Abstract
In contrast to Escherichia coli, where the 3′ ends of tRNAs are primarily generated by exoribonucleases, maturation of the 3′ end of tRNAs is catalysed by an endoribonuclease, known as RNase Z (or 3′ tRNase), in many eukaryotic and archaeal systems. RNase Z cleaves tRNA precursors 3′ to the discriminator base. Here we show that this activity, previously unsuspected in bacteria, is encoded by the yqjK gene of Bacillus subtilis. Decreased yqjK expression leads to an accumulation of a population of B.subtilis tRNAs in vivo, none of which have a CCA motif encoded in their genes, and YqjK cleaves tRNA precursors with the same specificity as plant RNase Z in vitro. We have thus renamed the gene rnz. A CCA motif downstream of the discriminator base inhibits RNase Z activity in vitro, with most of the inhibition due to the first C residue. Lastly, tRNAs with long 5′ extensions are poor substrates for cleavage, suggesting that for some tRNAs, processing of the 5′ end by RNase P may have to precede RNase Z cleavage.
Keywords: CCA/endonuclease/tRNA/RNase Z/3′ tRNase
Introduction
tRNA plays the role of decoding mRNAs during the process of translation. In general, bacterial tRNAs are transcribed from operons containing other components of the translational apparatus such as rRNAs, ribosomal proteins and aminoacyl-tRNA synthetases, although there are also numerous examples of isolated tRNA genes. In both examples, whether isolated or in operons, processing of the tRNA from a longer transcript is required to generate the functional molecule. Although tRNA maturation has been studied in most detail in Escherichia coli, the process is well documented, and is remarkably similar in eukaryota, bacteria and the archaea. Extensions on the 5′ side of the tRNA are removed by RNase P, a quasi-universal enzyme (Frank and Pace, 1998; Brown, 1999; Condon and Putzer, 2002), with Aquifex aeolicus being the only documented species in any of the three kingdoms not possessing RNase P activity (Willkomm et al., 2002). In bacteria, RNase P is a two-component enzyme consisting of a protein and an RNA subunit, with the RNA subunit providing the catalytic function (Guerrier-Takada et al., 1983). In eukaryota and archaea, the catalytic RNA subunit is bound by multiple proteins with no relationship to their bacterial counterpart (Hall and Brown, 2002).
Although, the enzymes responsible for the generation of the 3′ end of tRNAs appear to be more varied in the different systems examined, this perception may change over time. In E.coli, tRNAs are generally thought to be processed from the primary transcript by endonucleolytic cleavage downstream of the tRNA, followed by 3′ to 5′ exonucleolytic digestion of the RNA to produce the mature 3′ end (Deutscher, 1995). In many cases, the downstream endonucleolytic event is catalysed by RNase E, and this can account for the essential role of this enzyme for E.coli cell growth (Li and Deutscher, 2002; Ow and Kushner, 2002). The exonucleolytic degradation of the 3′ trailing sequences is catalysed by any one of an armoury of redundant exonucleases, RNase T, RNase D, RNase BN, RNase PH, RNase II and possibly PNPase (Li and Deutscher, 1996).
In the archaea, the maturation of the 3′ end of tRNAs occurs by endonucleolytic cleavage 3′ to the discriminator base (Schierling et al., 2002), the unpaired base immediately preceding the CCA motif and used as an identity element by many aminoacyl-tRNA synthetases. In yeast and higher eukaryota, two types of pathways have been characterized for tRNA 3′ maturation. In addition to endonucleolytic cleavage 3′ to the discriminator base (Garber and Gage, 1979; Hagenbuchle et al., 1979; Solari and Deutscher, 1983; Castano et al., 1985; Stange and Beier, 1987; Oommen et al., 1992; Papadimitriou and Gross, 1996; Han and Kang, 1997; Nashimoto, 1997; Mayer et al., 2000), a pathway of exonucleolytic degradation of 3′ trailer sequences also exists in some cases (Garber and Altman, 1979; Solari and Deutscher, 1983; Engelke et al., 1985; Papadimitriou and Gross, 1996). The enzyme responsible for cleavage 3′ to the discriminator base has been well characterized in potato (Kunzmann et al., 1998), wheat (Mayer et al., 2000) and the archaeon Haloferax volcanii, where it is called RNase Z, and in mammals and insects, where it is called 3′ tRNase (Nashimoto, 1997; Mohan et al., 1999) or 3′ pre-tRNase (Castano et al., 1985). RNase Z from all species examined recognizes the tRNA structure in precursor molecules, the T-arm and acceptor stem in particular (Levinger et al., 1998; Nashimoto et al., 1998, 1999a; Schiffer et al., 2001; Schierling et al., 2002). The gene encoding RNase Z was identified recently from Arabidopsis thaliana and the archaeon Methanococcus jannaschii (Schiffer et al., 2002), allowing an examination of its phylogenetic distribution. In addition to being well conserved in eukaryota and archaea, RNase Z is surprisingly also widely distributed among bacteria (Condon and Putzer, 2002). Orthologues are found in all of the major bacterial groups, although relatively poorly represented in the proteobacteria, where it is found only in E.coli and Salmonella species.
Following maturation of the 3′ end of tRNA in eukaryota and archaea, the CCA motif, found at the 3′ end of the amino acid accepting stem of all tRNAs, is added by an enzyme known as nucleotidyl-transferase (or CCase). In E.coli, however, the CCA motif is encoded directly by the tRNA gene in all cases. If exonucleolytic degradation of the 3′ trailer sequence were to remove the CCA motif, it can be replaced by nucleotidyl-transferase. Indeed a significant amount of recycling of the CCA motif is known to occur in E.coli (Deutscher et al., 1977). In Bacillus subtilis, the best studied of the Gram-positive bacteria, about two-thirds of tRNAs (59 out of 86) have the CCA motif encoded by their genes.
The maturation of the 3′ end of tRNAs has not been studied in B.subtilis up to now. Of the six exonucleases thought to participate in 3′ trailer degradation in E.coli, only RNase PH and PNPase have orthologues in B.subtilis. Bacillus subtilis does however have a gene, yqjK, whose product shows significant homology to M.jannaschii RNase Z. Here we show that the B.subtilis yqjK gene encodes a functional RNase Z, that can only process the 3′ end of tRNAs lacking an encoded CCA moiety. This raises the intriguing question of why E.coli, whose tRNAs all have an encoded CCA motif, has an RNase Z homologue.
Results
Identification of B.subtilis RNase Z homologue, YqjK
BLAST searches using M.jannaschii RNase Z as a query sequence revealed the presence of a surprisingly large number of potential orthologues of this enzyme throughout the bacterial kingdom. In B.subtilis, the yqjK gene shows 58% similarity (38% identity) to M.jannaschii RNase Z (Figure 1). The homology is highest at the N-terminus, concentrated around the metal-binding phosphodiesterase (PDE) and Zn2+ finger motifs within the β-lactamase domain (Schiffer et al., 2002).
Fig. 1. Sequence comparison of potential B.subtilis, E.coli and M.jannaschii RNase Z orthologues. Identical amino acids are marked with dark shading. Similar amino acids have lighter shading. The phosphodiesterase (PDE) domain (thick overline), proposed zinc finger motif (asterisks) and β-lactamase domain (thin overline) (Schiffer et al., 2002) are indicated.
