism of propagation. The same extent of movement was observed if silencing originated from a limited set of phloem companion cells (Figures 4–6) or from agro-infiltrated patches containing many primary silenced cells (Figures 1–3). Moreover, bombardment of GFP siRNAs in leaves of line 16c, which typically affects only a few cells, triggers the appearance of silencing foci comparable in diameter to the extent of silencing observed at the edge of much larger agro-infiltrated areas (Klahre et al., 2002). Based on these and other observations (Supplementary data 1) it is likely that silencing movement over 10–15 cells can be promoted from one single cell. Moreover, such movement occurs independently of the presence of homologous transcripts in adjacent cells (Figure 3).
Short-range movement of RbcS silencing was triggered by PVX derivatives with no DNA phase in their replication cycle (Figure 4F–H). Thus, the nucleic acid component of the signal involved is likely RNA. Synthesis of this RNA was not dependent upon SDE1 or SDE3 (Figures 5 and 6) and its spread was not influenced by homologous transcripts in recipient tissues (Figure 3). This makes the involvement of full-length target mRNA unlikely and excludes participation of the predicted RNA products of transitivity (i.e. de novo synthesized dsRNA, secondary siRNAs). Thus, primary siRNAs are good candidates for the short-distance signal molecule. Experiments with P1 (Figure 2) show that 21 nt siRNAs were sufficient for short-range movement of GFP silencing. Moreover, limited movement of RbcS silencing was also associated with production of siRNAs that were mainly 21 nt long (Figure 4I). Therefore, it is likely that movement of 21 nt siRNAs accounts for short distance spread of silencing. However, none of the experiments described here rules out the possibility that, although not necessary, the 25 nt siRNAs may also be sufficient for short-distance spread.
Limited silencing movement is likely through plasmodesmata because stomata guard cells, which are symplastically isolated from neighbouring cells by plasmodesmata occlusion (Wille and Lucas, 1984) were unaffected by the process in Arabidopsis (Figure 5F). Extensive cell-to-cell movement of silencing also likely occurs through plasmodesmata (Voinnet et al., 1998). In addition, it was manifested within the same tissues and was triggered by the same constructs that activated limited silencing movement. Extensive movement was linked to transitivity mediated by SDE1 and SDE3 because it correlated precisely with accumulation of secondary, but not primary, siRNAs. Strikingly, secondary siRNAs were almost exclusively of the 21 nt size class, the proposed nucleic acid component of the short-distance signal (Figure 6E).
A model for cell-to-cell movement of RNA silencing in plants
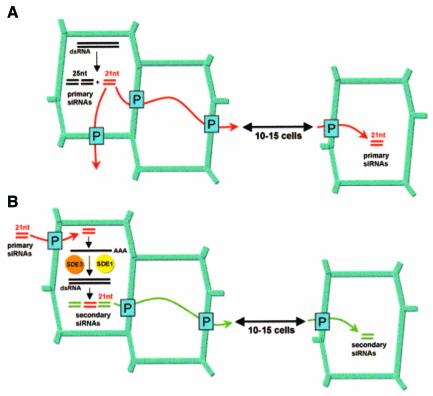
Limited and extensive cell-to-cell movement of silencing could be mediated by distinct mechanisms involving separate molecules. However, based on the results discussed above, the difference could be more simply explained in terms of a single movement process with varying intensities. In this model (Figure 7), local initiation of silencing would produce 21 nt and 25 nt primary siRNAs. The primary 21 nt siRNA would move to 10–15 adjacent cells, independently of the presence of homologous transcripts in those cells. This initial wave of movement could then have two possible outcomes. First, primary 21 nt siRNAs could initiate synthesis of secondary 21 nt siRNAs through the action of SDE1 and SDE3 using homologous transcripts as templates. As proposed for primary 21 nt siRNAs, the newly synthesized 21 nt siRNAs could then move over a further distance of 10–15 cells in which the same SDE1/SDE3-mediated process would be initiated. Such re-iterated short-distance signalling events would then eventually translate into extensive movement. The second possible outcome would be that silencing does not move any further because of a lack or inability of homologous transcripts to act as templates for SDE1 and SDE3 (as for the RbcS and sulphur mRNAs; Figure 4). This would preclude further production of 21 nt siRNAs (Figure 7) and movement would stop.
Fig. 7. Model for cell-to-cell movement of RNA silencing in plants. (A) Silencing can spread over 10–15 cells in the absence of amplification through movement of 21 nt primary siRNAs. (B) Extensive cell-to-cell movement requires 21 nt siRNA-induced de novo synthesis of dsRNA by the action of SDE1 and SDE3 using transgene mRNA as template. This leads to production of secondary 21 nt siRNAs that spread over a further 10–15 cells. P: plasmodesmata.
