Abstract
The Lactococcus lactis Ll.LtrB group II intron uses a major retrohoming mechanism in which the excised intron RNA reverse splices into one strand of a DNA target site, while the intron-encoded protein uses a C-terminal DNA endonuclease domain to cleave the opposite strand and then uses the cleaved 3′ end as a primer for reverse transcription of the inserted intron RNA. Here, experiments with mutant introns and target sites indicate that Ll.LtrB can retrohome without second-strand cleavage by using a nascent strand at a DNA replication fork as the primer for reverse transcription. This mechanism connecting intron mobility to target DNA replication may be used by group II intron species that encode proteins lacking the C-terminal DNA endonuclease domain and for group II intron retrotransposition to ectopic sites.
Keywords: DNA replication/retrotransposon/reverse transcriptase/ribozyme
Introduction
Mobile group II introns found in bacterial and organelle genomes insert site-specifically into DNA target sites by a process termed retrohoming (Lambowitz et al., 1999; Belfort et al., 2002). Retrohoming has been characterized in the greatest detail for the yeast mtDNA aI1 and aI2 introns and the Lactococcus lactis Ll.LtrB intron, which encode proteins having reverse transcriptase (RT), RNA splicing (maturase) and DNA endonuclease activities. First, the intron-encoded protein (IEP) promotes RNA splicing by stabilizing the catalytically active structure of the intron RNA, and it remains associated with the excised intron lariat RNA in an RNP particle that mediates intron mobility. For mobility, the intron RNA in this RNP reverse splices directly into one strand of the DNA target site, while the IEP cleaves the second strand and uses the cleaved 3′ end as a primer for reverse transcription of the inserted intron RNA. The resulting intron cDNA is then integrated into the recipient DNA via cellular recombination or repair mechanisms.
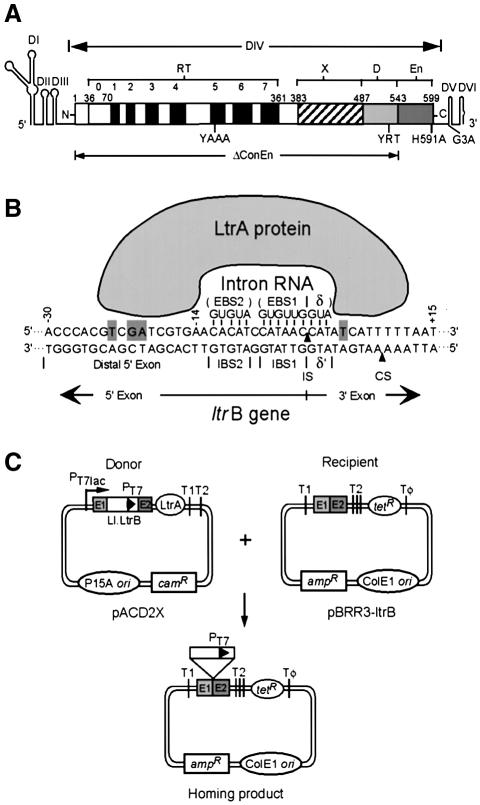
The yeast and lactococcal group II IEPs have four conserved domains: an N-terminal RT domain; domain X, a putative RNA-binding domain associated with maturase activity; a C-terminal DNA-binding domain (D); and a DNA endonuclease domain (En) (Figure 1A). The RNP particles formed during RNA splicing initiate mobility by using both the IEP and base-pairing of the intron RNA to recognize target sequences in double-stranded DNA (Figure 1B). RNPs bind DNA non-specifically and then search for target sites, with the IEP thought to first recognize a small number of specific bases in the distal 5′-exon region via major groove interactions (Singh and Lambowitz, 2001; Aizawa et al., 2003). These base interactions, bolstered by phosphate-backbone interactions, trigger local DNA unwinding, enabling the intron RNA’s EBS2, EBS1 and δ sequences to base pair to complementary DNA target sequences IBS2, IBS1 and δ′ for reverse splicing (EBS and IBS denote exon- and intron-binding sites, respectively). Second-strand cleavage occurs after a lag and requires additional interactions between the IEP and the 3′ exon. The DNA endonuclease domain, which carries out second-strand cleavage, contains conserved amino acid sequence motifs characteristic of the H-N-H family of DNA endonucleases, interspersed with two pairs of conserved cysteine residues (Gorbalenya, 1994; Shub et al., 1994). The H-N-H residues form part of a metal ion-binding fold containing a single catalytically essential Mg2+ ion, while the conserved cysteine residues likely function to stabilize the higher-order structure of the domain (San Filippo and Lambowitz, 2002).
Fig. 1. The Ll.LtrB group II intron, DNA target site interactions, and plasmid-based intron mobility assay. (A) Ll.LtrB intron. The intron is shown as a thin line, with RNA secondary structure domains DI–VI drawn schematically. The IEP, denoted LtrA protein, is shown as an open box. The protein contains an N-terminal RT domain, with conserved RT sequence motifs 0–7; domain X associated with maturase activity; and C-terminal DNA-binding (D) and DNA endonuclease (En) domains. The locations of mutations YAAA, YRT, H591A, G3A and ΔConEn are shown below. (B) Mechanism of DNA target site recognition. The DNA target site in the ltrB gene is recognized by an RNP complex containing the IEP (LtrA protein) and excised intron lariat RNA, with both protein interactions and base pairing of the intron RNA contributing to DNA target site recognition. Bases identified as being recognized directly by the LtrA protein (Singh and Lambowitz, 2001) are highlighted with gray backgrounds. The arrowheads pointing to the top and bottom strands indicate the intron-insertion site (IS) and second-strand cleavage site (CS), respectively. (C) Mobility assay. The intron-donor plasmid pACD2X is a CamR pACYC184-derivative (P15 replicon), which contains a 940-nt Ll.LtrB-ΔORF intron and flanking exons, with a phage T7 promoter inserted near the 3′ end of the intron. The intron is expressed from a T7lac promoter, with the IEP (LtrA protein) expressed from a position just downstream of the 3′ exon. The recipient plasmid pBRR3-ltrB is an AmpR pBR322-derivative, which contains the Ll.LtrB target site (ligated E1-E2 sequence of the ltrB gene) cloned upstream of a promoterless tetR gene. The insertion of the intron into the target site activates the expression of the tetR gene, yielding TetR+AmpR colonies. T1, T2 and Tφ are E. coli rrnB T1, T2 and phage T7 transcription terminators, respectively.
In contrast to the yeast mtDNA and L.lactis Ll.LtrB introns, many bacterial group II IEPs lack the H-N-H DNA endonuclease domain and some also lack cognates of the C-terminal DNA-binding domain (Martínez-Abarca and Toro, 2000b; Dai and Zimmerly, 2002; San Filippo and Lambowitz, 2002). Among the best characterized is the Sinorhizobium meliloti group II intron RmInt1, which encodes a protein having RT and X domains, but only a short (20 amino acid) C-terminal extension that appears to be unrelated to the DNA-binding domains of the yeast mtDNA and lactococcal IEPs (Martínez-Abarca et al., 1998; San Filippo and Lambowitz, 2002). Nevertheless, RmInt1 retrohomes efficiently, inserting into 20–48% of target plasmids introduced into intron-containing strains (Martínez-Abarca et al., 2000). Biochemical studies showed that the RmInt1 IEP has RT activity and the intron RNP can reverse splice into double- or single-stranded DNA substrates, but cannot carry out site-specific second-strand cleavage, as expected from the lack of DNA endonuclease domain (Muñoz-Adelantado et al., 2003). Like the yeast mtDNA and L.lactis Ll.LtrB introns, RmInt1 uses both the IEP and base pairing of the intron RNA to recognize its DNA target site, but recognition by the IEP appears more limited, with just two key nucleotide residues flanking the base-paired region being critical (Jiménez-Zurdo et al., 2003). Together, these findings suggest that retrohoming of group II introns that lack the C-terminal DNA endonuclease activity is similar to that of the yeast and lactococcal introns in involving reverse splicing into DNA target sites and reverse transcription of the inserted intron RNA, but requires an alternate mechanism to procure a primer for reverse transcription (Muñoz-Adelantado et al., 2003). Possible alternate mechanisms include priming by non-specific DNA nicks; de novo initiation, as found for the related Mauriceville retroplasmid RT (Wang et al., 1993); a protein primer, like hepatitis B virus RT (Wang and Seeger, 1992); or the use of a nascent strand generated during DNA replication.
