Abstract
The assembly of hepatitis C virus (HCV) is not well understood. We investigated HCV nucleocapsid assembly in vitro and the role of electrostatic/hydrophobic interactions in this process. A simple and rapid in vitro assay was developed in which the progress of assembly is monitored by measuring an increase in turbidity, thus allowing the kinetics of assembly to be determined. Assembly is performed using a truncated HCV core (C1-82), containing the minimal assembly domain, purified from E. coli. The increase in turbidity is linked to the formation of nucleocapsid-like particles (NLPs) in solution, and nucleic acids are essential to initiate nucleocapsid assembly under the experimental conditions used. The sensitivity of NLP formation to salt strongly suggests that electrostatic forces govern in vitro assembly. Mutational analysis of C1-82 demonstrated that it is the global positive charge of C1-82 rather than any specific basic residue that is important for the assembly process. Our in vitro assembly assay provides an easy and efficient means of screening for assembly inhibitors; we have identified several inhibitory peptides that could represent a starting point for drug design.
Keywords: Hepatitis C virus, nucleocapsid, in vitro assembly, kinetics, absorbance spectrometry
Introduction
Hepatitis C virus (HCV) is a major public health concern worldwide since more than 3% of the world’s population is infected with this virus [1]. HCV is a plus-strand RNA virus that causes acute and chronic liver disease and is associated with 95% of cases of post-transfusion hepatitis [2] and over 50% of non-A, non-B hepatitis. Current therapy is the use of pegylated interferon and ribavirin but outcomes are unsatisfactory since only 42% of patients infected with HCV genotype 1 (the most common genotype in North America) respond positively to treatment [3]. Therefore, there is an urgent need to identify new targets for the development of drugs to both cure, and prevent the spread of, the disease.
HCV core protein—a 191-amino-acid protein located at the N terminus of the HCV polyprotein—is the largest core protein in the family of Flaviviridae. The C-terminal region of HCV core is highly hydrophobic and targets the protein to the ER membranes. This hydrophobic region is further cleaved by a host protease, the signal peptide peptidase, to generate the mature protein of 177 or 179 amino acids [4, 5]. The core protein is also submitted to post-translational modifications such as ubiquitinylation [6] and phosphorylation [7, 8], which can potentially regulate its degradation in the infected cell. Little is known about HCV nucleocapsid assembly; however, based on what is known in other RNA viruses, assembly is expected to be initiated by the interaction between core protein and viral RNA. Electrostatic forces appear to be important in this phenomenon [9].
The core protein has been shown to package the viral RNA [10], possibly through specific interactions with the 5′ untranslated region (5′UTR) of the RNA genome [11, 12]. The region around the 5′ UTR is characterised by both secondary and tertiary structure [13-16], contributing to the internal ribosome entry site (IRES) required for initiation of cap-independent translation [17]. The N-terminal half (N-terminal 82 amino acids) of the protein is rich in positively charged residues (K, R). This domain is sufficient to trigger the formation of nucleocapsid-like particles (NLPs) in vitro when structured RNA is added to the purified protein [18].
A more effective assay for the detection of nucleocapsid formation is required to improve our understanding of the biochemistry of this process and to allow screening for inhibitors of assembly. Here we describe the development of a method that allows the kinetics of assembly to be followed in vitro by measurement of solution turbidity using a spectrophotometer. This system has revealed the critical role of electrostatic interactions in particle formation and has identified a domain within the core that could be sensitive to assembly inhibitors. This novel method can be used to perform high-throughput screening of potential assembly inhibitors such as inhibiting compounds or peptides.