Separate long-distance and cell-to-cell movement processes?
Since transitivity leads exclusively to 21 nt siRNAs (Figure 6E), the model in Figure 7 predicts that the 25 nt siRNAs would be progressively diluted away from their source of initiation in the course of extensive silencing movement. This could not be tested in Arabidopsis because the atSUC2-GF-FG construct produced high levels of both siRNA species in the vasculature of every leaf. However, previous work in line 16c (Hamilton et al., 2002) showed that the 21 nt GFP siRNAs are indeed vastly over-represented compared with the 25 nt siRNAs in systemic tissues undergoing extensive cell-to-cell silencing movement (as depicted in Figure 1F). In contrast, both siRNA classes accumulate to similar high levels in the agro-infiltrated leaves where silencing is initially triggered (Hamilton et al., 2002; Figure 1C, inlay).
The same work nevertheless showed a tight correlation between 25 nt siRNAs in agro-infiltrated leaves and the onset of silencing in systemic leaves (Hamilton et al., 2002). Since systemic silencing requires the prior phloem transport of the silenced state between infiltrated and systemic tissues, these observations could be reconciled if there are separate signal molecules for cell-to-cell and long-distance transport. Thus, abundant 25 nt siRNAs (or a derivative/precursor molecules) synthesized in infiltrated tissues could act as a phloem-specific silencing signal. Long-distance transport of such molecules could induce de novo production of 21 nt siRNAs in or near the vasculature of young leaves. Extensive cell-to-cell movement of silencing would then proceed from the veins independently of the 25 nt siRNA (Figure 7). Thus, the vascular restriction of the 25 nt siRNA and apparent lack of amplification (Figure 6E) would cause its progressive under-representation in systemic leaf tissues. This idea is consistent with previous findings that graft transmission of transgene silencing can occur through sections of nontransgenic plants, in which the scope for relay amplification is therefore extremely reduced or inexistent (Palauqui et al., 1997).
The proposal of separate mechanisms for cell-to-cell and phloem transport of RNA silencing is supported by two experimental evidences. First, some silencing suppressors have contrasted effects on each transport process. Hence, the P1 protein did prevent systemic silencing but not its limited movement at the edge of agro-infiltrated patches. Conversely, AC2 did not suppress long-distance spread but inhibited limited movement (Figure 2A; Supplementary data 4). The second evidence is from experiments involving cadmium. Indeed, treatments of transgenic tobacco and line 16c with non-toxic concentrations of cadmium prevented phloem transport but not cell-to-cell spread of silencing targeted against GUS and GFP, respectively (Ueki and Citovsky, 2001).
Transitive RNA silencing in plants
Our results indicate that SDE3, in addition to SDE1, is required for transitivity in plants. Other silencing-related proteins have been genetically positioned upstream or at the step of dsRNA synthesis and it will be interesting to determine whether they also influence transitivity and extensive movement of silencing. These factors include AGO-1 (Fagard et al., 2000), a member of the ARGONAUTE/ZWILLE/RDE superfamily, and SGS3/SDE2, a protein of unknown function (Mourrain et al., 2000).
Recent work in wheat germ extracts suggests that distinct Dicer-like enzymes generate the 21 and 25 nt siRNAs in plants (Tang et al., 2003). Our analysis of transitivity in Arabidopsis indicates that the putative dsRNA synthesized by SDE1 and SDE3 is preferentially cleaved by a Dicer that generates the 21 nt siRNA species. This finding contrasts with the observation that de novo dsRNA synthesis in wheat germ extracts is linked to production of siRNAs that are almost exclusively 25 nt long (Tang et al., 2003). The most straightforward explanation is that the RdRp responsible for transitivity in vitro is not an SDE1 wheat homologue. Alternatively, reactions in vivo may be only partially recapitulated in the wheat germ extracts. For instance, wheat extracts may lack cellular components that normally link transitivity products to specialized Dicer-like enzymes with distinct processing activities. Identification of the RdRp responsible for transitivity in vitro will help to address this issue.