In addition to retrohoming, group II introns retrotranspose to ectopic sites that resemble the normal homing site at low frequency (Lambowitz et al., 1999; Belfort et al., 2002). This process enables intron dispersal during evolution and may be relevant to the origin of nuclear pre-mRNA introns, which are thought to be decendents of mobile group II introns (Michel and Ferat, 1995). Work from several laboratories has shown that retrotransposition occurs predominantly by reverse splicing into DNA target sites, but most retrotransposition sites lack sequences required for efficient second-strand cleavage, again requiring an alternate mechanism to generate a reverse transcription primer (Yang et al., 1998; Martínez-Abarca and Toro, 2000a; Dickson et al., 2001; Ichiyanagi et al., 2002). Notably, retrotransposition sites generally have a good match for the IBS1 sequence recognized by base pairing of the intron RNA, but have more divergent IBS2 sequences and lack distal 5′-exon sequences recognized by the IEP (Ichiyanagi et al., 2002 and references therein). In addition, recent findings have shown that retrotransposition of the Ll.LtrB intron in L.lactis has a pronounced strand bias, consistent with the preferred use of the nascent lagging strand as a primer for reverse transcription (Ichiyanagi et al., 2002). At present, however, it remains uncertain whether retrotransposition occurs by reverse splicing into double- or single-stranded DNA sites, and whether the observed strand bias does in fact reflect use of nascent lagging-strand primers or has some other explanation.
Here, we used a combination of biochemical experiments and powerful Escherichia coli genetic assays for retrohoming of the Ll.LtrB intron to investigate how group II intron mobility can occur when second-strand cleavage is blocked by mutations. Our results indicate that the Ll.LtrB intron can retrohome in the absence of second-strand cleavage by using a nascent strand generated during DNA replication as a primer for reverse transcription, but with a strong preference for use of the leading over the lagging DNA strand, the opposite of the strand bias found for retrotransposition.
Results
Retrohoming of wild-type and mutant Ll.LtrB introns
To investigate how retrohoming occurs in the absence of second-strand cleavage, we used the Ll.LtrB mutant YRT, which has three point mutations in the C-terminal region (Y529A, R531A and T533A; Figure 1A). This mutant has no detectable DNA endonuclease activity in vitro (<1% wild type), but retains high RT and reverse splicing activity (117 and 59% wild type, respectively; San Filippo and Lambowitz, 2002). A second DNA endonuclease mutant used in some experiments, H591A, has a higher level of residual DNA endonuclease activity (∼2% wild type) along with somewhat decreased RT activity (64% wild type) and 90% wild-type reverse splicing activity (San Filippo and Lambowitz, 2002). In contrast, deletion of the conserved DNA endonuclease domain (ΔConEn) or other previously characterized mutations in the conserved H-N-H or cysteine motifs strongly inhibit RT activity in addition to second-strand cleavage, possibly reflecting a structural interaction between the DNA endonuclease and RT domains (San Filippo and Lambowitz, 2002). Consequently, these mutants cannot be used to identify effects due specifically to the absence of DNA endonuclease activity.
First, the mobility frequencies of the wild-type and mutant Ll.LtrB introns were determined by using an E.coli two-plasmid assay in which a donor intron with a phage T7 promoter inserted near its 3′ end integrates into a target site upstream of a promoterless tetR gene, thereby activating that gene (Figure 1C; Guo et al., 2000; Karberg et al., 2001). To be able to introduce mutations into the LtrA protein without affecting the structure of the intron RNA, we used an intron-donor plasmid, pACD2X, which contains a 940-nt Ll.LtrB-ΔORF intron and short flanking exons, with the IEP expressed from a position just downstream of the 3′ exon.
As summarized in Table I, under the conditions used, the wild-type Ll.LtrB intron had a mobility frequency of 52 ± 7% based on the proportion of TetR colonies, while the YRT mutant, which lacks detectable DNA endonuclease activity, had a mobility frequency of 0.54 ± 0.07%, ∼1% wild type but still relatively high by absolute standards. H591A, which has a mutation in the DNA-endonuclease active site, had a somewhat lower mobility frequency (0.11 ± 0.02%), possibly reflecting partial inhibition of RT activity in addition to DNA endonuclease activity. G3A, which has a point mutation in intron RNA domain V that inhibits both RNA splicing and reverse splicing, also showed a low level of residual mobility (0.071%; see also D’Souza and Zhong, 2002; Figure 1A). In contrast, the mutant YAAA, which lacks RT activity, and a C-terminal truncation mutant (ΔConEn) that is deficient in RT activity as well as second-strand cleavage activity, were not detectably mobile (mobility frequencies <0.001%). The double mutant YRT + YAAA also had no detectable mobility. Together, these findings show that mutants that are defective in second-strand cleavage, but retain reverse splicing and RT activity still show significant residual homing, which is dependent on the RT activity of the IEP.
Table I. Mobility frequencies of wild-type and mutant Ll.LtrB introns.
| Ll.LtrB intron | Mobility frequency (%) |
|---|---|
| WT | 52 ± 7 |
| YRT | 0.54 ± 0.07 |
| H591A | 0.11 ± 0.02 |
| G3A | 0.071 ± 0.002 |
| ΔConEn | <0.001 |
| YAAA | <0.001 |
| YRT + YAAA | <0.001 |
Mobility frequencies of wild-type and mutant Ll.LtrB introns were determined by using an E.coli two-plasmid assay in which an intron with a phage T7 promoter near its 3′ end inserts into a target site upstream of a promoterless tetR gene, thereby activating that gene. Mobility frequencies are the ratio of TetR + AmpR/AmpR colonies and are the mean ± SD of at least three independent experiments in each case.