Construction of strain affording conditional expression of YqjK
We were curious to know whether the B.subtilis yqjK gene encoded a functional RNase Z enzyme. Since this gene was predicted by Kobayashi et al. (2003) to be essential for B.subtilis viability, we chose to inactivate yqjK by Campbell-type insertion of the plasmid pMUTIN-Z (see Materials and methods) within the N-terminal portion of the yqjK coding sequence to give strain SSB320. In this strain, expression of yqjK is under isopropyl-β-d-thiogalactopyranoside (IPTG) control from the spac promoter present in the integrated plasmid. The removal of IPTG from the culture medium did not lead to an arrest of cell growth, however, and SSB320 cells grew with a doubling time of about twice that of the parental strain in the absence of IPTG (data not shown), suggesting that some leakiness occurs at the spac promoter. The addition of further copies of the LacI repressor from plasmid pMAP65 (Petit et al., 1998) permitted full growth inhibition in the absence of IPTG (data not shown), confirming that the gene is essential.
Effect of decreased YqjK expression on tRNA processing in vivo
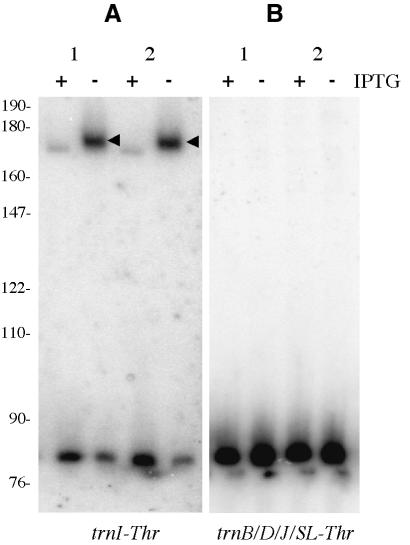
Strain SSB320 (without pMAP65) was tested initially for its ability to process the two B.subtilis tRNAThr isoacceptors correctly in vivo, in the presence and absence of IPTG. Specific oligonucleotide probes were designed for the minor threonine-isoaccepting species found at the end of the rrnI operon (trnI-Thr) and the major threonine-isoaccepting species encoded by the rrnB, rrnD and rrnJ operons (trnB, trnD and trnJ-Thr), and the isolated gene trnSL-Thr1. Northern blots of total RNA isolated from two independent clones of SSB320 showed significant accumulation of a precursor species for the trnI-Thr species in the absence of IPTG, while the major isoacceptor showed no such accumulation (Figure 2). This suggests that the product of the yqjK gene is involved, either directly or indirectly, in the maturation of the tRNA encoded by the trnI-Thr gene.
Fig. 2. Processing of B.subtilis threonine tRNAs under conditions of decreased yqjK expression in vivo. (A) Northern blot of RNA isolated from two independent clones (1 and 2) of SSB320 in the presence (+) and absence (–) of IPTG, probed for trnI-Thr with oligo HP262. (B) Northern blot of RNA isolated from the same clones, probed for trnB/D/J/SL-Thr with oligo HP263. Accumulating precursor species are marked with arrowheads. The positions of a DNA marker are indicated in nucleotides.
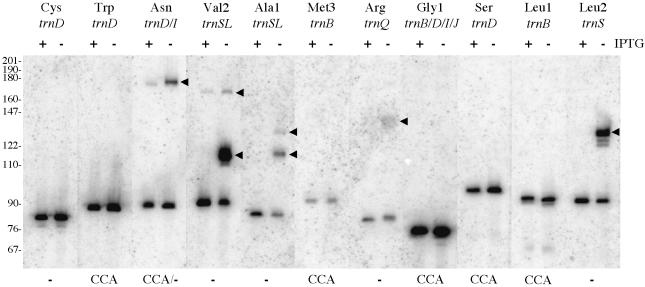
To get an idea of what proportion of B.subtilis tRNAs might be affected by the decrease in yqjK expression, specific oligonucleotide probes were designed for 11 more tRNAs. As can be seen from the northern blot in Figure 3, precursor species accumulate for five of these tRNAs in the RNA samples isolated from SSB320 growing in the absence of IPTG. Thus, the maturation of about half of the non-redundant tRNA species of B.subtilis is affected by the decrease in YqjK levels.
Fig. 3. Processing of 11 B.subtilis tRNAs in the absence of yqjK expression in vivo. The autoradiogram shows a northern blot of RNA isolated from SSB320 in the presence (+) and absence (–) of IPTG, probed for the tRNAs indicated. Some probes hybridize to redundant tRNA genes, e.g. trnB/D/I/J-Gly1. Precursor species (arrowheads) accumulate for five tRNA species. Those tRNAs with an encoded CCA motif are indicated under the autoradiogram. The trnD-Asn gene has an encoded CCA motif, while trnI-Asn does not. The positions of a DNA marker are indicated in nucleotides.
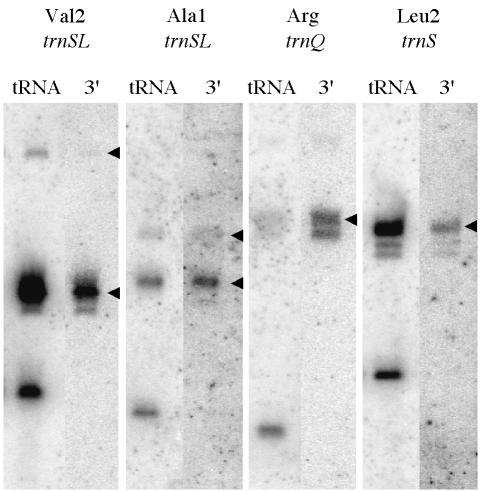
To confirm that the precursor species that accumulate with decreased yqjK expression had 3′, rather than 5′ extensions, the northern blots for four of these tRNA species (trnSL-Val2, trnSL-Ala1, trnQ-Arg and trnS-Leu2) were reprobed with oligonucleotides specific for their respective trailer sequences, close to their predicted 3′ termini. In each case, the identical precursor species was detected by the 3′ trailer probe (Figure 4), clearly showing that the decrease in YqjK levels leads to the accumulation of tRNA precursors with 3′ extensions.
Fig. 4. The tRNA precursors that accumulate under conditions of decreased yqjK expression have 3′ extensions. The northern blots for the Val2-trnSL, Ala1-trnSL, Arg-trnQ and Leu2-trnS tRNAs (tRNA) shown in Figure 3 were re-probed with oligos specific for the 3′ extensions (3′). Only the –IPTG samples are shown. The precursor species are indicated by arrowheads.
We noticed a relatively good correlation between whether or not a precursor RNA accumulated in the absence of IPTG and whether the corresponding tRNA gene had an encoded CCA motif. With one possible exception, complete maturation of tRNAs with an encoded CCA motif occurred in the absence of IPTG. The tRNAs encoded by trnD-Asn (encoded CCA) and trnI-Asn (no CCA) are identical, and thus we could not determine which of them showed a maturation defect due to the decrease in yqjK expression (Figure 3). Although the corollary is also generally true, i.e. most of the tRNAs lacking a CCA motif in their genes accumulated as precursor species in the absence of IPTG, no precursors were observed for the tRNAs encoded by trnD-Cys (Figure 3), trnD-Thr and trnSL-Thr1 (Figure 2). A likely explanation is discussed below. This experiment suggested that the presence of an encoded CCA motif might be inhibitory for YqjK-dependent tRNA maturation.
