Abstract
We previously showed that the availability of a nonamer peptide derived from certain HLA class I signal sequences is a necessary requirement for the stabilization of endogenous HLA-E expression on the surface of 721.221 cells. This led us to examine the ability of HLA-E to protect HLA class I transfectants from natural killer (NK) cell-mediated lysis. It was possible to implicate the CD94/NKG2A complex as an inhibitory receptor recognizing this class Ib molecule by using as target a .221 transfectant selectively expressing surface HLA-E. HLA-E had no apparent inhibitory effect mediated through the identified Ig superfamily (Ig-SF) human killer cell inhibitory receptors or ILT2/LIR1. Further studies of CD94/NKG2+ NK cell-mediated recognition of .221 cells transfected with different HLA class I allotypes (i.e., -Cw4, -Cw3, -B7) confirmed that the inhibitory interaction was mediated by CD94/NKG2A recognizing the surface HLA-E molecule, because only antibodies directed against either HLA-E, CD94, or CD94/NKG2A specifically restored lysis. Surface stabilization of HLA-E in cold-treated .221 cells loaded with appropriate peptides was sufficient to confer protection, resulting from recognition of the HLA class Ib molecule by the CD94/NKG2A inhibitory receptor. Consistent with the prediction that the ligand for CD94/NKG2A is expressed ubiquitously, our examination of HLA-E antigen distribution indicated that it is detectable on the surface of a wide variety of cell types.
Natural killer (NK) cells play an important role in the nonadaptive immune response, which is related to their ability to kill target cells lacking major histocompatibility (MHC) class I (1). NK cells express receptors that interact with MHC class I serving to inhibit cell-mediated cytotoxicity. In the mouse, these molecules are encoded by the Ly49 gene family, a group of type II membrane glycoproteins that are members of the C-type lectin superfamily (2, 3). The human killer cell inhibitory receptors (KIRs), which interact with HLA class I molecules, are type I glycoproteins, members of the Ig superfamily (Ig-SF) (4–6).
In addition, the human CD94/NKG2 C-type lectin receptor complex also plays a role as NK cell receptor for HLA class I (6, 7). The invariant CD94 molecule is expressed as a heterodimer covalently associated with members of the NKG2 family on most NK cells and a subset of T lymphocytes (8, 9). The CD94/NKG2A heterodimer constitutes an inhibitory receptor, whereas the association of CD94 with other NKG2 proteins lacking immunoreceptor tyrosine-based inhibitory motifs (i.e., NKG2C) may form receptors with triggering function (10, 11). CD94/NKG2A has been proposed to be involved in the recognition of a wide variety of HLA-A, -B, and -C allotypes (12, 13), although these studies did not provide a clear rationale for the subdivision among the HLA class I apparently recognized by CD94/NKG2A. Additional inhibitory receptors of the Ig-SF have recently been identified (14); among them, ILT2/LIR1 is broadly distributed in different leukocyte lineages and has also been proven to interact with several HLA class I molecules as well as with the UL18 glycoprotein, an HLA homologue of human cytomegalovirus (15, 16).
Recently, some studies have focused on NK recognition of nonclassical class I molecules, specifically HLA-G (17–20). Although conflicting results have been obtained on the role of Ig-SF KIRs and the CD94/NKG2 receptor recognition of cells expressing HLA-G, the general conclusion that this class Ib molecule can interact with at least some NK receptors to inhibit lysis in a manner similar to HLA class Ia antigens appears sound. Thus far, a formal proof for interaction with HLA-G has been reported only for the ILT2/LIR1 receptor (15).
In addition to HLA-G, the human nonclassical or class Ib genes that are expressed include HLA-E and F (21). We previously obtained results indicating that the availability of a nonamer peptide derived from certain HLA class I signal sequences is a necessary requirement for HLA-E expression on the surface of 721.221 cells (22). These data showed that a number of HLA class I transfectants of .221 were displaying not only the transfected class I allele on the surface, but also the endogenous HLA-E protein. This led us to form the hypothesis that HLA-E might play a role in the NK–HLA class I interactions in studies that used .221 transfectants. In addition, because HLA-E has been highly conserved and exhibits low allelic polymorphism (23), we extended this hypothesis to propose that an interaction with HLA-E and the invariant CD94/NKG2A receptor was likely. In this study, we present strong evidence in support of this hypothesis.
MATERIALS AND METHODS
Antibodies and Antisera.
mAb 3D12 (IgG1, ref. 22) reacts with the HLA-E free heavy chain as well as the heavy chain associated with β2-microglobulin and peptide. Anti-CD94 mAb HP-3B1 (IgG2a) and anti-HLA class I HP-1F7 (IgG1) have been described (18, 24). mAb Z199 (IgG2b) specific for the CD94/NKG2A heterodimer (9), anti-CD56 (C218), anti p58.1 KIR (EB6), and anti p58.2 KIR (GL183) mAbs were kindly provided by A. Moretta (University of Genova, Italy). HP-3E4 anti-KIR mAb was described previously (25). mAbs reacting with p70 KIRs (DX9 and 5.133) were kindly provided by L. Lanier (DNAX) and M. Colonna (Basel Institute for Immunology, Switzerland), respectively. The CD3-PE (UCHT1), anti-CD20-PE (2H7), anti-CD14-PE (M5E2) and anti-CD56-PE (B159) were purchased from PharMingen. The W6/32 mAb, which reacts with monomorphic determinants on HLA class I, was obtained from the American Type Culture Collection. HP-F1 mAb anti-ILT2/LIR1 has been recently described (15). Antibody 16G1 (IgG1, ref. 29) was used as a negative control for 3D12. M. Bonneville (Institut National de la Santé et de la Recherche Médicale U463, Nantes, France) kindly supplied the B1.23.2 anti-framework HLA B/C mAb (26).