Retrohoming of the YRT mutant occurs by reverse splicing into double-stranded DNA
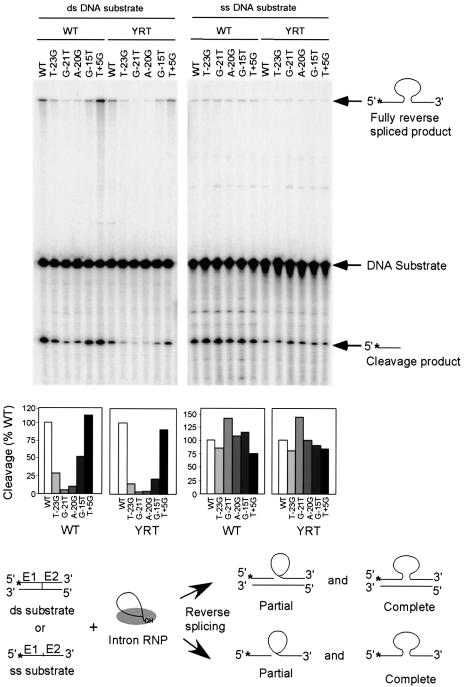
We next wanted to investigate whether the YRT mutant uses a double- or single-stranded DNA target for mobility. In the case of the yeast aI2 intron, we showed previously that deletion of the distal 5′-exon region of the DNA target site inhibits reverse splicing into double-stranded DNA target sites, but not otherwise identical single-stranded DNA target sites, suggesting that interactions between the IEP and this region are required primarily for DNA unwinding (Guo et al., 1997). To identify mutations at specific positions of the Ll.LtrB target site that similarly inhibit reverse splicing into double-stranded, but not single-stranded DNA, we carried out biochemical assays of reverse splicing with double- and single-stranded DNA substrates having nucleotide substitutions at the five positions found to be most critical for mobility of the wild-type Ll.LtrB intron (T-23, G-21, A-20, G-15 and T+5) (Figure 2; Guo et al., 2000; Zhong et al., 2003). In these assays, DNA substrates labeled at the 5′ end of the top strand were incubated with wild-type and YRT mutant RNP particles, and then analyzed by gel electrophoresis and autoradiography to detect labeled products resulting from partial and complete reverse splicing of the intron RNA into that strand (Figure 2). For both wild-type and the YRT mutant, all four distal 5′-exon mutations (T-23G, G-21T, A-20G and G-15T) inhibited reverse splicing into the double-stranded, but not otherwise identical single-stranded DNA substrates, whereas the 3′-exon mutation T+5G, which was shown previously to specifically inhibit second-strand cleavage (Mohr et al., 2000), did not significantly affect reverse splicing into either double- or single-stranded DNA.
Fig. 2. Reverse splicing assay with double- and single-stranded DNA target sites. Reverse splicing was assayed by incubating wild-type or YRT mutant RNP particles with 5′-top-strand labeled double- or single-stranded 70mer DNAs containing the wild-type target sequence (positions –35 to +35) or the indicated mutations. The products were analyzed in a denaturing 6% polyacrylamide gel, which was dried and quantitated with a PhosphorImager. Labeled products are indicated to the right of the gel, with the position of the label indicated by an asterisk. The schematic at the bottom diagrams reverse splicing of the intron RNA into double- or single-stranded DNA. Partial reverse splicing results in cleaved 5′-exon (the predominant product) and intron lariat linked to 3′-exon (not labeled), while complete reverse splicing results in insertion of linear intron RNA between the two DNA exons. The bar graphs show reverse splicing activity relative to the wild-type DNA target site based on quantitation of the partially and fully reverse spliced products.
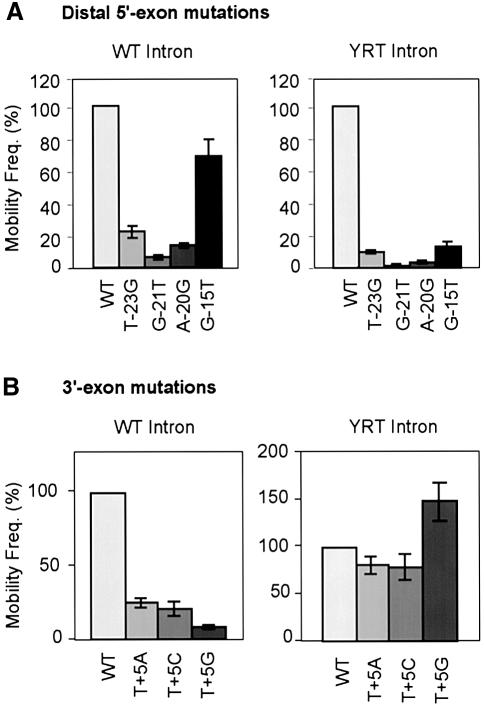
Figure 3A shows the effect of the same distal 5′-exon mutations in intron mobility assays. Significantly, all four mutations inhibited the mobility of both the wild-type and the YRT mutant introns, with the extent of inhibition roughly paralleling the effect of the mutation on reverse splicing into double-stranded DNA. The small amount of residual mobility with the mutant target sites could reflect incomplete inhibition of reverse splicing and/or a subset of events that occur by reverse splicing into transiently single-stranded DNA targets. These findings indicate that mobility of the YRT mutant occurs by a mechanism that is similar to wild type in involving reverse splicing into double-stranded DNA.
Fig. 3. Mobility assays with wild-type and mutant DNA target sites. Mobility assays were carried out as described in Figure 1 and Materials and methods, with donor plasmids expressing the wild-type (WT) or YRT mutant introns and recipient plasmids containing wild-type or indicated mutant Ll.LtrB target sites (positions –30 to +15). (A) Distal 5′-exon mutations T-23G, G-21T, A-20G and G-15T; (B) 3′-exon mutations T+5A, T+5C and T+5G. The bar charts show mobility frequencies for mutant DNA target sites relative to the wild-type DNA target site assayed in parallel. Values are the mean ± SD of three independent experiments.
Retrohoming of the YRT mutant does not require site-specific second-strand cleavage
Figure 3B compares the mobility of the wild-type and YRT mutant introns with DNA target sites having mutations at 3′-exon position T+5, which is not required for reverse splicing, but is required for second-strand cleavage (see above; Mohr et al., 2000). The data show that although all three nucleotide substitutions at this position inhibited mobility of the wild-type intron by 4- to 10-fold, they did not substantially inhibit the mobility of the YRT mutant. In fact, the T+5G mutation, which had the lowest mobility frequencies for the wild-type intron, gave increased mobility frequencies for the YRT mutant. These findings indicate that retrohoming of the YRT mutant differs from wild type in not depending on site-specific second-strand cleavage, as expected from the lack of DNA endonuclease activity.
YRT mutant RNPs cannot carry out targetDNA-primed reverse transcription (TPRT)
Although the YRT mutant showed no detectable second-strand cleavage at position +9 with DNA oligonucleotide substrates (San Filippo and Lambowitz, 2002), it remained possible that the mutant could cleave at a low level or at farther downstream sites, or cleave non-specifically at multiple sites and then use the cleaved 3′ ends to prime reverse transcription. It was also possible that the mutant could initiate cDNA synthesis de novo or by using the RT as a protein primer (see Introduction).
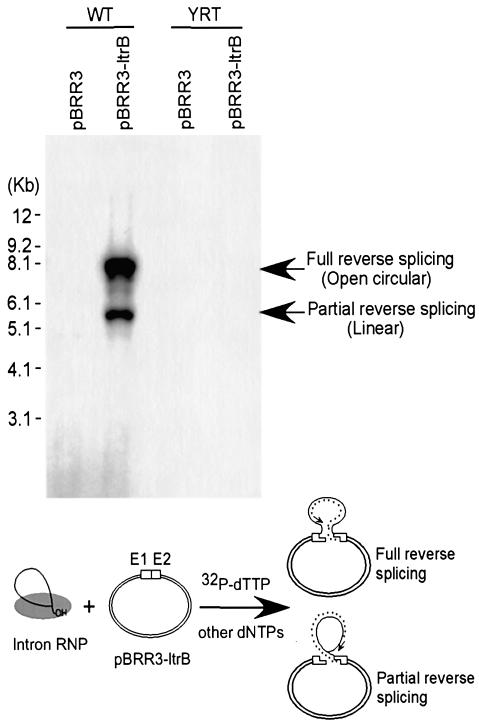
To assess these possibilities, we carried out target DNA-primed reverse transcription assays in which the recipient plasmid pBRR3-ltrB, shown to support mobility of the YRT mutant in vivo, was incubated with wild-type or mutant RNPs in the presence of [32P]dTTP and other unlabeled dNTPs. The products were then analyzed by gel electrophoresis and autoradiography to detect [32P]dTTP incorporated by reverse transcription of the inserted intron RNA. Figure 4 shows that the wild-type RNPs yielded the expected labeled bands with recipient plasmid pBRR3-ltrB, but not with control plasmid pBRR3, which lacks the Ll.LtrB target site. In contrast, although the YRT RNPs have high RT activity and can reverse splice into the target site, they gave no detectable TPRT product (<0.2% wild-type activity), presumably because they are unable to carry out second-strand cleavage to generate a primer for reverse transcription. These findings indicate that the YRT mutant IEP does not generate a primer by either specific or non-specific DNA cleavage and is not capable of de novo initiation or protein priming.