Possible biological roles for limited and extensive cell-to-cell movement of RNA silencing in plants
The discovery, in Arabidopsis, of abundant miRNAs with homology to transcription factor mRNAs indicates that RNA silencing, as well as an antiviral function, has roles in plant development (reviewed in Bartel and Bartel, 2003). Cell-to-cell movement of silencing could be relevant to both biological functions. Thus, extensive movement of an amplified virus-induced silencing signal could ensure that a large antiviral response is mounted in cells that are about to be infected. Limited silencing movement over a nearly constant number of cells could participate in non-cell-autonomous regulation of gene expression through miRNA trafficking. Indeed, many plant miRNAs are 21 nt long and are likely to function as siRNAs (Bartel and Bartel, 2003). For instance, movement of miRNAs over a set number of cells could generate gradients of gene expression in meristems and primordia. We are currently screening for Arabidopsis mutants with altered patterns of short distance spread of sulphur and GFP silencing. If cell-to-cell movement of RNA silencing has developmental functions, some of these mutants should exhibit aberrant or modified morphology.
Materials and methods
Transgenic plants and Agrobacterium
Nicotiana benthamiana lines 16c and Arabidopsis line GFP142 have been described previously (Ruiz et al., 1998; Dalmay et al., 2000). Grafting of N.benthamiana was performed according to Voinnet et al. (1998). The GFP142/sde1 and GFP142/sde3 plants result from an outcross of the Amp 243 locus (using wt C24 plants) that was present in the originally described sde1-1 and sde3-1 Arabidopsis mutants (Dalmay et al., 2000, 2001). Transformation of the atSUC2-GF-FG, atSUC2-SUL-LUS and atSUC2-RUB-RUB constructs with Agrobacterium strain GV3101 was performed as described previously (Bechtold et al., 1993). Selection was on medium containing 10 mg/l l-phosphinotricin. Agrobacterium-mediated transient expression of GFP, GUS and suppressors of silencing in N.benthamiana leaves was as described previously (Hamilton et al., 2002). Histochemical staining was as described previously (Jefferson et al., 1987).
GFP imaging
GFP expression was monitored with a hand-held UV light or under a Nikon SMZ1500 dissecting microscope coupled to a 100 W epifluorescence module (Nikon). A band-pass filter allowed removal of chlorophyll fluorescence. The LSM510 microscope (Zeiss) was used for confocal imaging. For transversal sections, the plant material was embedded in 1% low melting-point agarose and cut transversally with a razor blade.
DNA constructs
The cassettes for transient expression of GFP and silencing suppressors were as described previously (Hamilton et al., 2002). PVX-RbcS and the Rx construct have been described previously (Bendahmane et al., 1999; Jones et al., 1999). The atSUC2 promoter was PCR-amplified (Pfu turbo, Promega; primer sequences available on request) from plasmid pEP1 (Imlau et al., 1999). The resulting PCR product was inserted as a EcoRI-XhoI restriction fragment into pFGC5941 (www.ag.arizona.edu/chromatin/fgc5941.html), a binary vector containing a chalcone synthase (CHS) intron, designed to produce dsRNA in plants. The resulting plasmid was then used to clone the 400 bp PCR-amplified ‘GF’ fragment upstream of the CHS intron (primer sequences available on request). The antisense ‘GF’ fragment (referred to as ‘FG’) was then cloned downstream of the CHS intron. A similar approach was followed to create plasmids atSUC2-SUL-LUS and atSUC2-RUB-BUR. The Rbcs and sulphur fragments were 400 nt PCR products amplified from Arabidopsis cDNA (AGI no. At4g18480 for Sulphur and AGI no. At5g38410 for Rubisco small subunit 3b; primer sequences available upon request).
RNAs analysis
Total RNA extraction was performed with Tri-Reagent (Sigma). Analysis of high and low molecular weight RNA was as described except that the Perfect-Hyb buffer (Sigma) was used for hybridization (Hamilton et al., 2002). 32P-labelled RNA oligonuclotides of 21 and 24 nt were used as standards. Probes were DNA fragments labelled by random priming incorporation of [32P]dCTP (Amersham). The full-length GFP cDNA was used for the GFP-specific probe. The ‘GF’-specific probe was based on the ‘GF’ fragment of atSUC2-GF-FG. The non-overlapping ‘P’-specific probe was as described (Voinnet et al., 1998). The RbcS-specific probe corresponded to the insert carried by PVX-RbcS. All hybridization signals were detected by phosphorimaging.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank N.Sauer for plasmid pEP1, and D.Baulcombe and T.Dalmay for seeds of sde1 and sde3. We thank A.Maule, C.Lecellier and E.Parizotto for fruitful discussions, and R.Wagner and his team for excellent plant care. Work in our laboratory is supported by an ATIP from the CNRS.