Purification and assay of his-tagged YqjK in vitro
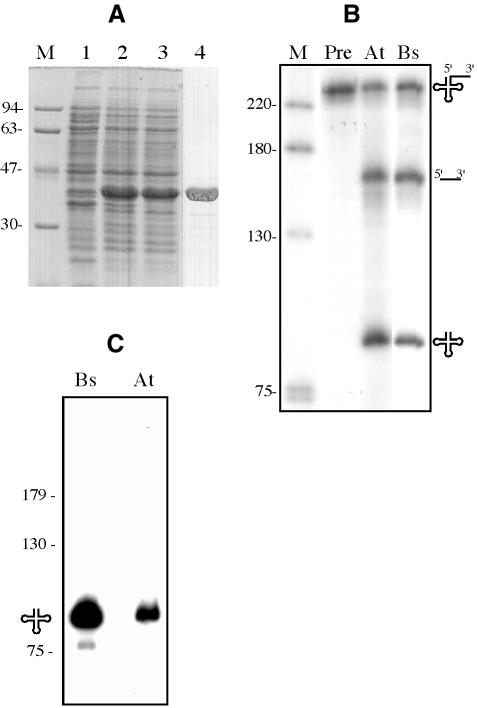
To determine whether the product of the yqjK gene was directly responsible for tRNA maturation in B.subtilis and whether the CCA motif was indeed inhibitory for cleavage, we cloned the gene in-phase with the coding sequence for a C-terminal histidine tag and purified YqjK from overexpressing E.coli cells (Figure 5A). The activity of the YqjK protein was tested initially on a tRNATyr precursor (pTyrI) from Oenothera berteriana (evening primrose). Incubation of this pre-tRNA with the YqjK protein yielded the same bands as those generated by a previously characterized nuclear RNase Z enzyme, or nuz (Schiffer et al., 2002), from A.thaliana (Figure 5B).
Fig. 5. (A) Purification of His-tagged YqjK from B.subtilis. Lane 1, extract from uninduced culture; lane 2, extract from culture induced with 0.5 mM IPTG; lane 3, soluble fraction from extract of induced culture; lane 4, purified His-tagged YqjK (RNase Z). A molecular weight marker (in kDa) is shown in the lane marked M. (B) Processing of Oenothera berteriana mitochondrial pre-tRNATyr (Pre) by 2.5 ng/µl Arabidopsis thaliana (At) nuclear RNase Z (nuz) and 3.5 ng/µl B.subtilis YqjK (Bs). A molecular weight DNA marker is shown in the lane marked M. The different precursor molecules and products are symbolized at the sides of the autoradiogram. (C) Labelling of RNase Z-processed tRNATyr with [α-32P]pCp. tRNATyr, produced by maturation of pTyr by B.subtilis YqjK (Bs) and A.thaliana nuz (At), was incubated with [α-32P]pCp and RNA ligase.
The cleavage products of plant and mammalian RNase Z are a mature tRNA with a 3′-OH and a trailer fragment bearing a 5′-monophosphate residue. To determine whether YqjK also produced a mature tRNA with a 3′-OH, we cleaved unlabelled pTyr1 in parallel reactions with B.subtilis YqjK and A.thaliana nuz as a control, and eluted the respective mature tRNA species from acrylamide gels (Kunzmann et al., 1998). We could label the tRNA by ligation of 32P-labelled pCp with RNA ligase, showing that the mature tRNA has a 3′-OH residue (Figure 5C) and is ready for addition of the CCA moiety. From the above experiments, we conclude that B.subtilis yqjK indeed encodes a bacterial RNase Z and propose that the gene be renamed rnz.
An encoded CCA motif inhibits RNase Z activity in vitro
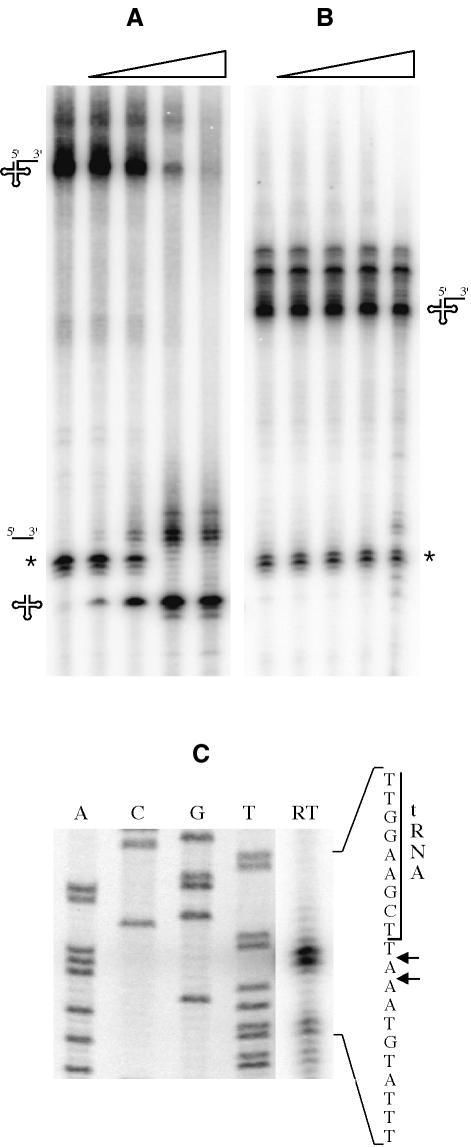
Previous experiments with partially purified mammalian and Drosophila 3′ tRNase (Nashimoto, 1997; Mohan et al., 1999) and our in vivo data suggested that tRNAs without an encoded CCA sequence may be better substrates for RNase Z. To test this hypothesis, we synthesized the two threonine-isoaccepting species, trnI-Thr and trnB-Thr, with 83 and 47 nucleotide 3′ trailing sequences, respectively, in vitro from PCR-amplified templates. These tRNA precursors were then incubated with increasing concentrations of B.subtilis RNase Z (Figure 6A and B). Each lane shown represents a 10-fold increase in enzyme concentration. While the trnI-Thr precursor was cleaved by RNase Z in vitro at low concentrations of enzyme, the trnB-Thr precursor, which has an encoded CCA motif, was not cleaved at all, consistent with the in vivo data. The site of cleavage of the trnI-Thr precursor was mapped precisely by reverse transcription of the 3′ trailer sequence following cleavage of the precursor by RNase Z in vitro (Figure 6C). Two cleavage sites of similar intensity were mapped to one and two nucleotides downstream of the discriminator base for this particular tRNA in vitro. Previous studies have shown that the cleavage specificity of RNase Z can vary from one tRNA precursor to the next in vitro, but that processing is thought to occur accurately 3′ to the discriminator base in vivo (Kunzmann et al., 1998; Mayer et al., 2000; Kruszka et al., 2003).
Fig. 6. Processing of trnI-Thr (A) and trnB-Thr tRNA (B) precursors by RNase Z in vitro. Precursor tRNAs with 83 and 47 nucleotide trailer sequences, respectively, were synthesized by T7 RNA polymerase in vitro (see Materials and methods). The right-angled triangle indicates the direction of increasing enzyme concentration: 0, 0.15, 1.5, 15 and 150 ng/µl RNase Z. An RNA species (asterisk), resulting from premature transcription arrest of T7 RNA polymerase approximately six nucleotides downstream of both tRNA structures, is also a substrate for RNase Z. The different precursor molecules and products are symbolized at the sides of the autoradiograms. The double band corresponding to the trailer species in (A) is likely to be due to the tendency of T7 RNAP to add one or two uncoded nucleotides to the 3′ end of transcripts. (C) Mapping of the RNase Z cleavage site on trnI-Thr pre-tRNA. The autoradiogram shows the reverse transcriptase reaction (RT) using primer HP62 and the corresponding sequencing reaction (lanes A, C, G and T). A portion of the sequence is given to the right of the gel. The two short arrows indicate the sites of RNase Z cleavage. The 3′ end of the mature tRNA sequence is indicated.