Fig. 4. Target DNA-primed reverse transcription assay. Wild-type and YRT mutant RNP particles were incubated with recipient plasmid pBRR3-ltrB, which contains the Ll.LtrB target site (positions –30 to +15), or control plasmid pBRR3, which lacks the Ll.LtrB target site, in the presence of [α-32P]dTTP and other dNTPs. Products were analyzed in a 0.7% agarose gel, which was dried and scanned with a PhosphorImager. DNA marker positions are shown to the left, and the products are identified to the right based on previous characterization (Saldanha et al., 1999). The schematic at the bottom shows the TPRT reaction, with products resulting from cDNA synthesis after partial or complete reverse splicing of the intron RNA into the DNA target site. The small asterisks indicate cDNA with the arrow indicating the direction of reverse transcription.
Mobility of YRT displays a replicationorientation bias
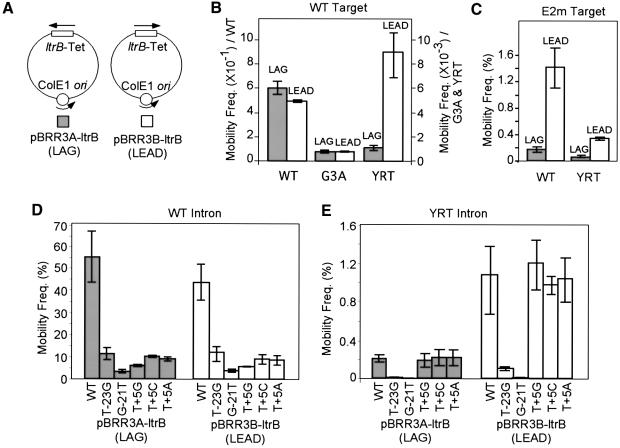
One possible mechanism for mobility of the YRT mutant in the absence of second-strand cleavage is to use a nascent strand generated during target DNA replication as a primer for reverse transcription of the inserted intron RNA. To test this possibility, we carried out mobility assays with recipient plasmids in which the Ll.LtrB target site and tetR reporter gene were cloned in opposite orientations relative to the direction of DNA replication, so that reverse transcription of the inserted intron RNA could potentially use either a nascent leading (LEAD) or lagging (LAG) strand DNA primer (Figure 5A). As a control, we also included mutant G3A, which has a point mutation in intron RNA domain V that inhibits RNA splicing and reverse splicing, but still shows a low level of residual mobility comparable to that of YRT (Table I).
Fig. 5. Mobility assays with DNA target sites cloned in opposite orientations relative to the direction of plasmid replication. (A) Recipient plasmids pBRR3A-ltrB and pBRR3B-ltrB. The plasmids contain the ltrB target site/tetR cassette cloned in opposite orientations denoted LAG or LEAD, depending on whether the nascent lagging or leading DNA strands could be used as a primer for reverse transcription of the inserted intron RNA. (B) Mobility assays with the wild-type (WT), G3A or YRT mutant introns and recipient plasmids pBRR3A-ltrB and pBRR3B-ltrB. The scale on the left is for the wild-type intron, and the scale on the right is for the G3A and YRT mutant introns. (C) Mobility assays with the wild-type Ll.LtrB intron and recipient plasmids containing different orientations of mutant DNA target site E2m, which has 3′-exon mutations that inhibit second-strand cleavage but not reverse splicing (see Materials and methods). (D and E) Mobility assays with the wild-type (D) or YRT mutant (E) introns and recipient plasmids containing different orientations of mutant DNA target sites having distal 5′-exon (T-23G and G-21T) or 3′-exon (T+5G, T+5C and T+5A) mutations. Mobility assays were carried out as described in Figure 1 and Materials and methods. Values are the mean ± SD of three independent experiments.
Strikingly, the wild-type Ll.LtrB intron and the G3A mutant displayed little if any replication orientation bias, whereas the YRT mutant showed a pronounced (8-fold) replication orientation bias, consistent with the preferred use of the nascent leading DNA strand as a primer for reverse transcription of the inserted intron RNA (Figure 5B; Table II). The endonuclease-deficient mutant (H591A) showed the same strand bias, but it was less pronounced (2- to 3-fold), likely reflecting its residual DNA endonuclease activity, which permits replication-orientation-independent mobility (not shown). We confirmed that there was no differential loss of the recipient plasmid with either target site orientation in these experiments (not shown). The strand bias for the YRT mutant was additionally confirmed by mixing experiments in which cells containing the YRT donor and either orientation recipient plasmid were mixed 1:1 before induction and plating. PCR screening of TetR colonies showed that ∼90% resulted from intron homing into the recipient plasmid that permits use of the nascent leading strand as reverse transcription primer (not shown).
Table II. Mobility assays with target sites cloned in high and low copy number plasmids.
| Donor intron/target site | Orientation | Mobility frequency (%) |
Fold decrease | |
|---|---|---|---|---|
| pBR322 | pSC101 | |||
| WT/WT | LEAD | 49 ± 1 | 9.8 ± 0.3 | 5 |
| LAG | 59 ± 5 | 9.2 ± 0.3 | 6 | |
| WT/G-21T | LEAD | 4.6 ± 0.7 | 0.65 ± 0.04 | 7 |
| LAG | 4.1 ± 0.9 | 0.59 ± 0.17 | 7 | |
| WT/T-23G | LEAD | 12.6 ± 3.4 | 1.5 ± 0.4 | 9 |
| LAG | 11.6 ± 2.8 | 1.5 ± 0.3 | 8 | |
| G3A/WT | LEAD | 7.6 ± 0.4 × 10–2 | 1.7 ± 0.2 × 10–2 | 4 |
| LAG | 7.7 ± 1.4 × 10–2 | 1.9 ± 0.2 × 10–2 | 4 | |
| YRT/WT | LEAD | 0.89 ± 0.16 | 4.1 ± 0.7 × 10–3 | 215 |
| LAG | 0.11 ± 0.02 | 8.0 ± 2.3 × 10–4 | 138 | |
| WT/E2m | LEAD | 1.4 ± 0.3 | 2.0 ± 0.2 × 10–2 | 69 |
| LAG | 0.17 ± 0.04 | 8.2 ± 1.4 × 10–4 | 205 | |
| YRT/E2m | LEAD | 0.33 ± 0.02 | 3.2 ± 0.2 × 10–3 | 105 |
| LAG | 0.050 ± 0.017 | 4.2 ± 0.2 × 10–4 | 118 | |
Mobility of wild-type, G3A and YRT mutant Ll.LtrB introns was assayed using high copy number (pBR322-based) and low copy number (pSC101-based) recipient plasmids in which the wild-type (WT) or mutant Ll.LtrB intron target sites were cloned in opposite orientations relative to the direction of DNA replication. Mobility frequencies are the mean ± SD for at least three independent experiments in each case.