Materials and methods
Cloning and expression of HCV core proteins in E. coli
The nucleotide sequence of HCV core protein was optimized with most abundant codons for translation in bacteria [19]. This optimized sequence was used to generate the construct C1-82 as well as the other mutant forms (Fig. 5A). All forms of C1-82 were cloned into the pET3d expression vector (New England Biolabs), which harbours a 6-histidine-tag at the C-terminal end to ease the purification process on a nickel affinity column (Qiagen). The mutated forms RR39,40AA, RR43,47AA, RK50,51AA, RR55,59AA, E54A, RR61,62AA, Δ8-23, Δ39-62 and C1-71 were amplified from the C1-82 clone with the following primers: K23A (forward): 5′- and K23A (reverse): 5′-; RR39,40AA (forward): 5′-GCGGGTCCGCGTCTGGGTGTTCG-3′ and RR39,40AA (reverse): 5′-CGCCGGCAGCAGGTAAACACCACC-3′; RR43,47AA (forward): 5′-GGTGTTGCGGCGACCCGTAAAACCTCTGAAC-3′ and RR43,47AA (reverse): 5′-CAGCGCCGGACCACGACGCGGCAGCAGG-3′; RK50,51AA (forward): 5′-GCGACCTCTGAACGTTCTCAGCCG-3′ and RK50,51AA (reverse): 5′-CGCGGTCGCACGAACACCCAGACG-3′; E54A (forward): 5′-CGTTCTCAGCCGCGTGGTCGTC-3′ and E54A (reverse): 5′-CGCAGAGGTTTTACGGGTCGCAC-3′; RR55,59AA (forward ): 5′-CAGCCGGCGGGTCGTCGTCAGCCGATCCCG-3′ and RR55,59AA (reverse): 5′-AGACGCTTCAGAGGTTTTACGGGTCGC-3′; RR61,62AA (forward ): 5′-GCGCAGCCGATCCCGAAAGCGCG-3′ and RR61,62AA (reverse): 5′-CGCACCACGCGGCTGAGAACGTTC-3′; Δ8-23 (forward:) 5′-TTCCCGGGTGGCGGTCAG-3′ and Δ8-23 (reverse): 5′-CTGCGGTTTCGGGTTGG-3′; Δ39-62 (forward): 5′-CAGCCGATCCCGAAAGCG-3′ and Δ39-62 (reverse): 5′-CGGCAGCAGGTAAACACC-3′; C1-71 8H (forward): 5′-CACCACCATCACCACCATCACCACTAA-3′ and C1-71 (reverse): 5′-CGGACGACGCGCTTTCGGGATCGGCTG-3′. Clones were circularized by ligation of the PCR products. The sequences of the clones were confirmed by DNA sequencing.
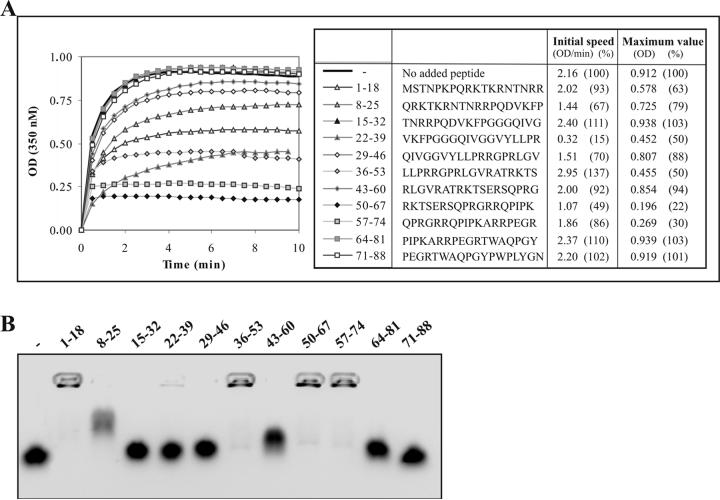
Figure 5.
Peptide-based inhibition of in vitro assembly of C1-82. (A) Kinetics of in vitro assembly with C1-82 (760 pmol) initiated with 38 pmol of IRES alone or together with 3.8 nmol of various peptides. (B) EMSA (electrophoresis mobility shift assay) (0.8% agarose) of 16 nmol of each peptide mixed with 16 pmol of IRES. Mixing was performed 10 minutes prior to loading the gel. The gel was stained with ethidium bromide and photographed using a digital camera under UV254nm.
Purification of HCV-C proteins in E. coli
The expression and purification of the recombinant proteins were performed using a Ni NTA resin (Qiagen) as previously described [18]. HCV-C proteins were eluted in assembly buffer (1.7 mM magnesium acetate, 100 mM potassium acetate, 25 mM Hepes (pH 7.4), and 500 mM imidazol). In order to eliminate NLPs pre-assembled in E. coli, purified HCV-C proteins were ultracentrifuged for 3 hours at 100,000g at 4°C (Optima L-90K ultracentrifuge Beckman Coulter). The purity of the protein was estimated by 10 % SDS-PAGE and confirmed by western blot. Core proteins were quantified using a BCA protein assay kit (Pierce).