Cells and Cell Lines.
Peripheral blood was obtained from healthy donors, and peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll–Hypaque centrifugation. NK clones were established as described previously (27). NKL is an NK cell line kindly provided by M. Robertson (Dana–Farber Cancer Institute, Boston, MA) (28). The HLA-class I-deficient 721.221 and derivative transfected cells used here were described in Lee et al. (22). LCL .221-AEH cells (which express the E*0101 allele) were cultured in the presence of hygromycin B (Calbiochem) in the medium at 200 milliunits/ml. LCL 721.221 cells transfected with HLA-B*0702, B*2705, C*0401, C*0302, and B*5101 were kindly provided by P. Parham (Stanford University, CA) and R. Biassoni (Centro Biotecnologie Avanzate, Genoa, Italy).
Flow Cytometry.
PBMCs were stained with biotin-conjugated 3D12 mAb and visualized by fluorescein isothiocyanate (FITC)-conjugated avidin and phycoerythrin-conjugated anti-CD3, CD20, CD14, or CD56 mAbs. Cells were analyzed on a FACScan flow cytometer (Becton Dickinson). NK clones were phenotyped by indirect immunofluorescence staining conducted as described previously (24).
Peptide Binding to HLA-E.
Peptide-induced stabilization of HLA-E molecules on .221 cells was carried out as described (22). Briefly, HLA-E transfected .221 cells were cultured at 25.5°C for 32 h. Peptides solubilized in DMSO were added into the culture to a final concentration of 200 μM and incubated at the same temperature for another 15 h. After 5 h at 37°C, cells were pelleted, washed, and incubated with 3D12 followed by FITC-conjugated goat anti-mouse Ig-specific antibody (BioSource International, Camarillo, CA).
Cytotoxicity Assays.
NKL cells or NK clones were tested in a 4-hr 51Cr release assay against the .221 cell line and the transfectants expressing different HLA class I. For peptide specificity assays, .221-E cells were incubated at 25.5°C for 7 h before peptides were added. Peptides (SynPep) at final concentration 200 μM were added to the culture for 15 h at the same temperature. One million cells, incubated with or without peptides, were labeled with 200 μCi of 51Cr for 1 h at 37°C and used as targets. In experiments testing the antagonistic effect of mAbs, cells were preincubated at 4°C for 30 min with either antireceptor reagents (for effector cells) or anti-HLA mAbs (for target cells) before performing the assay. Specific lysis was calculated as previously described (27), and in every case spontaneous release was below 20% of the maximum lysis.
RESULTS
HLA-E Is a Broadly Expressed Class I Antigen.
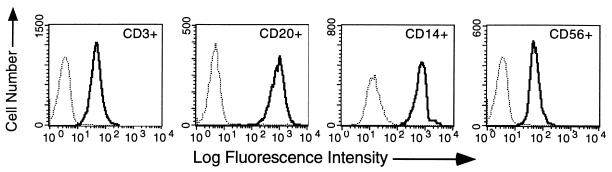
Although HLA-E mRNA expression has been detected in virtually all cells and tissues examined (30), a demonstration of protein expression has been difficult because of the lack of specific reagents. However, we recently generated mAbs that specifically detect HLA-E in the background of other HLA class I (22) and have used these to begin a study of HLA-E cell surface expression in vivo. The results presented in Fig. 1 represent a subset of the analyses carried out with antibody 3D12, which specifically reacts with the HLA-E complex. These data indicated that HLA-E is expressed in virtually all peripheral blood mononuclear cells and that levels varied over an order of magnitude depending on the cell type (compare B cells with T or NK cells). In a limited survey of protein expression we have detected HLA-E in all tissues examined including liver, skin, lung, and notably placental cells where only HLA-G had previously been detected (A.I. and D.E.G., data not shown).
Figure 1.
Distribution of HLA-E in peripheral blood mononuclear cells. Peripheral blood lymphocytes were stained with phycoerythrin-labeled anti-CD3 (UCHT1, T cells), anti-CD20 (2H7, B cells), anti-CD14 (M5E2, monocytes), and anti-CD56 (B159, NK cells) mAbs and FITC-labeled anti-HLA-E (3D12) and FITC-labeled isotype-matched negative control (16G1). Cells were stained and analyzed as described in Materials and Methods, and relative fluorescent intensity was measured. Histograms of the FITC dimension of the four subpopulations are displayed separately as indicated in the upper right corner of each. Bold traces correspond to 3D12 and light traces correspond to the isotype-matched negative control.
HLA-E Inhibits NK Lysis via the CD94/NKG2A Receptor.