To determine whether there are differences in the retrohoming mechanism with DNA target sites cloned in different orientations, we tested the effects of mutations in different regions of the DNA target site (Figure 5D and E). Retrohoming of the wild-type intron with both target site orientations was inhibited by both distal 5′-exon mutations (T-23 and G-21T) and 3′-exon mutations (T+5A, C or G), while retrohoming of the YRT intron with both target site orientations was sensitive to the distal 5′-exon mutations, but not the 3′-exon mutations. These findings indicate that although the leading-strand orientation is preferred, retrohoming of the YRT mutant with both target site orientations occurs predominantly by reverse splicing into double-stranded DNA without site-specific second-strand cleavage.
The wild-type Ll.LtrB intron shows a replication origin orientation bias with DNA target sites that do not support second-strand cleavage
The above results suggest that the YRT mutant, which is unable to carry out second-strand cleavage, might use a nascent DNA strand at a replication fork as an alternate primer for reverse transcription. Previous experiments in which a wild-type intron with randomized EBS and δ sequences integrated at randomly selected sites in the E.coli chromosome showed a similar leading-strand bias for the subset of integration sites lacking T+5, suggesting that the wild-type Ll.LtrB intron can use a similar mechanism (Zhong et al., 2003). To test this directly, we carried out mobility assays using the wild-type Ll.LtrB intron with a DNA target site (E2m) that could support reverse splicing, but has multiple 3′-exon changes (see Materials and methods) that abolish second-strand cleavage (not shown). As shown in Figure 5C, the wild-type intron now behaves similarly to the YRT mutant in showing the expected leading-strand bias. This finding indicates that the replication orientation bias is due to inhibition of second-strand cleavage and not to the YRT mutation.
Dependence of intron mobility on DNA replication
Additional evidence that mobility of the Ll.LtrB intron in the absence of second-strand cleavage is dependent on DNA replication was obtained by comparing the mobility frequencies with DNA target sites cloned in high copy number (pBR322-based) and low copy number (pSC101-based) recipient plasmids. As summarized in Table II, the mobility frequency of the wild-type intron decreased 5- or 6-fold with the low copy number target site in either orientation. Furthermore, the decrease was similar for the G3A mutant, which is defective in the initial reverse splicing step, and only slightly greater (7- to 9-fold) for the wild-type intron with mutant DNA target sites having distal 5′-exon mutations (G-21T and T-23G) that affect specific binding of the RNPs. In contrast, both the YRT mutant with the wild-type target site and the wild-type intron with the E2m target site, the combinations for which mobility occurs in the absence of second-strand cleavage, showed a much more dramatic 69- to 215-fold decrease, presumably reflecting a greater dependence on the number of DNA replication events per cell cycle. This number is equal to plasmid copy number, but stochastically divided so that some plasmids undergo multiple replication events in each cycle (Rownd, 1969; Bazaral and Helinski, 1970). We also used real-time PCR to assay mobility of the wild-type and YRT mutant introns with recipient plasmids the replication of which is temperature sensitive (Hamilton et al., 1989). Although the YRT mutant was mobile at the plasmid’s permissive temperature (30°C), but not the restrictive temperature (37°C), as expected, the wild-type Ll.LtrB intron was also mobile at the permissive but not the restrictive temperature (J.Zhong and A.M.Lambowitz, unpublished data). These findings indicate that mobility of the wild-type intron is also dependent on DNA replication for some step, perhaps second-strand DNA synthesis.
Discussion
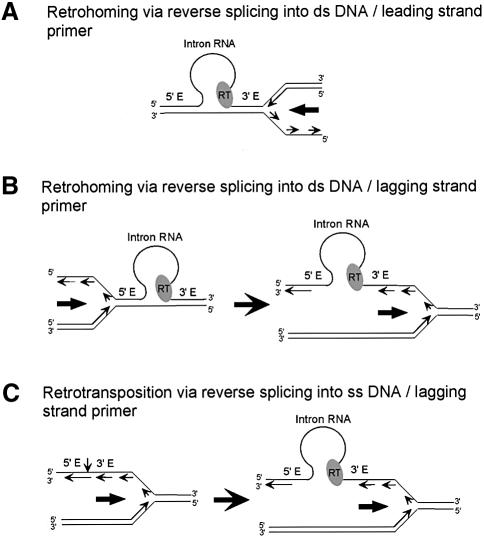
Our results indicate that retrohoming of the Ll.LtrB intron in the absence of second-strand cleavage occurs predominantly by reverse splicing of the intron RNA into double-stranded DNA, followed by use of a nascent leading DNA strand as a primer for reverse transcription of the inserted intron RNA (Figure 6A). First, we found that retrohoming of the YRT mutant, which lacks detectable DNA endonuclease activity, is similar to that of the wild-type intron in being sensitive to DNA target site mutations that inhibit reverse splicing into double- but not single-stranded DNA and in requiring the RT activity of the IEP. It differs, however, in being insensitive to mutations at 3′-exon positions required for second-strand cleavage and in showing a pronounced replication orientation bias consistent with the preferred use of the leading DNA strand as a primer for reverse transcription. Furthermore, we found the same preference for leading-strand DNA primers for the wild-type intron with both plasmid and chromosomal DNA target sites that support reverse splicing but not second-strand cleavage (this sudy; Zhong et al., 2003). Finally, comparison of mobility frequencies with high and low copy number recipient plasmids indicates that mobility in the absence of second-strand cleavage is strongly dependent on the frequency of DNA replication events. The YRT mutant also shows a lower level of mobility with target sites in the opposite orientation, which responds similarly to mutations or recipient plasmid copy number, and thus likely occurs by reverse splicing into double-stranded DNA followed by use of a nascent lagging strand to prime reverse transcription (Figure 6B). It remains possible that priming via non-specific opposite strand nicks makes a smaller contribution to mobility in both target sites orientations. As discussed below, combining our results with those of Ichiyanagi et al. (2002), we can also infer that retrotransposition of the Ll.LtrB intron to ectopic sites in L.lactis likely occurs by reverse splicing into single-stranded DNA followed by use of a nascent lagging DNA strand as a reverse transcription primer (Figure 6C).
Fig. 6. Models for group II intron mobility mechanisms using nascent leading and lagging-strand DNA primers at a replication fork. (A and B) Retrohoming via reverse splicing into double-stranded DNA, followed by use of a nascent leading strand (A) or lagging strand (B) as a primer for reverse transcription of the inserted intron RNA. After reverse splicing into the leading-strand template, the intron RNP is positioned to directly use the nascent leading strand as a primer prior to passage of the replication fork. In contrast, after reverse splicing into the lagging-strand template, the DNA replication machinery must traverse the inserted intron RNP prior to use of a lagging-strand primer. The observed leading-strand preference may reflect interference between the replication machinery and inserted intron RNP during passage of the replication fork in the lagging-strand orientation. (C) Retrotransposition of the Ll.LtrB intron in L.lactis. The group II intron RNP reverse splices into single-stranded DNA behind the replication fork and uses a nascent lagging strand to prime reverse transcription (see also Ichiyanagi et al., 2002). The models account for the opposite strand preferences of group II intron retrohoming and retrotransposition and for the finding that most group II intron retrotransposition sites lack distal 5′-exon sequences shown here to be required for efficient reverse splicing into double-stranded DNA. Some retrotransposition events also occur via reverse splicing into the leading-strand template and may use a nascent leading strand or non-specifically nicked DNA strand as primer.