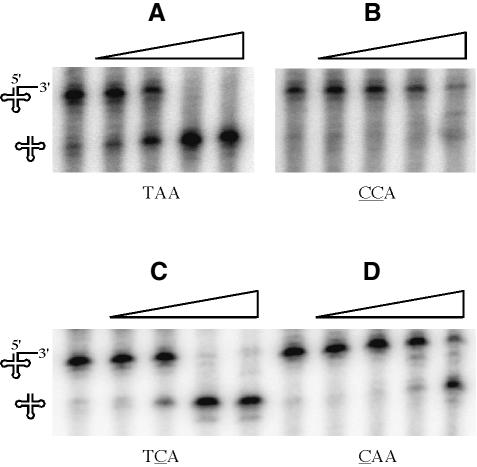
It was possible that sequence variation outside of the CCA motif in the two tRNA precursors could account for the difference in their susceptibility to maturation by RNase Z. To test whether the CCA motif was directly responsible, we made a variant of the trnI-Thr precursor where the native TAA sequence 3′ to the discriminator base was replaced by CCA, a two-nucleotide change. At least 100-fold more RNase Z was required for processing of the CCA-containing tRNA precursor compared with the wild-type equivalent (Figure 7A versus B). This proves conclusively that it is the CCA motif and no other sequence element that inhibits RNase Z cleavage in B.subtilis. To determine which of the two C residues was primarily responsible for the inhibitory effect, precursor tRNAs were made where only one C residue was introduced at a time (i.e. TAA→TCA and TAA→CAA). The TCA mutant was processed with a similar efficiency to the precursor with the native TAA sequence, while the CAA mutant required ∼10-fold more RNase Z for maturation (Figure 7C and D). Thus, as in mammalian systems, the first cytosine of the CCA motif is the primary anti-determinant for RNase Z cleavage, although in the double C context, the second cytosine clearly makes an additional contribution. The Km and Vmax of each of these substrates for RNase Z is given in the legend to Figure 7. The inhibitory effect seems to be primarily at the level of the Vmax.
Fig. 7. Effect of the CCA motif on processing of trnI-Thr precursor tRNA by RNase Z. Native and mutant precursor tRNAs with six nucleotide 3′ trailer sequences were synthesized by T7 RNA polymerase, taking advantage of the natural transcription termination at this site. RNase Z processing assays of trnI-Thr precursor with native (A) TAAATG and mutant (B) CCAATG, (C) TCAATG, (D) CAAATG trailer sequences downstream of discriminator base. The Vmax and Km values of each of the substrates for RNase Z are as follows: TAAATG (Vmax 52.4 fmol/min; Km 0.97 µM), CCAATG (Vmax 0.23 fmol/min; Km 2.4 µM), TCAATG (Vmax 33.8 fmol/min; Km 0.58 µM), CAAATG (Vmax 0.34 fmol/min; Km 2.2 µM). The direction of increasing enzyme concentration (0, 0.15, 1.5, 15 and 150 ng/µl RNase Z) is given by the right-angled triangles. The precursor molecule and mature tRNA are symbolized at the sides of the autoradiograms. In (A), a band the same size as the cleaved product, visible in the absence of added RNase Z, is due to transcription arrest/pausing at this site by T7 RNA polymerase.
Long 5′ extensions inhibit RNase Z activity in vitro
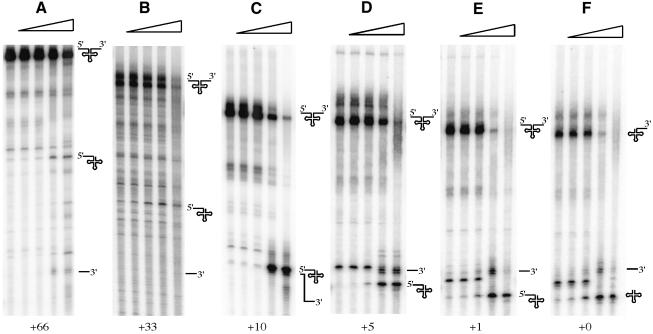
In potato mitochondria, processing of the 5′ end of the tRNA by RNase P must precede cleavage of the 3′ end by RNase Z (Kunzmann et al., 1998) and, in H.volcanii, 5′ extensions lead to reduced efficiency of RNase Z cleavage (Schierling et al., 2002). Pig liver 3′ tRNase is also highly sensitive to 5′ extensions of ≥9 nucleotides (Nashimoto et al., 1999b). To test the effect of 5′ extensions on RNase Z processing in B.subtilis, we prepared precursor trnI-Thr tRNAs with the same 83 nucleotide 3′ trailer and with 66, 33, 10, 5 and 1 nucleotide extensions on their 5′ ends and compared their maturation with that of the same pre-tRNA with a mature 5′ end (Figure 8). Ten- to 100-fold more RNase Z was required for maturation of the precursor with the 33 or 66 nucleotide 5′ extensions, suggesting that for tRNAs with long 5′ extensions, the hierarchy of cleavage events, RNase P before RNase Z, must be respected. However, with the shorter 5′ extensions, in the 1–10 nucleotide range, only a minor decrease in the efficiency of RNase Z cleavage was observed compared with the precursor with the mature 5′ end. Thus, in such cases, the order of the RNase P and RNase Z cleavages would be predicted to be significantly less important.
Fig. 8. Effect of 5′ extensions on processing of trnI-Thr precursor tRNA by RNase Z. Precursor tRNAs were synthesized with 83 nucleotide 3′ trailers and 5′ extensions of (A) 66, (B) 33, (C) 10, (D) 5, (E) 1 nucleotide or (F) no extension. The various precursor molecules, mature tRNA and 83 nucleotide 3′ trailer are symbolized to the right of each autoradiogram. The direction of increasing enzyme concentration (0, 0.15, 1.5, 15 and 150 ng/µl RNase Z) is given by the right-angled triangles. In (E) and (F), a band the same size as the cleaved product, visible in the absence of added RNase Z, is due to transcription arrest/pausing at this site by T7 RNA polymerase.
Discussion
In this study, we have shown that the yqjK gene of B.subtilis encodes a functional RNase Z (3′ tRNase) enzyme. This is the first demonstration of endonucleolytic maturation of the 3′ end of tRNAs in bacteria, a process generally assumed to be exonucleolytic from studies in E.coli. We showed that B.subtilis RNase Z has a preference for tRNA precursors which do not possess an encoded CCA sequence, both in vivo and in vitro. Lastly, long 5′ extensions are inhibitory to B.subtilis RNase Z activity, suggesting that as in plants, cleavage of such tRNA precursors by RNase P must precede that by RNase Z. For tRNAs with shorter 5′ extensions, however, the order of cleavage in B.subtilis is likely to be less important.