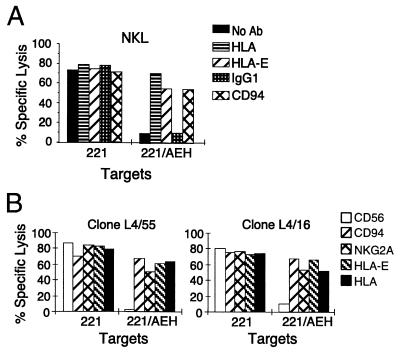
Considering the wide tissue distribution of HLA-E protein and our work on HLA-E expression in LCL .221 (22), we predicted that HLA-E might be involved in the protection from lysis by NK cells via the CD94/NKG2A heterodimer. To approach the issue, we first used as effector the NKL cell line (CD94/NKG2+, IgSF KIR− ILT2/LIR1+) and tested the effect the presence or absence of HLA-E on the surface of LCL .221 might have on cytolysis. In these experiments we employed as targets the .221 line, which expresses endogenous HLA-E intracellularly but not on the surface (31), and .221-AEH, which expresses relatively high levels of surface HLA-E loaded with the HLA-A2 signal sequence-derived nonamer (22). As shown in Fig. 2A, strong inhibition of NKL-mediated cytolysis was apparent when HLA-E was present on the surface of .221 cells (.221-AEH). Further, the addition of pan class I antibody or of anti-HLA-E specific antibody 3D12 nearly completely restored lysis. The inhibitory effect of HLA-E appeared mediated through the CD94/NKG2 receptor complex, as evidenced by the restoration of lysis when anti-CD94 reagent was added.
Figure 2.
The presence of HLA-E on the surface of .221 cells inhibits NK lysis involving the CD94/NKG2 receptor. (A) Chromium release assay using line NKL (CD94/NKG2+, IgSF KIR− ILT2/LIR1+) against 721.221 and .221-AEH. Bars indicate the extent of lysis and are coded according to the presence of antibodies included in the respective assay (see key). (B) Chromium release assay using Z199+ (CD94/NKG2A+) NK clones against 721.221 and .221-AEH. Bars indicating extent of lysis are coded according to the antibody added (see key).
To obtain further support for this conclusion, we tested NK clones expressing the CD94/NKG2A receptor (Z199+) against the same target cells. Again, for a number of Z199+ NK clones, nearly complete protection from lysis was observed when HLA-E was present on the surface of .221 target cells. Fig. 2B shows the results obtained with two representative NK clones, demonstrating that the inhibition of lysis was specific both for HLA-E, because protection was reversed by the 3D12 mAb, and for the CD94/NKG2A complex, because cytotoxicity was reconstituted by CD94 (HP-3B1) or CD94/NKG2A (Z199) specific mAbs.
HLA-E Does Not Functionally Interact with Known Ig-SF NK Inhibitory Receptors.
To carefully examine the possibility that HLA-E might also be recognized by a Ig-SF KIR, and to obtain further support for the CD94/NKG2A-HLA-E interaction, a panel of 165 NK clones derived from 7 different donors was studied. Phenotypic analyses of microcultures were carried out with a panel of Ig-SF KIR-specific mAbs (GL183, EB6, HP-3E4, 5.133, and DX9), together with ILT2/LIR1 receptor (HP-F1), CD94 (HP-3B1), and CD94/NKG2A (Z199) specific mAbs. All clones were tested in cytotoxicity assays against the .221 and .221-AEH target cells. Without exception, clones inhibited by .221-AEH were all Z199+ and no relation was found with their differing patterns of expression of the Ig-SF NK receptors. It is of note that some Z199+ clones were able to efficiently kill the .221-AEH transfectant and corresponded to cells that were also not inhibited upon crosslinking of the receptor by anti-CD94 mAbs in rADCC assays, as described previously (27). The basis for the apparent unresponsiveness of this subset of NK clones is still unclear.
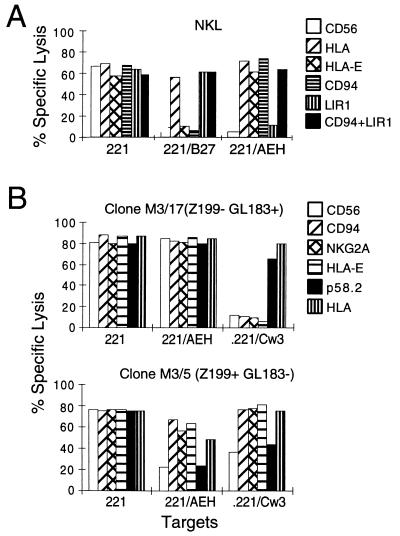
Remarkably, none of the Z199− clones tested (n = 49) was inhibited by .221-AEH cells, regardless of their pattern of reactivity with mAbs specific for KIRs and ILT2/LIR1, thus indicating that the Ig-SF NK inhibitory receptors were not involved in recognition of HLA-E. This was further confirmed by detailed functional analyses of clones, selected according to their Ig-SF KIR, ILT2/LIR1, and CD94/NKG2A phenotype. First, we tested the ability of NKL cells to lyse the LCL .221-B27 transfectant, which expresses HLA-B*2705 but not surface HLA-E (22). As shown in Fig. 3A, NKL-directed cytolysis was strongly inhibited by the presence of HLA-B*2705 and by .221-AEH. However, the receptor molecules present on NKL cells mediating the inhibition of these respective transfectants appeared clearly distinct. With .221-B27 targets, cytolysis was restored by the addition of anti-HLA antibody and with HP-F1 mAb specific for ILT2/LIR1, whereas antibodies reactive with CD94/NKG2 had no effect, consistent with results from previous studies (12). A striking difference was obtained with .221-AEH cells, which also strongly inhibited NKL-mediated cytolysis. In this case, however, the effect appeared exclusively mediated through the CD94/NKG2A receptor complex. The specificity of this response to HLA-E was confirmed by the restoration of lysis upon addition mAb 3D12. These results ruled out a functional involvement of the ILT2/LIR1 receptor in recognition of HLA-E.