The finding that retrohoming of the YRT mutant is abolished by a mutation that inhibits the RT activity of the IEP suggests that the host DNA polymerase does not simply copy through the inserted intron RNA, but instead that the nascent strand generated during DNA replication is used to prime reverse transcription by the IEP. We speculate that the host DNA polymerase disengages upon encountering the highly structured intron or tightly bound IEP, leaving the nascent DNA strand in position to be used as a primer by the IEP. Since the LtrA protein has very low processive DNA-dependent DNA polymerase activity (J.Zhong and A.M.Lambowitz, unpublished data), it is likely that a host replicative DNA polymerase resumes DNA synthesis after reverse transcription of the intron RNA. The situation may be analogous to the mode of action of a lesion bypass DNA polymerase, which acts at the site of a DNA lesion after the host replicative polymerase disengages and then dissociates to permit resumption of replication. In this case, the continuity of replication is thought to be mediated by the interaction of the two DNA polymerases with the β-clamp processivity factor, which remains associated with the template DNA during the polymerase switches (Pham et al., 2001). The possibility of a direct interaction between Ll.LtrB RNPs and the DNA replication machinery was raised previously by the finding that an Ll.LtrB intron with randomized target-site recognition sequences (EBS and δ sequences) shows a strong bias for integration at sites near the E.coli chromosomal DNA replication origin (Zhong et al., 2003).
The preference found here for leading-strand DNA primers can be explained simply by the geometry at the replication fork. After reverse splicing into the leading-strand template, the intron RNA is positioned to directly use a nascent leading-strand primer prior to passage of the replication fork (Figure 6A). In contrast, if the intron RNA reverse splices into the lagging-strand template, the replication fork must traverse the inserted intron RNP prior to use of a lagging-strand primer (Figure 6B). In that case, the inserted intron RNP may interfere with or be disrupted by the passage of the replication fork, leading to a reduced efficiency for the use of lagging-strand primers. Other mechanisms, including direct interaction of the group II intron RNPs with the DNA replication machinery, could also contribute to the observed strand bias.
In addition to providing insight into mobility mechanisms, our results show that mutations at all critically recognized positions in the distal 5′-exon region of the DNA target site inhibit reverse splicing of the Ll.LtrB intron into double-stranded but not otherwise identical single-stranded DNA target sites, suggesting that recognition of these positions by the IEP is required primarily for DNA unwinding, as expected for an early step in DNA target site recognition. These findings extend previous work with the yeast aI2 intron based on deletion of different segments of the DNA target site (Guo et al., 1997). The latter studies also showed that the target specificity for reverse splicing into single-stranded DNA is lower than that for double-stranded DNA, with the IBS and δ′ sequences being sufficient for the reaction, but that the IEP also plays a role, since deletion of the C-terminal DNA binding domain inhibited reverse splicing into single-stranded DNA by ∼90% (Guo et al., 1997). The finding here that none of the critical bases recognized by the IEP is required for efficient reverse splicing into single-stranded DNA suggests that this additional role involves largely non-sequence specific phosphate–backbone interactions, which occur on both DNA strand (Singh and Lambowitz, 2001).
The mobility mechanisms found here using nascent leading or lagging strand DNA primers may be used by group II introns that encode proteins lacking a DNA endonuclease domain. Indeed, the use of a nascent DNA strand as a primer for reverse transcription is a possible explanation for the previously puzzling observation that retrohoming of the S.meliloti RmInt1 group II intron appeared limited to initial generations after the introduction of a target plasmid into an intron-containing strain, saturating with ∼50% of the recipient plasmid. This 50% upper limit could reflect that the replication-dependent mobility process always generates a copy of wild-type recipient plasmid in addition to a homing product. In addition, dependence on DNA replication may explain why some group II introns insert into conjugating plasmids (e.g. Yeo et al., 1997). Group II introns that encode proteins lacking DNA endonuclease activity may have additional adaptations that facilitate efficient mobility via a DNA replication-generated primer. Such adaptations could again include specific interactions with the host replication machinery, perhaps permitting more efficient targeting of single-stranded regions, or encoding RTs that have higher processive DNA-dependent DNA polymerase activity, thereby enabling the RT to continue to replicate the host chromosomal DNA for longer periods after passage through the intron RNA. Ichiyanagi et al. (2002) noted that a number of group II introns that lack a C-terminal DNA endonuclease domain (mainly bacterial class C introns whose orientation was known), are found on the lagging template strand, a bias suggesting that these introns might retrohome preferentially by reverse splicing into single-stranded DNA (see above). In addition, priming via non-specific nicks on the opposite strand may be used in some cases.
The use of a nascent DNA strand as a reverse transcription primer is also likely a major mechanism used for the retrotransposition of group II introns to ectopic sites, which typically lack sequences required for site-specific second-strand cleavage. As noted previously, Ichiyanagi et al. (2002) studying retrotransposition of the Ll.LtrB intron in L.lactis found a replication orientation bias, consistent with the preferred use of lagging-strand DNA primers, opposite that found here for retrohoming. Furthermore, retrotransposition sites generally lack critical distal 5′-exon sequences, whose recognition by the IEP is shown here to be required for efficient reverse splicing into double-stranded but not single-stranded DNA (see Figure 2). Together, these findings support the hypothesis that these retrotransposition events occurred predominantly by reverse splicing of the intron RNA into single-stranded DNA at a replication fork, followed by the use of a nascent lagging strand as a primer (Figure 6C; Ichiyanagi et al., 2002). The low frequency of Ll.LtrB intron retrotransposition in L.lactis (1.8 × 10–4) presumably reflects the infrequent occurrence of single-stranded sites, which permit intron integration with relaxed target specificity. The implication is that inaccurate reverse splicing at double-stranded DNA sites occurs at much lower frequency than that dictated by the availability of single-stranded sites. Nevertheless, a fraction of Ll.LtrB intron retrotransposition sites were found on the leading-strand template, suggesting that retrotransposition by inaccurate reverse splicing into double-stranded DNA sites may also occur in some cases. The balance between these two retrotransposition mechanisms may differ for different group II introns or even for the same intron under different conditions depending on the target specificity at double-stranded sites and the availability of single-strand sites.
With respect to evolution, it has been suggested that ancestral mobile group II introns were similar to S.meliloti RmIntI in encoding proteins that had RT and X domains but lacked the C-terminal DNA-binding and DNA endonuclease domains (Martínez-Abarca and Toro, 2000b; Belfort et al., 2002). Our results suggest that this hypothesized ancestral group II intron could have retrohomed or retrotransposed by reverse splicing into DNA target sites and then using a nascent DNA strand as a reverse transcription primer. At first, reverse splicing may have been restricted to single-stranded DNA target sites recognized primarily by base pairing of the intron RNA, with the protein gradually playing a greater role in DNA target site recognition. The less stringent target specificity of the ancestral intron would have favored its spread by retrotransposition. Acquisition of the C-terminal DNA-binding domain, which is thought to interact with the distal 5′-exon region to promote DNA unwinding (Guo et al., 1997), would both increase DNA target specificity and permit efficient reverse splicing into double-stranded DNA target sites, while simultaneous or separate acquisition of the DNA endonuclease domain provides a means of generating a reverse transcription primer independent of DNA replication. The multiply layered mobility mechanisms now increase the evolutionary fitness of mobile group II introns by enabling them to adapt to different conditions. Finally, by connecting mobility to DNA replication, the intron insures that it will insert only into viable genomes. For the same reason, a connection to DNA replication may be a general feature of the mobility mechanisms of transposable elements.