Two pathways for tRNA processing in B.subtilis
Although significant quantities of precursor species accumulate for CCA-less tRNAs in the absence of IPTG, in each case most of the tRNA is still processed correctly. Further experiments are required to determine whether this can be attributed to the residual levels of RNase Z in strain SSB320 or whether a second pathway of tRNA maturation is responsible. Clearly, an RNase Z-independent tRNA processing pathway must exist for those tRNAs that are not substrates for the enzyme. In E.coli, an exonucleolytic pathway of tRNA 3′ maturation has been characterized in some detail employing any one of six redundant exonucleases (Li and Deutscher, 1996). RNase PH and PNPase are the only two of these enzymes to have orthologues in B.subtilis. It will be interesting to determine whether these enzymes are components of an exonucleolytic tRNA 3′ maturation mechanism in B.subtilis.
RNase Z cleavage of the trnI-Thr precursor occurs one or two nucleotides downstream of the discriminator base in vitro. Thus, while it is possible that an exonuclease activity may be necessary to trim off extra nucleotides in vivo before addition of the CCA can proceed, differences between in vitro and in vivo specificities of RNase Z have been noted previously (Kunzmann et al., 1998; Mayer et al., 2000; Kruszka et al., 2003), and it is thought that RNase Z cleavage is restricted to a single site directly 3′ to the discriminator base in vivo. One possible explanation for the apparent difference between in vitro and in vivo specificity is that processing is likely to occur co-transcriptionally in vivo. Alternatively, additional factors or tRNA modifications may guide RNase Z to the correct cleavage site in vivo.
Nature of trnI-Thr precursor
We were curious as to the nature of the large trnI-Thr precursor that accumulates in the absence of RNase Z. Two of the probes used in the northern blot shown in Figure 3 were designed to hybridize to the tRNAs lying four nucleotides upstream and 56 nucleotides downstream of this tRNA, trnI-Asn and trnI-Gly1. The tRNAAsn probe hybridizes to a precursor species very similar in size to that detected by the trnI-Thr probe (Figure 2). If we assume that this precursor comes from trnI-Asn, rather than trnD-Asn (which has an encoded CCA), this suggests that this precursor includes two tRNAs, both of which are substrates for RNase Z. In some cases, including trnI-Thr, precursor tRNAs can be seen even in the presence of IPTG. Either these precursors are only slowly converted to mature tRNA, even in wild-type cells, or the fully-induced spac promoter does not provide sufficient RNase Z to process these precursors fully.
Are trnD-Cys, trnD-Thr and trnSL-Thr1 precursors substrates of RNase Z?
tRNAs with an encoded CCA are not substrates for RNase Z in vivo and are very poor substrates in vitro. This does not mean, however, that all pre-tRNAs lacking a CCA motif are processed by the RNase Z-dependent pathway in vivo. Indeed, three examples of CCA-less tRNA precursors, trnD-Cys, trnD-Thr and trnSL-Thr1, were identified in the in vivo screen that appeared correctly processed in the absence of RNase Z. Curiously, all five tRNAs that accumulate as precursors in the absence of RNase Z expression in vivo have a potential terminator hairpin just downstream. The size of the bands observed in Figure 3 is consistent with a precursor molecule extending to this terminator structure in each case: trnI-Asn-Thr, 195 nucleotides; trnSL-Val2, 104 nucleotides; trnSL-Ala1, 106 nucleotides; trnQ-Arg, 133 nucleotides; and trnS-Leu2, 123 nucleotides. It is thus possible that precursors are not observed for the trnD-Cys, trnD-Thr and trnSL-Thr1 tRNAs simply because there is no secondary structure downstream to protect the trailer sequence from exonucleolytic degradation. In these three cases, processing at the 5′ side of the tRNA just downstream by RNase P would be predicted to leave short unprotected 3′ trailers of 7, 22 and 23 nucleotides, respectively, that may be rapidly degraded by exonucleases. Consistent with this hypothesis, we have seen that the trnD-Cys and trnD-Thr1 tRNA precursors are good substrates for RNase Z processing in vitro (data not shown).
Inhibition of RNase Z activity by CCA motif
The replacement of the native TAA sequence by CCA was sufficient essentially to abolish cleavage of the trnI-Thr precursor by RNase Z, showing that the incompatibility lay with the pair of cytosine residues rather than the adenosine of the CCA moiety. While the major contribution to the inhibitory effect came from the first cytosine, inhibition was not as complete as when two C residues were present, suggesting that the second C also plays a role in this context. This is consistent with results obtained in mammals and insects, where 3′ trailers beginning with C, CC or CCA significantly inhibited 3′ tRNase activity (Nashimoto, 1997; Mohan et al., 1999). Interestingly, neither the archaeal nor plant enzymes appear to be inhibited by trailers beginning with CCA (Schiffer et al., 2003). Bacillus subtilis trnI-Thr terminating in a CCA motif is completely recalcitrant to RNase Z activity, whereas addition of just one extra 3′ nucleotide is sufficient to permit a low level of processing at very high enzyme concentrations (data not shown). It makes physiological sense that fully functional tRNAs would not be substrates for RNase Z to prevent endless cycles of processing and CCA addition. At least two kinds of ‘proof-reading’ mechanisms could have been imagined. The CCA could be bound by an inhibitor that would prevent access by RNase Z. Alternatively, RNase Z could possess amino acid residues at or near the active site that are incompatible with the CCA sequence. The fact that in vitro purified RNase Z has the discriminatory capacity suggests that the latter is the more likely explanation. It will be interesting to discover which amino acids of RNase Z are incompatible with the cytosine pair and to learn the structural basis for this incompatibility. Escherichia coli and Salmonella typhi are the only two sequenced members of the proteobacteria with a potential orthologue of RNase Z, encoded by the elaC gene, which gave its name to the human tumour susceptibility gene ELAC2 (Tavtigian et al., 2001). Indeed, it has been confirmed recently that the human ELAC2 gene encodes an enzyme with 3′ tRNase activity (Takaku et al., 2003). Since all of the tRNA genes of E.coli and Salmonella have an encoded CCA, it will be interesting to know whether the CCA motif is also inhibitory in these cases. The E.coli elaC gene product has been shown to have zinc-dependent phosphodiesterase (ZiPD) activity (Vogel et al., 2002), but its potential role in tRNA maturation was not suspected at the time.
Inhibition of RNase Z activity by long 5′ extensions
Our data suggest that for tRNA precursors with long 5′ extensions, ≥33 nucleotides, cleavage by RNase P must precede that by RNase Z, whereas for extensions of ≤10 nucleotides, the order of the RNase P and RNase Z cleavage reactions is probably less important. Of the 27 non-CCA-encoding tRNAs of B.subtilis, 11 are predicted to have 5′ extensions of ≤10 nucleotides (Supplementary figure available at The EMBO Journal Online). The length of the 5′ extensions was calculated from the 3′ end of the upstream tRNA or rRNA, or predicted from the position of an upstream promoter. Six B.subtilis tRNAs have CCA-less precursors with a predicted 5′ extension of >33 nucleotides. We believe that for these tRNAs at least, the order of cleavages must be respected. The 5′ extensions of two CCA-less tRNA precursors could not be predicted due to the lack of obvious promoter motifs, and the eight remaining 5′ extensions range in size from 11 to 29 nucleotides, with an average length of 20 nucleotides. Further experiments are required to determine whether these tRNA precursors are likely to have order-of-cleavage preference.