Figure 3.
HLA-E does not prevent lysis through Ig-SF NK inhibitory receptors. (A) Line NKL was used in a chromium release assay against 721.221, .221-B27, and .221-AEH cells; the effects of anti-NK receptor mAbs, including the anti-ILT2/LIR1 antibody HP-F1, were tested. (B) Clones M3/17 and M3/5 were selected for their expression pattern of CD94/NKG2A and p58.2 (phenotype indicated in parentheses), determined by FACS analysis. Z199− cells not expressing CD94/NKG2A were not inhibited by HLA-E despite the presence of KIR-type receptor p58.2. In contrast, .221/AEH and .221-Cw3 cells inhibited the CD94/NKG2A+ p58.2- NK clones (Lower). In both cases, anti-HLA-E mAb reconstituted lysis comparably to anti-CD94/NKG2A. In both A and B, target cells are indicated beneath each and bars (keyed on right) indicate the extent of lysis.
We next extended these studies by testing NK clones that were selected for their expression of the CD94/NKG2A complex or the p58.2 KIR (GL183+) using as a target .221-Cw3, which expresses both HLA-C*0301 and HLA-E on the surface. Representative results are shown in Fig. 3B. Similar studies with CD94/NKG2A NK clones displaying functional p58.1 or p70 KIRs also ruled out their involvement in recognition of HLA-E (data not shown). On the other hand, from the results obtained with CD94/NKG2A+ (Z199+) GL183− NK clones (Fig. 3B Lower) it appeared that the protection of .221-Cw3 cells was a result of the presence of endogenous HLA-E on their surface, because only the addition of anti-HLA-E mAb (3D12) or anti-CD94/NKG2A reagents restored lysis. Furthermore, addition of the B.1.23.2 mAb specific for a framework determinant shared by HLA-B and -C molecules (26) but unreactive with HLA-E (M.L., unpublished data), did not reconstitute lysis of Z199+GL183− NK clones against .221-Cw3 cells (data not shown). In agreement with another report (32), the B1.23.2 efficiently antagonized specific KIR-mediated recognition of .221-Cw3.
HLA-E Bound to Appropriate Peptide Is Sufficient for Inhibition of NK Lysis via CD94/NKG2A.
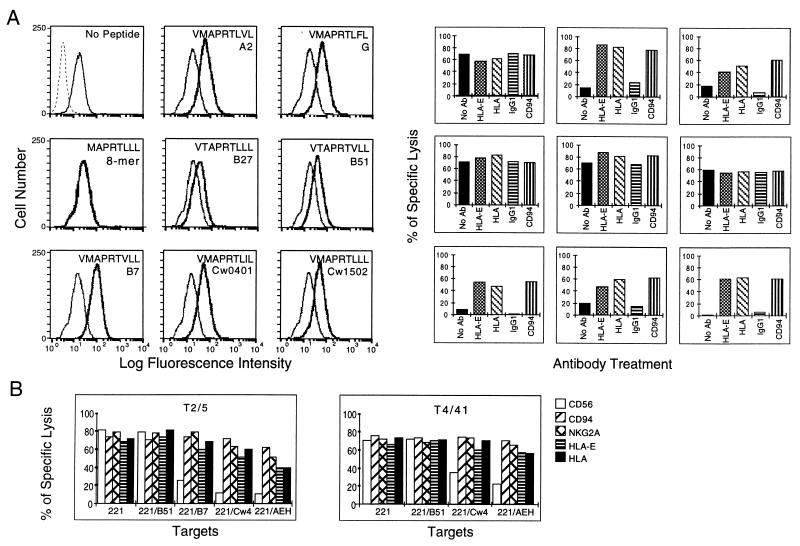
Our previous work showed that cold-treated .221 cells, which express HLA-E associated with β2-microglobulin but devoid of peptide, could be effectively loaded with certain HLA class I signal sequence-derived nonamers (22). Therefore, to further support the notion that a properly assembled HLA-E-peptide complex was necessary and sufficient for inhibition of NK lysis, we used LCL .221-E cold-treated with (and without) the addition of various HLA class I signal sequence-derived nonamers. For these experiments, we chose three classes of peptides: (i) those strongly binding and that induced surface HLA-E expression on class I transfectants (HLA-A*0201, G, B*0701, C*0401, C*1502), (ii) those weakly binding and that did not induce surface HLA-E in transfectants (HLA-B*2705, B*5101), and (iii) an 8-mer signal sequence derivative that showed no binding in the peptide addition assay. We also tested whether surface-expressed HLA-E without bound peptide could affect cytolysis, because cold treatment does increase surface levels of HLA-E plus β2-microglobulin without peptide (22). A representative example of these experiments is shown in Fig. 4; flow cytometry of cold-treated .221-E cells before and after the addition of the indicated peptides confirmed that peptide addition stabilized the HLA-E complex on the surface of .221 as predicted. Thus, although peptides derived from HLA-B*2705 or B*5101 induced some degree of HLA-E stabilization, it was clearly weaker than that apparent from the remaining five class I-specific nonamers tested.