Materials and methods
Escherichia coli strains and growth conditions
Escherichia coli HMS174(DE3)[F– recA hsdR RifR] (Novagen, Madison, WI) was used for intron mobility assays, and DH5α was used for cloning. Bacteria were grown in LB medium with antibiotics added as follows: ampicillin 100 µg/ml; chloramphenicol 25 µg/ml; tetracycline 25 µg/ml.
Recombinant plasmids
The wild-type Ll.LtrB intron donor plasmid pACD2X contains a 940-nt Ll.LtrB-ΔORF intron with a phage T7 promoter inserted near its 3′ end (San Filippo and Lambowitz, 2002). The intron and short flanking exons are cloned behind a T7lac promoter in a CamR pACYC184-derivative, with the LtrA ORF cloned just downstream of the 3′ exon. Donor plasmids containing mutant Ll.LtrB introns were constructed by PCR or recloning segments of previously constructed mutant introns into pACD2X as follows: pACD2X-YRT, 170-bp BamHI–AatII fragment of pIMP-YRT (San Filippo and Lambowitz, 2002); pACD2X-YAAA (D308A, D309A), 1.8-kb MfeI–AatII fragment of pACD-YAAA (Guo et al., 2000); pACD2X-H591A, 340-bp BamHI–XhoI-digested DNA generated by PCR of pIMP-H591A (San Filippo and Lambowitz, 2002) with primers 5′-GGTGCTCGAGATTCACTTGTGTTTATGAATCAC GTGACGAGCACAATG and 5′-TGAACTCCGCGGGATTTGTAATT ACTAC; pACD2X-ΔConEn, 600-bp BglII–XhoI-digested DNA generated by PCR of pACD2X with primers 5′-ACATAGCAGTCAACC CGCTC and 5′-GCCCTCGAGTTATCAACATTTAGCTTTTAACCT. pACD2X-G3A was as described previously (D’Souza and Zhong, 2002).
Recipient plasmids pBRR3A-ltrB and pBRR3B-ltrB are AmpR pBR322-derivatives that contain the Ll.LtrB target site/tetR gene cassette cloned in opposite orientations relative to the direction of DNA replication. To construct these plasmids, the target site/tetR region of pBRR3-ltrB (San Filippo and Lambowitz, 2002) was amplified by PCR with primers 5′-TCCCCCGGGATAGCTGAAACGCCGTAGC and 5′-TCCCCCGGGTAGTTTATCACAGTTAAATTG, and the resulting 2.2-kb DNA was digested with SmaI and cloned in either orientation between the blunted AatII and AvaI sites of pBR322.
pB101A-ltrB and pB101B-ltrB contain the target site/tetR cassette cloned in opposite orientations in pSC101. These plasmids were constructed as follows. First, the replication origin and rep101 ORF were amplified from pSC101 by PCR with primers 5′-AGTACGC GTACAGTAAGACGGGTAAGCC and 5′-CGGGATCCACCGTTTTC ATCTGTGCATA, and an ampR cassette was amplified from pBR322 by PCR with primers 5′-AGTACGCGTTTATTTTTCTAAATACATTCAA and 5′-CGGGATCCAAAAAGGATCTTCACCTAGAT. The PCR products were digested with MluI–BamHI and ligated to generate plasmid pA101, then a 170-bp HincII–FspI fragment containing the pUC19 polylinker was inserted into the blunted MluI site of pA101 to generate pB101. Finally, the Ll.LtrB target site/tetR cassette (2.2-kb PCR product amplified from pBRR3-ltrB; see above) was cloned in both orientations in the PvuII site of the pB101 polylinker to generate pB101A-ltrB and pB101B-ltrB.
Mutations were introduced at different positions of the Ll.LtrB target site by annealing two 5′ phosphorylated oligonucleotides, which correspond to DNA target site positions –30 to +15 with appropriate sequence changes and termini that create ‘cleaved’ AatII and EcoRI sites, then ligating into AatII–EcoRI-digested recipient plasmid. The E2m target site has top strand 3′-exon positions +1 to +15 changed to CAGCGGCAATATTTG. All constructs were verified by DNA sequencing.
Group II intron mobility assays
Intron donor and recipient plasmids were co-transformed into E.coli HMS174(DE3), which contains an IPTG-inducible phage T7 RNA polymerase (Guo et al., 2000; Karberg et al., 2001). After induction with 100 µM IPTG for 1 h at 37°C, mobility events were detected by plating cells on LB medium containing tetracycline and ampicillin, and mobility frequencies were calculated as the ratio of AmpR + TetR/AmpR colonies. For mixing experiments, cells containing the intron donor and either recipient plasmid pBRR3A-ltrB or pBRR3B-ltrB, which contain the Ll.LtrB target site in opposite orientations, were grown to OD595 of 0.2 and mixed in a 1:1 ratio before induction. After plating as above, AmpR + TetR colonies were analyzed by colony PCR to determine the orientation of the inserted intron.
Reconstitution of group II intron RNPs
RNPs were reconstituted by incubating purified LtrA protein with in vitro-synthesized Ll.LtrB lariat RNA (Saldanha et al., 1999). The LtrA protein was expressed in E.coli from pIMP-1P and purified as described previously (Saldanha et al., 1999). Ll.LtrB RNA containing the 940-nt Ll.LtrB-ΔORF intron with short flanking exons was transcribed from a gel-purified double-stranded DNA template generated by PCR of pACD2X with primers 5′-CCATCGGTGATGTCGGCGA and 5′-CTAGCAGCACGCCATAGTG, using a MEGAscript™ T7 in vitro transcription kit (Ambion, Austin, TX). The Ll.LtrB RNA was then self-spliced for 3 h at 37°C in 1.5 M NH4Cl, 50 mM MgCl2, 40 mM Tris–HCl, pH 7.5, and the resulting intron lariat RNA was purified in a denaturing 4% polyacrylamide gel.
Reverse splicing assays
Reverse splicing was assayed by incubating reconstituted Ll.LtrB RNP particles (4 µl; 0.025 OD260 units) with 32P-labeled DNA oligonucleotide or PCR-generated DNA substrates (150 000 c.p.m.; 1.25 nmol) for 20 min at 37°C in 20 μl of 10 mM KCl, 10 mM MgCl2, 50 mM Tris–HCl, pH 7.5 (Saldanha et al., 1999). The reaction time was verified to be in the linear range for reverse splicing into double- or single-stranded DNA substrates. Single-stranded DNA substrates were 70mers, corresponding to the wild-type or mutant Ll.LtrB target sequence (top strand positions –35 to +35). For 5′ labeling, the gel-purified 70mer (5 pmol) was incubated with 25 µCi [γ-32P]ATP (3000 Ci/mmol; NEN-DuPont, Boston, MA) and 10 U of T4 polynucleotide kinase (Invitrogen, Carlsbad, CA) for 1 h at 37°C, then purified using a nucleotide removal kit (Qiagen, Valencia, CA). 5′-labeled double-stranded DNA substrates were made by PCR of the same 70mers with an unlabeled primer complementary to positions +14 to +35, and a 5′-labeled primer corresponding to positions –35 to –16. DNA substrates were purified by electrophoresis in a 2% agarose gel. Reverse splicing reactions were terminated by phenol-CIA extraction and ethanol precipitation, and the products were analyzed in a denaturing 6% polyacrylamide gel, which was dried and scanned with a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Target DNA-primed reverse transcription (TPRT) assays
TPRT was assayed by incubating gel-purified supercoiled plasmid DNA substrate (0.5 µg) with reconstituted RNPs (2 µl; 0.0125 OD260 units) in 10 µl of reaction medium containing 10 mM KCl, 50 mM MgCl2, 50 mM Tris–HCl, pH 7.5, 200 µM dATP, dGTP and dCTP, and 10 µCi [α-32P]dTTP (3000 Ci/mmol; NEN-DuPont). The reaction was initiated by addition of RNPs, incubated for 20 min at 37°C, chased with 200 µM dTTP for an additional 10 min, and then terminated by adding 1 µl of 200 mM EDTA. After column purification (QIAquick; Qiagen), TPRT products were analyzed in a 0.7% agarose gel, which was dried and scanned with a PhosphorImager (Molecular Dynamics).