Materials and methods
Construction of conditional RNase Z mutant
The N-terminal portion of the yqjK gene, including the Shine–Dalgarno and initiation codon, was amplified from B.subtilis W168 chromosomal DNA using oligonucleotides HP519 and HP520 (Table I). The PCR product was cleaved with EcoRI and BamHI and cloned between the same sites in pMUTIN-4m (Vagner et al., 1998) to yield plasmid pMUTIN-Z. Concatamers of this plasmid (isolated from E.coli recA+ strain JM101) were used to transform B.subtilis W168 with selection for MLS resistance in the presence of 1 mM IPTG. Integration in the yqjK gene was confirmed by PCR and the strain was named SSB320. In this strain, expression of yqjK from the spac promoter is under IPTG control.
Table I. Oligonucleotides used in this study.
| Oligo | Sequence (5′→3′) |
|---|---|
| HP61 | AACACATCTAGATCGTAGGTTCGAATC |
| HP62 | AACACAGCATGCCTGAACTACTTCCGC |
| HP262 | CGCTGACCTCTTCCTTACCATGGA |
| HP263 | CCCCCAACCTACTGATTACAAGTCAGT |
| HP293 | ACCTGCGAAGCTTTTTCATTATACATATCGGTTTTAC |
| HP294 | GGTAATAGTCGACTTACAATAGACTTGAATATTATATC |
| HP396 | AATTTAATACGACTCACTATAG |
| HP519 | TTCAGAATTCCCGCTTGATCCTGTATTCTTGTAGGTATCTG |
| HP520 | GTGTGGATCCGAAGGCTTCCACTCCATGAATAACAGA |
| HP521 | TCCGAAGCTTGCCTCGCGGGACGTTTACCTCTAAGAAATC |
| HP536 | CGGGGATAAAGGTTTTGCAGACCTC |
| HP537 | CCACACCGGAGGTTTTGGAGACCTC |
| HP538 | CTACGACCGATCGGTTAACAGCCGA |
| HP539 | CAGCGACCTCTACCCTGTCAAGGTA |
| HP540 | CCCTGACCTCTACGCTGCCAGCGTA |
| HP541 | CTACGACCGGACGGTTATGAGCCGT |
| HP542 | CCCCGACAGACGTGGTACCGGAAAC |
| HP543 | CCGCGACCCCCACCTTGGCAAGGTG |
| HP544 | CCTTGAGACGGAGTTGACCGCCTAC |
| HP545 | CCCCACGTCTGTAAAGACACTAGAG |
| HP546 | CTCCACGGGTTATTCACCCACTAGG |
| HP552 | AATACATTGGAGCTTCCAAGCGGGCTCGAACCGCTGACCTCTTCCTTACCATGGAAGTGCTCTACCTGCTGAGCTATGGAAGCTATAGTGAGTCGTATTAGGATCC |
| HP553 | AATACATTTAAGCTTCCAAGCGGGCTCGAACCGCTGACCTCTTCCTTACCATGGAAGTGCTCTACCTGCTGAGCTATGGAAGCTATAGTGAGTCGTATTAGGATCC |
| HP560 | ATTAATACGACTCACTATAGCTTCCATAGCTCAGCAGGTAG |
| HP561 | GGATCCTAATACGACTCACTATAGCTTCCATAGCTCAGCAGGTAGAGCACTTCCATGGTAAGGAAGAGGTCAGCGGTTCGAGCCCGCTTGGAAGCTCCAATGTATT |
| HP562 | GGATCCTAATACGACTCACTATAGCTTCCATAGCTCAGCAGGTAGAGCACTTCCATGGTAAGGAAGAGGTCAGCGGTTCGAGCCCGCTTGGAAGCTTAAATGTATT |
| HP563 | ATTAATACGACTCACTATAGCCGGTGTAGCTCAATTGGTAG |
| HP564 | ATTAATACGACTCACTATAGCGGAGCATTGCTTCCATAGCTC |
| HP565 | ATTAATACGACTCACTATAGCATTGCTTCCATAGCTCAGCAG |
| HP566 | ATTAATACGACTCACTATAGGCTTCCATAGCTCAGCAGGTAG |
| HP567 | AATACATTGAAGCTTCCAAG |
| HP568 | AATACATTTGAGCTTCCAAG |
| HP600 | ATTAATACGACTCACTATAGATCGTAGGTTCGAATCCTACCT |
| HP608 | GCTGCGCAAGGGTTTAAGCTATGATT |
| HP609 | ATCGAATAAGGGTTTAAAGGTATGGAG |
| HP610 | GCCAAACCCTTGTCTTTCCTGTTGTTCTATTGGACG |
| HP611 | GTGTCAAAACCCTTGGTATCATTGATGC |
RNA preparation and northern blots
SSB320 cells were grown in 25 ml of 2× YT medium + 0.5% glucose to an OD600 of 1.0, in the presence or absence of 1 mM IPTG. A 20 ml aliquot of culture was pelleted and frozen at –20°C. RNA was isolated from cell pellets by the glass bead method of Mayford and Weisblum (1989). Five micrograms of total RNA were migrated on a 5% sequencing gel and electro-blotted to a Hybond-N membrane (Amersham) in TAE buffer (10 mM Tris pH 7.8, 5 mM sodium acetate, 0.5 mM EDTA) at 7 V for 2 h at 4°C. RNA was fixed by imbibing the membrane with 50 mM NaOH for 2 min on each side. For tRNA probes, the membrane was pre-hybridized in filtered (0.2 µm) 5× SSPE, 1% SDS, 5× Denhardt’s (Maniatis et al., 1982), 100 µg/ml sonicated herring sperm DNA (denatured for 10 min at 100°C) for 1 h at 60°C. 5′-32P-labelled oligonucleotide probe (1 pmol) was added and incubation continued for 1 h at 60°C. The temperature of the hybridization oven was then decreased by 5°C every 30 min to 30°C. The membrane was washed twice in 6× SSPE at room temperature for 15 min and twice with pre-heated 3× SSPE, 0.5% SDS at 56°C for 15 min. The membrane was then exposed to a PhosphorImager screen (Molecular Dynamics). The oligonucleotide probes used were the following from Table I: trnI-Thr, HP262; trnB/D/J/SL-Thr, HP263; trnD-Cys, HP536; trnD-Trp, HP537; trnD/I-Asn, HP538; trnSL-Val2, HP539; trnSL-Ala1, HP540; trnB-Met3, HP541; trnQ-Arg, HP542; trnB/D/I/J-Gly1, HP543; trnD-Ser, HP544; trnB-Leu1, HP545; and trnS-Leu2, HP546.
For the northerns with the 3′ extension probes for trnSL-Val2 (HP608) and trnQ-Arg (HP610), hybridization was overnight at 56°C and the two heated wash steps were perfomed at the same temperature. For trnSL-Ala1 (HP609) and trnS-Leu2 (HP611), these steps were performed at 42°C.
Overproduction and purification of histidine-tagged RNase Z
The entire yqjK gene was amplified by PCR using oligonucleotides HP519 and HP521 (Table I), cut with EcoRI and HindIII and cloned in plasmid pET21a (Novagen) digested with the same enzymes. The resulting plasmid was called pET21-Z and contains the yqjK gene fused in-frame to a C-terminal histidine tag.