Figure 4.
Appropriate peptide binding to HLA-E is necessary for inhibition of NK lysis mediated through CD94/NKG2A. (A Left) Synthesized peptides were added to cold-treated .221-E cells, and the respective synthetic peptide was added as described in Materials and Methods. Histograms of FACS-analyzed cold-treated .221-E and .221-E plus peptide are presented. Bold traces correspond to the cells treated with peptide and semi-bold correspond to cells with no peptide added. The dotted trace in the experiment where no peptide was added (Top Left) corresponds to .221-E cells that were not cold-treated and thus lacking detectable HLA-E surface expression. The sequence of the peptide added and the name of a representative HLA allotype from which that peptide could be derived are indicated in the upper portion of each histogram. (Right) Chromium release assays using line NKL against the untreated and peptide-treated cells. The results are placed in the position corresponding to the respective FACS histogram on the left. Bars indicate the extent of lysis, and the respective antibody treatment is indicted beneath each bar. (B) Chromium release assays using NK clones (Z199+ EB6− ILT2/LIR1−) against .221 and .221 transfectants as targets. Clone names are indicated in the upper part of each box, and target cell names are indicated beneath each set of bars. Bars indicating extent of lysis are keyed (right) according to the antibody added.
Next, we tested the susceptibility of .221 cells carrying each peptide–HLA-E complex to lysis by NKL cells (Fig. 4). The first group of peptides apparently did stabilize an HLA-E complex that could function to interact with the CD94/NKG2A receptor as evidenced by the substantial protection against cytolysis in those samples. In all cases, lysis was restored by the addition of anti-HLA-E and anti-CD94 antibodies, again supporting the notion that the inhibitory effect was mediated through an HLA-E-CD94 interaction. By contrast, although treatment with the HLA-B*2705- and B*5101-derived nonamers did lead to the stabilization of detectable levels of HLA-E complex, no reduction of cytolytic activity was observed. In addition, although detectable levels of surface HLA-E were also apparent in cold-treated cells in the absence of peptide, no inhibitory effect was detected.
To further support this idea, additional studies were carried out with Z199+ EB6− ILT2/LIR1− NK clones tested against .221-Cw4 and .221-B7, two allotypes previously reported to be recognized by CD94/NKG2A and .221-B51, which was reported not to confer protection. As shown in representative experiments (Fig. 4B), lysis was comparably reconstituted by antibodies specific for either HLA-E, CD94, or the CD94/NKG2A complex when NK cells were directed against HLA-C*0401 or HLA-B*0701. The B1.23.2 anti-framework HLA-B/C had no effect in these assays (not shown), as described above for the recognition of .221-Cw3 cells. It is of note that the use of Z199+ NK clones that did not express the ILT2/LIR1 receptor, reacting with HLA-B7 (15), was important in these experiments to avoid the complex overlapping inhibitory effects of both types of receptors. In no case did HLA-B*5101 confer protection via CD94/NKG2-mediated inhibition, as predicted by the lack of surface HLA-E expression in .221-B51 cells. Altogether, these results supported the notion that HLA-E is interacting with the CD94/NKG2A inhibitory receptor and, moreover, that the apparent CD94-mediated recognition of different HLA class I allotypes can be explained by the induction of HLA-E surface expression upon transfection of HLA molecules.
DISCUSSION
Current models of NK cell function have supposed that the CD94/NKG2A heterodimer is interacting with an epitope common to classical HLA class I (12) or, alternatively, that CD94 might be a coreceptor involved in signaling rather than directly binding class I (6). The data that have led to these hypotheses are almost entirely based on experiments that used LCL .221 cells transfected with classical class I. In our initial studies of HLA-E expression and peptide binding, we discovered that LCL .221 expressed endogenous HLA-E on the cell surface once other class I molecules were transfected (22). This prompted us to review experiments that used LCL .221 in testing the specificity of NK receptors. We had observed that not all class I provide a suitable peptide for HLA-E binding (e.g., HLA-B*2705) and compared this data with studies that implicated an undefined epitope common to many but not all classical class I as interacting with the CD94/NKG2A heterodimer (13, 19). Upon finding a concordance between the HLA allotypes that protected .221 transfectants from lysis and those that provided HLA-E with a suitable nonamer and thus induced surface expression, we reasoned that perhaps the NK cell receptor was in fact interacting with HLA-E itself rather than with classical class I molecules.