RNA transcripts
The plasmid used for the transcription of the IRES, corresponding to the full-length 5′UTR of HCV H77c (nt 1-374) (genotype 1a generously provided by J. Bukh, NIH) [20], was amplified by PCR with primers 5′ end IRES (5′-ACTGAACTAGTGCCAGCCCCCTGATGGGGGCG-3′) and 3′ end IRES and cloned downstream of a T7 promoter of pBlueScript II KS (+/-) vector (Stratagene). PCR product with primers 5′ end T7 promoter (5′-AGTGAGCGCGCGTAATACGACTCA-3′) and 3′ end IRES 5′-CGATGGATCCGGTTTTTCTTTGAGGTTTAGGTATCGTGCTCATGGTGCACGGTGTACGAGACC-3′ was used as a template for in vitro transcription (T7 RiboMAX™ Express Large Scale RNA Production System of Promega) of IRES. Ribosomal NTP (Promega), yeast tRNA (Sigma) and polynucleotides (polyU, polyA, polyC) (Amersham) were commercial preparations. All nucleic acids were dissolved in assembly buffer.
Kinetic analysis of in vitro assembly reactions
For kinetic analysis, 8 μg (760 pmol) of C1-82 was diluted in 50 μL of assembly buffer in a microcuvette (Eppendorf). Optical density was monitored at 350 nm in a spectrometer (Biochrom Ultrospec 2100pro UV/Vis) at 25°C. Different amounts of nucleic acids in a volume of 50μL were added to the solution, and mixed vigorously in the microcuvette. Assembly was monitored with the spectrophotometer. Approximately 6 s elapsed before the first time point was measured. Optical density was recorded by SWIFT II 2.0 software every 2 s for 10 min. The maximum value represent the maximum OD obtained in this 10 min of analysis. The initial speed constant was deduced from the slope of the kinetic curve for the first 10s of the reaction. Each slope is drawn based on 5 points measured at 2 second intervals.
When potassium chloride 1M, potassium nitrate 1M, potassium acetate 1M, sodium chloride 1M (Sigma), 4,4′-dianilino-1,1′-binaphtyl-5,5′-disulfonic acid dipotassium salt (Bis-ANS) (Sigma), ethanol, methanol or glycerol were added for kinetic analysis, each compound was mixed with C1-82 (760 pmol) in the microcuvette (maximum volume of 70 μL) prior to RNA addition. The optimal amount of IRES (38 pmol) in a final reaction volume of 100 μL was then added as previously described.
Electron microscopy
Samples collected after in vitro assembly were directly absorbed on 400 mesh carbo-Formvar grids (Canemco) for 5 min. The grids were washed once with filtered PBS and stained for 10 min with filtered 2 % (w/v) uranyl acetate. Grids were then dried on filter paper before being examined under an electron microscope with an acceleration voltage of 60 kV at a magnification of 150 000X. Images were captured and treated with Digital Micrograph™ (Gatan version 3.8.2).
Northwestern dot blot
The recombinant protein C1-82 was deposited on a nitrocellulose membrane (BioRad) using a vacuum to insure homogeneity of the dots. The membrane was blocked with 5% milk in assembly buffer at room temperature for 1 hour. The membrane was washed 3 times with assembly buffer followed by addition of 2.5 μg of radiolabelled IRES. The blots were incubated overnight at room temperature. After 3 washes, data acquisition was preformed on a phosphoimager (ImageQuant).
Electrophoresis mobility shift assay
The RNA (IRES) was incubated with recombinant proteins at room temperature for 10 min. We used 16 pmol of RNA and 16 nmol of peptides for each reaction in in vitro assembly buffer. The final volume of the reaction was 20 μL; 4 μL loading dye were added to the sample before loading onto a 0.8% agarose gel. Electrophoresis was performed in Tris / acetic /EDTA buffer for 30 min at 90V. The gel was stained with ethidium bromide and photographed with a digital camera (Bio-Rad).
Peptide library
The 11 peptides of the tested library were 18 amino acids in length. They were synthetic peptides corresponding to the first 88 amino acids of HCV strain H77 (NIH AIDS Research and Reference Reagent Program). Each peptide was added to IRES solution at a ratio of 1000 peptides for one molecule of IRES. This solution was then vigorously mixed with C1-82 solution in the microcuvette to initiate assembly. The ratio of C1-82:IRES was 20:1.