A 400 ml culture (2× YT + 0.5% glucose) of BL21-CodonPlus λ(DE3)-RIL (Stratagene) carrying pET21-Z was grown to an OD600 of 0.6 at 37°C and RNase Z expression was induced by the addition of 0.5 mM IPTG. The culture was harvested after 3 h, pelleted and frozen at –80°C until further use. The frozen cells were resuspended in 10 ml of buffer containing 20 mM Tris–HCl pH 7.8, 0.5 M NaCl, 10% glycerol and 0.1% Triton X-100 at 4°C. The suspension was passed twice through a French press (20 000 p.s.i.) and the lysate centrifuged for 30 min at 15 000 g. Imidazole-HCl pH 7.8 was added to the supernatant to give a final concentration of 1 mM, and the resultant suspension was applied to a 1 ml Ni2+-NTA column (Qiagen). The resin was then washed sequentially with ∼15 ml of: (i) 20 mM Tris–HCl pH 7.8, 0.3 M NaCl, 20 mM imidazole and (ii) 50 mM sodium phosphate pH 6.0, 0.3 M NaCl. RNase Z was eluted in buffer containing 20 mM Tris–HCl pH 7.8, 0.3 M NaCl and 250 mM imidazole. The fractions containing RNase Z were determined initially by measuring the protein concentration (Bio-Rad), pooled and immediately dialysed in buffer containing 20 mM Tris–HCl pH 7.8, 5 mM MgCl2 and 10% glycerol. Protein purity was then verified by SDS–PAGE analysis (12.5%) and estimated at >95%. The final yield was ∼8 mg of RNase Z. The protein was conserved at –20°C.
Synthesis of tRNA precursors in vitro
Labelled precursor tRNAs were prepared by in vitro transcription of either PCR templates or annealed oligonucleotides, with the upper primer containing a T7 RNA polymerase promoter sequence in each case. Synthesis of the tRNATyr precursor (pTyrI) from O.berteriana (evening primrose) mitochondria has been described previously (Kunzmann et al., 1998). The template for the trnI-Thr precursor with an 83 nucleotide 3′ trailer sequence was made by PCR amplification of plasmid pHMT1 using oligos HP560 and HP62. pHMT1 (Luo, 1998) contains the trnI-Thr gene amplified by PCR using oligos HP61 and HP62, cleaved by SphI and XbaI and cloned between the equivalent sites of pT7T318U (USB). The template for the trnB/D/J/SL-Thr precursor with a 47 nucleotide 3′ trailer was synthesized by PCR amplification of plasmid pHMT8 using oligos HP563 and HP294. pHMT8 (Luo, 1998) contains the trnB-Thr gene amplified by PCR using oligos HP293 and HP294, cleaved by HindIII and SalI and cloned between the equivalent sites of pDG148 (Stragier et al., 1988). The template for the trnI-Thr precursor with a six nucleotide 3′ trailer was created by annealing oligos HP553 and HP562. The mutant variant of this template (TAA→CCA) was created by annealing oligos HP552 and HP561. The TAA→TCA and TAA→CAA variants were synthesized by PCR amplification of pHMT1 using oligos HP560/HP567 and HP560/HP568, respectively. All of these templates were predicted to encode tRNA precursors with 10 nucleotide 3′ trailers. However, the tRNA seems to behave as a transcription terminator for T7 RNA polymerase, resulting in premature transcript arrest six nucleotides downstream. The template for the trnI-Thr precursor with a 66 nucleotide 5′ extension was synthesised by PCR from plasmid pHMT1 using oligos HP396 and HP62. The precursors with 10, 5 and 1 nucleotide 5′ extensions were also amplified by PCR from plasmid pHMT1 using the following oligo pairs HP564/HP62, HP565/HP62 and HP566/HP62. The T7 RNA polymerase in vitro transcription reactions were carried out using 0.1 µg of template according to the manufacturer’s instructions (Promega), except that 4 mM GMP was included in the transcription mix. In this way, the majority of transcripts begin with a 5′ mono-phosphorylated residue, mimicking the effect of RNase P cleavage at the 5′ side of the tRNA. Template DNA was removed by addition of RQ DNase (Promega). Unincorporated nucleotides were removed by G50 spin columns (Amersham).
RNase Z assays
RNase Z activity was assayed in either 5 or 100 µl reaction volumes in 40 mM Tris pH 8.4 and 2 mM each MgCl2, KCl and dithiothreitol (DTT), and incubated for 15–20 min at 37°C. About 40 000 Cerenkov counts (30–40 ng) of uniformly labelled precursor tRNA were used per assay, with varying concentrations of purified histidine-tagged RNase Z. The 5 µl reactions were mixed directly with 5 µl of loading buffer (95% formamide, 20 mM EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol) and applied to 5% polyacrylamide–urea gels. The 100 µl reactions were phenolized and ethanol precipitated before loading.
Kinetic analysis
Each substrate (10 nM) was incubated with varying concentrations of RNase Z (from 125 mM to 8 µM) for 1 min for the wild-type (TAA) and TCA variants of trnI-Thr, and for 18 min for the CAA and CCA variant, at 37°C in 5 µl reaction volumes as described above. The initial velocities of the CAA and CCA variant were still in the linear range at these incubation times. The reaction was stopped by addition of 5 µl of loading buffer and the products resolved on a 5% sequencing gel. The data were fitted to the following equation Vi = Vmax [E]/Km + [E] to calculate Km and Vmax values.
Mapping of RNase Z cleavage site
Non-radiolabelled trnI-Thr precursor with the 83 nucleotide 3′ trailer was prepared as described above. pre-tRNA (360 ng) was incubated with 0.7 µg of RNase Z in a final reaction volume of 50 µl for 20 min at 37°C. The reaction was phenol extracted, ethanol precipitated and resuspended in 3 µl of water. To this was added 1 µl of 5× reverse transcriptase buffer (250 mM Tris pH 8.3, 50 mM MgCl2, 400 mM KCl) and 0.6 pmol of 5′-32P-end-labelled oligo HP62. The mix was denatured at 75°C for 4 min, then incubated for 2 min in a dry ice/ethanol bath. A 5 µl aliquot of dNTP/DTT mix (2 mM each dNTP, 8 mM DTT) and 4 U of AMV reverse transcriptase (Finnzymes) were added and the mix incubated at 45°C for 30 min. Then 5 µl of loading buffer was added and half of the sample was applied to a 5% sequencing gel alongside a sequencing reaction of pHMT1 primed with oligo HP62.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank F.Dardel and V.Arluison for help with the kinetic analysis, and N.Ogasawara and P.Noirot for sharing strains and plasmids. This work was supported by funds from the CNRS (UPR 9073), MRE (Contract 92C0315), Université Paris VII-Denis Diderot (Contract DRED) and PRFMMIP from the Ministère de l’Education Nationale.