Our first experiments tested the CD94/NKG2A+ NKL cell line comparing as targets .221 cells, which do not express detectable levels of HLA-E on the surface, with .221-AEH, which expresses relatively high levels of surface E (Fig. 1A). From this type of experiment it became obvious that HLA-E could in fact play a role in the protection of cells from NK lysis. That protection was apparently mediated through an interaction of HLA-E with the CD94/NKG2A heterodimer because antibodies reactive with these molecules restored lytic ability when added to the assay. Moreover, the analysis of the CD94/NKG2A-mediated recognition of different HLA class Ia transfectants (i.e., .221-Cw3, .221-Cw4, .221-B7), where the effects of anti-HLA and antireceptor mAbs were evaluated, supported the idea that CD94/NKG2A interacted with the endogenous peptide-stabilized HLA-E rather than with the transfected HLA molecules. A further formal proof for a specific interaction of CD94/NKG2A with HLA-E has been provided by the work of Carretero et al. (43). In this study we showed that engagement of CD94/NKG2A with HLA-E molecules expressed on .221-AEH cells specifically promoted both tyrosine phosphorylation of NKG2A and coprecipitation of SHP in NK cells. More important, CD94/NKG2A transfected into heterologous cells (rat basophil leukemia cells), thus segregated from other NK receptors, specifically recruited SHP-1 upon treatment with .221-AEH cells.
In previous studies (13, 44) the effect of HLA class I expression on prevention of lysis via the CD94/NKG2A receptor divided the tested allotypes into two groups, i.e., protective or not. Considering what we now know about HLA-E peptide binding (22) and the results presented in this report, we can find nearly complete agreement between the apparent ability of the introduced HLA allele to protect via CD94/NKG2A and its ability to provide a suitable nonamer peptide for HLA-E complex formation and subsequent surface expression. The HLA-C*1503 molecule, which does not provide protection through CD94/NKG2 when transfected into .221, constitutes the single unknown because we have not tested the HLA-C*1503-encoded nonamer (VMTPRTLLL, which may be unique among HLA class I) for HLA-E binding. Thus, the data presented here and the resulting prediction that HLA-E is a major if not the sole ligand for the CD94/NKG2A inhibitory receptor are in fact supported by preexisting data. Also, observations about the level of HLA having differing effects on inhibition via CD94 (13) could be explained by differing levels of HLA-E expression controlled by the expression of the classical class I molecule.
Altogether, these results point out that HLA-E may play a fundamental role in the immune response, central to the function of NK immune surveillance; as such, several important areas of study might be affected by these results. Although HLA-E-bound peptides are derived from HLA class I signal sequences, peptide binding is nonetheless dependent on a functional TAP heterodimer (22). Although apparently most signal sequence-derived peptides that bind to class I are TAP-independent (33), there are a few exceptions (34). Such exceptionality suggests that TAP dependence for HLA-E peptide binding may have evolved as a selective advantage against viral infection. With HLA-E as the major ligand for the CD94/NKG2A heterodimer, TAP-dependent peptide loading affords control over HLA-E surface expression and function by a functional TAP complex. If it were otherwise, viral infections that lead to a down-regulation of classical class I expression by interfering with TAP-dependent peptide loading (35) would not reduce the level of HLA-E expression because class I synthesis and thus availability of signal peptide-derived nonamer would not be limited. These cells therefore would not only be able to avoid cytotoxic T lymphocyte recognition of viral peptides, but also would avoid lysis by NK cells expressing the CD94/NKG2 molecules because HLA-E would still be expressed. In addition, viral mechanisms that down-regulate class I protein synthesis by interfering with classical class I transcriptional mechanisms (36) or heavy chain stability (37) also predictably down-regulate HLA-E because of a lack of appropriate peptide. Indeed, these considerations suggest that viral mechanisms that do interfere with classical class I expression but do not alter HLA-E peptide binding and expression may have successfully evolved. In this regard, studies on the human cytomegalovirus UL18 glycoprotein providing protection against NK cells mediated through CD94 (38) might present an interesting case in point. However, on examination of the UL18 sequence for suitable HLA-E-binding peptides, none were apparent. Along very similar rationale, these data also have implications for studies of altered HLA class I phenotypes in human tumors (39).
A broad function for HLA-E as the major ligand for the CD94/NKG2A heterodimer in humans raises the question of which molecule might play a similar role in other mammals. The work of Boyson et al. (40) suggested that the codons encoding the amino acids in the peptide-binding region had been conserved in macaques and humans because they had last shared a common ancestor, suggesting the conservation of HLA-E function in primates. In the mouse, the Qa-1 antigen may be a likely candidate for a parallel function because it also binds a very similar nonamer peptide derived from the leader sequence of murine classical class I, peptide binding depends on a functional TAP molecule, and both Qa-1 and HLA-E have essentially similar qualitative and quantitative expression patterns (22, 30, 41). Such a possibility raises the question of what the receptor for Qa-1 might be. In this regard, evidence for the existence of murine CD94/NKG2 gene homologues has recently been obtained (W. M. Yokoyama, personal communication).