Acknowledgments
Acknowledgements
We thank Marlene Belfort for comments on the manuscript and Joe San Filippo for preparation of YRT protein and construction of recipient plasmids T-23G, G-21T, A-20G and T+5N. This work was supported by NIH grant GM37949.
References
- Aizawa Y., Xiang,Q., Lambowitz,A.M. and Pyle,A.M. (2003) The pathway for DNA recognition and RNA integration by a group II intron retrotransposon. Mol. Cell, 11, 795–805. [DOI] [PubMed] [Google Scholar]
- Bazaral M. and Helinski,D.R. (1970) Replication of a bacterial plasmid and an episome in Escherichia coli. Biochemistry, 9, 399–406. [DOI] [PubMed] [Google Scholar]
- Belfort M., Derbyshire,V., Parker,M.M., Cousineau,B. and Lambowitz,A.M. (2002) Mobile introns: pathways and proteins. In Craig,N.L., Craigie,R., Gellert,M. and Lambowitz,A.M. (eds), Mobile DNA II. ASM Press Publishers, Washington, DC, pp. 761–783. [Google Scholar]
- Dai L. and Zimmerly,S. (2002) Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res., 30, 1091–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson L., Huang,H.R., Liu,L., Matsuura,M., Lambowitz,A.M. and Perlman,P.S. (2001) Retrotransposition of a yeast group II intron occurs by reverse splicing directly into ectopic DNA sites. Proc. Natl Acad. Sci. USA, 98, 13207–13212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza L.M. and Zhong,J. (2002) Mutations in the Lactococcus lactis Ll.LtrB group II intron that retain mobility in vivo. BMC Mol. Biol., 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A.E. (1994) Self-splicing group I and group II introns encode homologous (putative) DNA endonucleases of a new family. Protein Sci., 3, 1117–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1997) Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA. EMBO J., 16, 6835–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Karberg,M., Long,M., Jones,J.P.,III, Sullenger,B. and Lambowitz,A.M. (2000) Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science, 289, 452–457. [DOI] [PubMed] [Google Scholar]
- Hamilton C.M., Aldea,M., Washburn,B.K., Babitzke,P. and Kushner,S.R. (1989) New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol., 171, 4617–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyanagi K., Beauregard,A., Lawrence,S., Smith,D., Cousineau,B. and Belfort,M. (2002) Retrotransposition of the Ll.LtrB group II intron proceeds predominantly via reverse splicing into DNA targets. Mol. Microbiol., 46, 1259–1272. [DOI] [PubMed] [Google Scholar]
- Jiménez-Zurdo J.I., García-Rodríguez,F.M., Barrientos-Durán,A. and Toro,N. (2003) DNA target site requirement for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J. Mol. Biol., 326, 413–423. [DOI] [PubMed] [Google Scholar]
- Karberg M., Guo,H., Zhong,J., Coon,R., Perutka,J. and Lambowitz,A.M. (2001) Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol., 19, 1162–1167. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–484. [Google Scholar]
- Martínez-Abarca F. and Toro,N. (2000a) RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res., 28, 4397–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Abarca F. and Toro,N. (2000b) Group II introns in the bacterial world. Mol. Microbiol., 38, 917–926. [DOI] [PubMed] [Google Scholar]
- Martínez-Abarca F., Zekri,S. and Toro,N. (1998) Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol. Microbiol., 28, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Martínez-Abarca F., García-Rodríguez,F.M. and Toro,N. (2000) Homing of a bacterial group II intron with an intron-encoded protein lacking a recognizable endonuclease domain. Mol. Microbiol., 35, 1405–1412. [DOI] [PubMed] [Google Scholar]
- Michel F. and Ferat,J.L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Mohr G., Smith,D., Belfort,M. and Lambowitz,A.M. (2000) Rules for DNA target-site recognition by a lactococcal group II intron enable retargeting of the intron to specific DNA sequences. Genes Dev., 14, 559–573. [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Adelantado E., San Filippo,J., Martínez-Abarca,F., García-Rodríguez,F.M., Lambowitz,A.M. and Toro,N. (2003) Mobility of the Sinorhizobium meliloti group II intron RmInt1 occurs by reverse splicing into DNA, but requires an unknown reverse transcriptase priming mechanism. J. Mol. Biol., 327, 932–943. [DOI] [PubMed] [Google Scholar]
- Pham P., Bertram,J.G., O’Donnell,M., Woodgate,R. and Goodman,M.F. (2001) A model for SOS-lesion-targeted mutations in Escherichia coli. Nature, 409, 366–370. [DOI] [PubMed] [Google Scholar]
- Rownd R. (1969) Replication of a bacterial episome under relaxed control. J. Mol. Biol., 44, 387–402. [DOI] [PubMed] [Google Scholar]
- Saldanha R., Chen,B., Wank,H., Matsuura,M., Edwards,J. and Lambowitz,A.M. (1999) RNA and protein catalysis in group II intron splicing and mobility reactions using purified components. Biochemistry, 38, 9069–9083. [DOI] [PubMed] [Google Scholar]
- San Filippo J. and Lambowitz,A.M. (2002) Characterization of the C-terminal DNA-binding/DNA endonuclease region of a group II intron-encoded protein. J. Mol. Biol., 324, 933–951. [DOI] [PubMed] [Google Scholar]
- Shub D.A., Goodrich-Blair,H. and Eddy,S.R. (1994) Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem. Sci., 19, 402–404. [DOI] [PubMed] [Google Scholar]
- Singh N.N. and Lambowitz,A. (2001) Interaction of a group II intron ribonucleoprotein endonuclease with its DNA target site investigated by DNA footprinting and modification interference. J. Mol. Biol., 309, 361–386. [DOI] [PubMed] [Google Scholar]
- Wang G.H. and Seeger,C. (1992) The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell, 71, 663–670. [DOI] [PubMed] [Google Scholar]
- Wang H. and Lambowitz,A.M. (1993) The Mauriceville plasmid reverse transcriptase can initiate cDNA synthesis de novo and may be related to reverse transcriptase and DNA polymerase progenitor. Cell, 75, 1071–1081. [DOI] [PubMed] [Google Scholar]
- Yang J., Mohr,G., Perlman,P.S. and Lambowitz,A.M. (1998) Group II intron mobility in yeast mitochondria: target DNA-primed reverse transcription activity of aI1 and reverse splicing into DNA transposition sites in vitro. J. Mol. Biol., 282, 505–523. [DOI] [PubMed] [Google Scholar]
- Yeo C.C., Tham,J.M., Yap,M.W. and Poh,C.L. (1997) Group II intron from Pseudomonas alcaligenes NCIB 9867 (P25X): entrapment in plasmid RP4 and sequence analysis. Microbiology, 143, 2833–2840. [DOI] [PubMed] [Google Scholar]
- Zhong J., Karberg,M. and Lambowitz,A.M. (2003) Targeted and random bacterial gene disruption using a group II intron (targetron) vector containing a retrotransposition-activated selectable marker. Nucleic Acids Res., 31, 1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]