References
- Bartel B. and Bartel,D.P. (2003) MicroRNAs: At the root of plant development. Plant Physiol., 132, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D.C. (1999) Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol., 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Bechtold N., Ellis,J. and Pelletier,G. (1993) In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. III, Sci. Vie, 316, 1194–1199. [Google Scholar]
- Bendahmane A., Kanyuka,K. and Baulcombe,D.C. (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell, 11, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Hamilton,A.J., Rudd,S., Angell,S. and Baulcombe,D.C. (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell, 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay T., Horsefield,R., Braunstein,T.H. and Baulcombe,D.C. (2001) SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J., 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard M., Boutet,S., Morel,J.-B., Bellini,C. and Vaucheret,H. (2000) AGO1, QDE-2 and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi and RNA interference in animals. Proc. Natl Acad. Sci. USA, 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu,S., Montgomery,M.K., Kostas,S.A., Driver,S.E. and Mello,C.C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature, 391, 806–811. [DOI] [PubMed] [Google Scholar]
- Hamilton A.J., Voinnet,O., Chappell,L. and Baulcombe,D.C. (2002) Two classes of short interfering RNA in RNA silencing. EMBO J., 21, 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein,E., Beach,D. and Hannon,G. (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cell extracts. Nature, 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Imlau A., Truernit,E. and Sauer,N. (1999) Cell-to-cell and long distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell, 11, 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh,T.A. and Bevan,M.W. (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J., 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelkmann W., Maiss,E. and Martin,R.R. (1992) The nucleotide sequence and genome organization of strawberry mild yellow edge-associated virus. J. Gen. Virol., 73, 475–479. [DOI] [PubMed] [Google Scholar]
- Jones L., Hamilton,A.J., Voinnet,O., Thomas,C.L., Maule,A.J. and Baulcombe,D.C. (1999) RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell, 11, 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U., Crete,P., Leuenberger,S.A., Iglesias,V.A. and Meins,F.J. (2002) High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl Acad. Sci. USA, 99, 11981–11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P. et al. (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell, 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Palauqui J.-C., Elmayan,T., Pollien,J.-M. and Vaucheret,H. (1997) Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J., 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin-Hernandez,A.M. and Baulcombe,D.C. (2001) Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J., 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Roberts A.G., Santa Cruz,S., Roberts,I.M., Prior,D.A.M., Turgeon,R. and Oparka,K.J. (1997) Phloem unloading in sink leaves of Nicotiana benthamiana: Comparison of a fluorescent solute with a fluorescent virus. Plant Cell, 9, 1381–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M.T., Voinnet,O. and Baulcombe,D.C. (1998) Initiation and maintenance of virus-induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa Cruz S., Roberts,A.G., Prior,D.A.M., Chapman,S. and Oparka,K.J. (1998) Cell-to-cell and phloem-mediated transport of potato virus X: The role of virions. Plant Cell, 10, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijen T., Fleenor,J., Simmer,F., Thijssen,K.L., Parrish,S., Timmons,L., Plasterk,R.H.A. and Fire,A. (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell, 107, 465–476. [DOI] [PubMed] [Google Scholar]
- Tang G., Reinhart,B.J., Bartel,D.P. and Zamore,P.D. (2003) A biochemical framework for RNA silencing in plants. Genes Dev., 17, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki S. and Citovsky,V. (2001) Inhibition of systemic onset of postranscriptional gene silencing by non-toxic concentrations of cadmium. Plant J., 28, 283–291. [DOI] [PubMed] [Google Scholar]
- Vaistij F.E., Jones,L. and Baulcombe,D.C. (2002) Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell, 14, 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O. (2001) RNA silencing as a plant immune system against viruses. Trends Genet., 17, 449–459. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Vain,P., Angell,S. and Baulcombe,D.C. (1998) Systemic spread of sequence-specific transgene RNA degradation is initiated by localised introduction of ectopic promoterless DNA. Cell, 95, 177–187. [DOI] [PubMed] [Google Scholar]
- Voinnet O., Pinto,Y.M. and Baulcombe,D.C. (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses. Proc. Natl Acad. Sci. USA, 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O., Lederer,C. and Baulcombe,D.C. (2000) A viral movement protein prevents systemic spread of the gene silencing signal in Nicotiana benthamiana. Cell, 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Wille A. and Lucas,W.J. (1984) Ultrastructural and histochemical changes on guard cells. Planta, 160, 129–142. [DOI] [PubMed] [Google Scholar]
- Winston W.M., Molodowitch,C. and Hunter,C.P. (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science, 295, 2456–2459. [DOI] [PubMed] [Google Scholar]