In summary, our results support that HLA-E is the major if not the sole ligand for the CD94/NKG2A heterodimer. One prediction regarding the ligand for the CD94/NKG2 receptor is that it be expressed ubiquitously, consistent with our preliminary data. Our preliminary data also suggest that HLA-E may play a broader role than simply conferring protection to cells from NK lysis, because a limited number of NK clones appeared to be specifically activated by HLA-E-transfected cells; all of these clones were Z199−, raising the possibility that other CD94/NKG2 heterodimer(s) (i.e., CD94/NKG2C) may be involved as triggering receptors (M.L.-B. and D.E.G., unpublished data). If HLA-E is confirmed to constitute the only ligand for the CD94/NKG2 receptor family, the presence of these receptors on a subset of T cells suggests the possibility that HLA-E might be involved in peripheral tolerance (6, 42).
Acknowledgments
We are grateful to Teresa Bellón and J. J. Pérez-Villar for helpful discussions. The expert assistance of Natalia Dorofeeva and Laura Braun is gratefully acknowledged. We also thank R. Biassoni, M. Bonneville, L. Lanier, A. Moretta, M. Robertson, and P. Parham for gifts of antibodies and cell lines. This work was supported by National Institutes of Health Grant AI38508 to D.E.G. N.L. was supported by National Research Service Award fellowship AI08911. The work at the Hospital de la Princesa was supported by Grants SAF96/0335 (Plan Nacional I+D) and PL950062 (European Commission). M.C. is supported by a fellowship from Fondo de Investigaciones Sanitarias.
ABBREVIATIONS
- Ig-SF
Ig superfamily
- NK cells
natural killer cells
- FITC
fluorescein isothiocyanate
- KIR
human killer cell inhibitory receptor
Footnotes
A commentary on this article begins on page 4791.
References
- 1.Ljunggren H G, Karre K. J Exp Med. 1985;162:1745–1759. doi: 10.1084/jem.162.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan J, Mager D, Jefferies W, Takei F. J Exp Med. 1994;180:2287–2295. doi: 10.1084/jem.180.6.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniels B F, Karlhofer F M, Seaman W E, Yokoyama W M. J Exp Med. 1994;180:687–692. doi: 10.1084/jem.180.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonna M, Samaridis J. Science. 1995;268:405–408. doi: 10.1126/science.7716543. [DOI] [PubMed] [Google Scholar]
- 5.Wagtmann N, Rajagopalan S, Winter C C, Peruzzi M, Long E O. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 6.Lanier L L. Immunity. 1997;6:371–378. doi: 10.1016/s1074-7613(00)80280-0. [DOI] [PubMed] [Google Scholar]
- 7.López-Botet M, Pérez-Villar J J, Carretero M, Rodriguez A, Melero I, Bellon T, Llano M, Navarro F. Immunol Rev. 1997;155:165–174. doi: 10.1111/j.1600-065x.1997.tb00949.x. [DOI] [PubMed] [Google Scholar]
- 8.Lazetic S, Chang C, Houchins J P, Lanier L L, Phillips J H. J Immunol. 1996;157:4741–4745. [PubMed] [Google Scholar]
- 9.Carretero M, Cantoni C, Bellon T, Bottino C, Biassoni R, Rodriguez A, Pérez-Villar J J, Moretta L, Moretta A, López-Botet M. Eur J Immunol. 1997;27:563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 10.Pérez-Villar J J, Carretero M, Navarro F, Melero I, Rodriguez A, Bottino C, Moretta A, López-Botet M. J Immunol. 1996;157:5367–5374. [PubMed] [Google Scholar]
- 11.Houchins J P, Lanier L L, Niemi E C, Phillips J H, Ryan J C. J Immunol. 1997;158:3603–3609. [PubMed] [Google Scholar]
- 12.Moretta A, Vitale M, Sivori S, Bottino C, Morelli L, Augugliaro R, Barbaresi M, Pende D, Ciccone E, López-Botet M. J Exp Med. 1994;180:545–555. doi: 10.1084/jem.180.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivori S, Vitale M, Bottino C, Marcenaro E, Sanseverino L, Parolini S, Moretta L, Moretta A. Eur J Immunol. 1996;26:2487–2492. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama W M. J Exp Med. 1997;186:1803–1808. doi: 10.1084/jem.186.11.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colonna M, Navarro F, Bellon T, Llano M, Garcia P, Samaridis J, Angman L, Cella M, López-Botet M. J Exp Med. 1997;186:1809–1818. doi: 10.1084/jem.186.11.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosman D, Fanger N, Borges L, Kubin M, Chin W, Peterson L, Hsu M L. Immunity. 1997;7:273–282. doi: 10.1016/s1074-7613(00)80529-4. [DOI] [PubMed] [Google Scholar]