References
- Brown J.W. (1999) The ribonuclease P database. Nucleic Acids Res., 27, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano J.G., Tobian,J.A. and Zasloff,M. (1985) Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNA-Met(i) (3′ pre-tRNase). J. Biol. Chem., 260, 9002–9008. [PubMed] [Google Scholar]
- Condon C. and Putzer,H. (2002) The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res., 30, 5339–5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher M.P. (1995) tRNA processing nucleases. In Söll,D. and RajBhandary,U.L. (eds), tRNA: Structure, Biosynthesis and Function. American Society for Microbiology, Washington, DC, pp. 51–65. [Google Scholar]
- Deutscher M.P., Lin,J.J. and Evans,J.A. (1977) Transfer RNA metabolism in Escherichia coli cells deficient in tRNA nucleotidyltransferase. J. Mol. Biol., 117, 1081–1094. [DOI] [PubMed] [Google Scholar]
- Engelke D.R., Gegenheimer,P. and Abelson,J. (1985) Nucleolytic processing of a tRNAArg–tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J. Biol. Chem., 260, 1271–1279. [PubMed] [Google Scholar]
- Frank D.N. and Pace,N.R. (1998) Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem., 67, 153–180. [DOI] [PubMed] [Google Scholar]
- Garber R.L. and Altman,S. (1979) In vitro processing of B.mori transfer RNA precursor molecules. Cell, 17, 389–397. [DOI] [PubMed] [Google Scholar]
- Garber R.L. and Gage,L.P. (1979) Transcription of a cloned Bombyx mori tRNA2Ala gene: nucleotide sequence of the tRNA precursor and its processing in vitro. Cell, 18, 817–828. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner,K., Marsh,T., Pace,N. and Altman,S. (1983) The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell, 35, 849–857. [DOI] [PubMed] [Google Scholar]
- Hagenbuchle O., Larson,D., Hall,G.I. and Sprague,K.U. (1979) The primary transcription product of a silkworm alanine tRNA gene: identification of in vitro sites of initiation, termination and processing. Cell, 18, 1217–1229. [DOI] [PubMed] [Google Scholar]
- Hall T.A. and Brown,J.W. (2002) Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA, 8, 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S.J. and Kang,H.S. (1997) Purification and characterization of the precursor tRNA 3′-end processing nuclease from Aspergillus nidulans. Biochem. Biophys. Res. Commun., 233, 354–358. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. et al. (2003) Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA, 100, 4678–4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruszka K., Barneche,F., Guyot,R., Ailhas,J., Meneau,I., Schiffer,S., Marchfelder,A. and Echeverria,M. (2003) Plant dicistronic tRNA-snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J., 22, 621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzmann A., Brennicke,A. and Marchfelder,A. (1998) 5′ End maturation and RNA editing have to precede tRNA 3′ processing in plant mitochondria. Proc. Natl Acad. Sci. USA, 95, 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L., Bourne,R., Kolla,S., Cylin,E., Russell,K., Wang,X. and Mohan,A. (1998) Matrices of paired substitutions show the effects of tRNA D/T loop sequence on Drosophila RNase P and 3′-tRNase processing. J. Biol. Chem., 273, 1015–1025. [DOI] [PubMed] [Google Scholar]
- Li Z. and Deutscher,M.P. (1996) Maturation pathways for E.coli tRNA precursors: a random multienzyme process in vivo. Cell, 86, 503–512. [DOI] [PubMed] [Google Scholar]
- Li Z. and Deutscher,M.P. (2002) RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA, 8, 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D. (1998) The threonyl- and valyl-tRNA synthetases of Bacillus subtilis: studies on the regulation of their expression and the structure of their leader regions. PhD Thesis, Université de Paris XI, Orsay.
- Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Mayer M., Schiffer,S. and Marchfelder,A. (2000) tRNA 3′ processing in plants: nuclear and mitochondrial activities differ. Biochemistry, 39, 2096–2105. [DOI] [PubMed] [Google Scholar]
- Mayford M. and Weisblum,B. (1989) Conformational alterations in the ermC transcript in vivo during induction. EMBO J., 8, 4307–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A., Whyte,S., Wang,X., Nashimoto,M. and Levinger,L. (1999) The 3′ end CCA of mature tRNA is an antideterminant for eukaryotic 3′-tRNase. RNA, 5, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M. (1997) Distribution of both lengths and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflects properties of 3′ processing endoribonuclease. Nucleic Acids Res., 25, 1148–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M., Geary,S., Tamura,M. and Kaspar,R. (1998) RNA heptamers that direct RNA cleavage by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res., 26, 2565–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M., Tamura,M. and Kaspar,R.L. (1999a) Minimum requirements for substrates of mammalian tRNA 3′ processing endoribonuclease. Biochemistry, 38, 12089–12096. [DOI] [PubMed] [Google Scholar]
- Nashimoto M., Wesemann,D.R., Geary,S., Tamura,M. and Kaspar,R.L. (1999b) Long 5′ leaders inhibit removal of a 3′ trailer from a precursor tRNA by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res., 27, 2770–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oommen A., Li,X.Q. and Gegenheimer,P. (1992) Cleavage specificity of chloroplast and nuclear tRNA 3′-processing nucleases. Mol. Cell. Biol., 12, 865–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C. and Kushner,S.R. (2002) Initiation of tRNA maturation by RNase E is essential for cell viability in E.coli. Genes Dev., 16, 1102–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou A. and Gross,H.J. (1996) Pre-tRNA 3′-processing in Saccharomyces cerevisiae. Purification and characterization of exo- and endoribonucleases. Eur. J. Biochem., 242, 747–759. [DOI] [PubMed] [Google Scholar]
- Petit M.-A., Dervyn,E., Rose,M., Entian,K.-D., McGovern,S., Ehrlich,D.S. and Bruand,C. (1998) PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol., 29, 261–273. [DOI] [PubMed] [Google Scholar]
- Schierling K., Rosch,S., Rupprecht,R., Schiffer,S. and Marchfelder,A. (2002) tRNA 3′ end maturation in archaea has eukaryotic features: the RNase Z from Haloferax volcanii. J. Mol. Biol., 316, 895–902. [DOI] [PubMed] [Google Scholar]
- Schiffer S., Helm,M., Theobald-Dietrich,A., Giege,R. and Marchfelder,A. (2001) The plant tRNA 3′ processing enzyme has a broad substrate spectrum. Biochemistry, 40, 8264–8272. [DOI] [PubMed] [Google Scholar]
- Schiffer S., Rosch,S. and Marchfelder,A. (2002) Assigning a function to a conserved group of proteins: the tRNA 3′-processing enzymes. EMBO J., 21, 2769–2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer S., Rosch,S. and Marchfelder,A. (2003) Recombinant RNase Z does not recognize CCA as part of the tRNA and its cleavage efficiency is influenced by acceptor stem length. Biol. Chem., 384, 333–342. [DOI] [PubMed] [Google Scholar]
- Solari A. and Deutscher,M.P. (1983) Identification of multiple RNases in Xenopus laevis oocytes and their possible role in tRNA processing. Mol. Cell. Biol., 3, 1711–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stange N. and Beier,H. (1987) A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J., 6, 2811–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Bonamy,C. and Karmazyn-Campelli,C. (1988) Processing of a sporulation σ factor in Bacillus subtilis: how morphological structure could control gene expression. Cell, 52, 697–704. [DOI] [PubMed] [Google Scholar]
- Takaku H., Minagawa,A., Takagi,M. and Nashimoto,M. (2003) A candidate prostate cancer susceptibilty gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res., 31, 2272–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavtigian S.V. et al. (2001) A candidate prostate cancer susceptibility gene at chromosome 17p. Nat. Genet., 27, 172–180. [DOI] [PubMed] [Google Scholar]
- Vagner V., Dervyn,E. and Ehrlich,S.D. (1998) A vector for systematic gene inactivation in Bacillus subtilis. Microbiology, 144, 3097–3104. [DOI] [PubMed] [Google Scholar]
- Vogel A., Schilling,O., Niecke,M., Bettmer,J. and Meyer-Klaucke,W. (2002) ElaC encodes a novel binuclear zinc phosphodiesterase. J. Biol. Chem., 277, 29078–29085. [DOI] [PubMed] [Google Scholar]
- Willkomm D.K., Feltens,R. and Hartmann,R.K. (2002) tRNA maturation in Aquifex aeolicus. Biochimie, 84, 713–722. [DOI] [PubMed] [Google Scholar]