- 17.Pazmany L, Mandelboim O, Vales-Gomez M, Davis D M, Reyburn H T, Strominger J L. Science. 1996;274:792–795. doi: 10.1126/science.274.5288.792. [DOI] [PubMed] [Google Scholar]
- 18.Pérez-Villar J J, Melero I, Navarro F, Carretero M, Bellon T, Llano M, Colonna M, Geraghty D E, López-Botet M. J Immunol. 1997;158:5736–5743. [PubMed] [Google Scholar]
- 19.Söderstrom K, Corliss B, Lanier L L, Phillips J H. J Immunol. 1997;159:1072–1075. [PubMed] [Google Scholar]
- 20.Pende D, Sivori S, Accame L, Pareti L, Falco M, Geraghty D, Le, Bouteiller P, Moretta L, Moretta A. Eur J Immunol. 1997;27:1875–1880. doi: 10.1002/eji.1830270809. [DOI] [PubMed] [Google Scholar]
- 21.Geraghty D E. Curr Opin Immunol. 1993;5:3–7. doi: 10.1016/0952-7915(93)90073-2. [DOI] [PubMed] [Google Scholar]
- 22.Lee, N., Goodlett, D. R., Ishitani, A., Marquardt, H. & Geraghty, D. E. (1998) J. Immunol., in press. [PubMed]
- 23.Geraghty D E, Stockschleader M, Ishitani A, Hansen J A. Hum Immunol. 1992;33:174–184. doi: 10.1016/0198-8859(92)90069-y. [DOI] [PubMed] [Google Scholar]
- 24.Aramburu J, Balboa M A, Ramirez A, Silva A, Acevedo A, Sanchez M, De Landazori M, López-Botet M. J Immunol. 1990;144:3238–3247. [PubMed] [Google Scholar]
- 25.Melero I, Salmeron A, Balboa M A, Aramburu J, López-Botet M. J Immunol. 1994;152:1662–1673. [PubMed] [Google Scholar]
- 26.Rebai N, Malissen B. Tissue Antigens. 1983;22:107–117. doi: 10.1111/j.1399-0039.1983.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 27.Pérez-Villar J J, Melero I, Rodriguez A, Carretero M, Aramburu J, Sivori S, Orengo A M, Moretta A, López-Botet M. J Immunol. 1995;154:5779–5788. [PubMed] [Google Scholar]
- 28.Robertson M J, Cochran K J, Cameron C, Le J M, Tantravahi R, Ritz J. Exp Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 29.Lee N, Malacko A R, Ishitani A, Chen M C, Bajorath J, Marquardt H, Geraghty D E. Immunity. 1995;3:591–600. doi: 10.1016/1074-7613(95)90130-2. [DOI] [PubMed] [Google Scholar]
- 30.Koller B H, Geraghty D E, Shimizu Y, DeMars R, Orr H T. J Immunol. 1988;141:897–904. [PubMed] [Google Scholar]
- 31.Shimizu Y, Geraghty D E, Koller B H, Orr H T, DeMars R. Proc Natl Acad Sci USA. 1988;85:227–231. doi: 10.1073/pnas.85.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain J F, de Smet C, Chambost H, Vitale M, Moretta A, Boon T, Coulie P G. Immunity. 1997;6:199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 33.Wei M L, Cresswell P. Nature (London) 1992;356:443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 34.Williams D B, Aikaterini V, Woong-Kyung S. Cell Biol. 1996;6:267–269. [Google Scholar]
- 35.Hengel H, Koopmann J O, Flohr T, Muranyi W, Goulmy E, Hammerling G J, Koszinowski U H, Momburg F. Immunity. 1997;6:623–632. doi: 10.1016/s1074-7613(00)80350-7. [DOI] [PubMed] [Google Scholar]
- 36.Proffitt J L, Sharma E, Blair G E. Nucleic Acids Res. 1994;22:4779–4788. doi: 10.1093/nar/22.22.4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beersma M F, Bijlmakers M J, Ploegh H L. J Immunol. 1993;151:4455–4464. [PubMed] [Google Scholar]
- 38.Reyburn H T, Mandelboim O, Vales-Gomez M, Davis D M, Pazmany L, Strominger J L. Nature (London) 1997;386:514–517. doi: 10.1038/386514a0. [DOI] [PubMed] [Google Scholar]
- 39.Garrido F, Ruiz-Cabello F, Cabrera T, Pérez-Villar J J, López-Botet M, Duggan-Keen M, Stern P L. Immunol Today. 1997;18:89–95. doi: 10.1016/s0167-5699(96)10075-x. [DOI] [PubMed] [Google Scholar]
- 40.Boyson J E, McAdam S N, Gallimore A, Golos T G, Liu X, Gotch F M, Hughes A L, Watkins D I. Immunogenetics. 1995;41:59–68. doi: 10.1007/BF00182314. [DOI] [PubMed] [Google Scholar]
- 41.Connolly D J, Cotterill L A, Hederer R A, Thorpe C J, Travers P J, McVey J H, Dyson J, Robinson P J. J Immunol. 1993;151:6089–6098. [PubMed] [Google Scholar]
- 42.Ware R, Jiang H, Braunstein N, Kent J, Wiener E, Pernis B, Chess L. Immunity. 1995;2:177–184. doi: 10.1016/s1074-7613(95)80066-2. [DOI] [PubMed] [Google Scholar]
- 43.Carretero, M., Palmieri, G., Llano, M., Tullio, V., Santoni, A., Geraghty, D. E. & López-Botet, M. (1998) Eur. J. Immunol., in press. [DOI] [PubMed]
- 44.Phillips JH, Chang C, Mattson J, Gónperz J E, Parham P, Lanier LL. Immunity. 1996;5:163–172. doi: 10.1016/s1074-7613(00)80492-6. [DOI] [PubMed] [Google Scholar]