Results and Discussion
In vitro assembly of HCV NLPs as monitored by an increase in solution turbidity
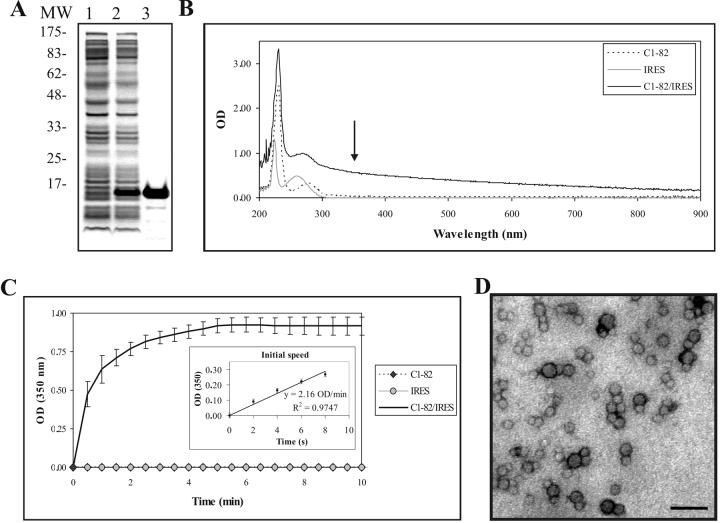
The region of the core protein corresponding to amino acids 1-82 has been shown to be the minimal domain required for assembly [18]; we have previously shown that C1-82 protein forms NLPs when tRNA is added to the protein [18]. Recombinant C1-82 protein was purified by affinity chromatography (Fig. 1A) using a 6xH tag located at the C-terminus of the protein. The RNA used in the assembly assay was derived from the full-length 5′ UTR (nt 1-374) of HCV genome H77c [20], hereafter referred to as IRES. Purified C1-82 and the IRES RNA were mixed at a molar ratio of 20:1 (protein: RNA) with 760 pmol of protein (8 μg) and 38 pmol of RNA (4.4 μg RNA) in 100 μl of low salt buffer. The absorbance of the two constituents and newly formed NLPs were measured using a spectrophotometer at different wavelengths after 10 min of reaction. Unlike the unassembled constituents (RNA and protein), the turbid assembly solution absorbs light at 350 nm (Fig. 1B). The assembly process was thus monitored at 350 nm for 10 minutes. As RNA was added to the solution, we observed a rapid increase in absorbance associated with the turbidity of the sample. One-half of the final absorbance value was reached in less then 30 sec, indicating that assembly of C1-82 is very rapid and efficient. The in vitro assembly assay was repeated 5 times with different preparations of proteins and RNA. The average value of the plateau was 0.926 OD with a standard deviation of 0.070 OD, showing that the assay is reliable and reproducible (Fig. 1C). As expected, neither the IRES nor the C1-82 protein alone exhibited any increase in turbidity under the experimental conditions used.
Figure 1.
(A) HCV C1-82 expression and purification. SDS-PAGE (10%) of lysed bacterial cell pellets before induction (1), after 3h induction with 1 mM IPTG (2), and C1-82 purified on a Ni2+ affinity column (3). Molecular markers in kDa (Sigma) are shown on the left. (B) Absorbance spectra of C1-82 (760 pmol), IRES (38 pmol) and a mixed solution of C1-82:IRES (ratio 20:1). The arrow indicates the wavelength selected for the kinetic assay. (C) Kinetics of in vitro assembly. Data points are the means (± SD) of 5 independent experiments performed with 760 pmol of C1-82 mixed with 38 pmol IRES. The kinetics of the first 10s of the reaction and the initial speed estimation are shown in the inset. (D) Electron micrograph of negatively stained NLPs produced in the in vitro assembly assay. Bar =100 nm.
To measure the speed at which the assembly reaction occurs, we determined the slope of the curve in the first 10 seconds of the reaction. The mean value for this constant (referred to as initial speed constant) for 5 repetitions using separate batches of purified C1-82 protein was estimated at 2.16 OD/min with a standard deviation of only 0.09 OD/min. The standard deviation between experiments was acceptable, confirmed that this is a robust and reliable method for measurement of NLP formation. The initial speed is a good indicator of the efficiency of in vitro assembly. The in vitro assembly of alphavirus core protein, a virus structurally similar to HCV, was also shown to be a rapid process [21].
NLP formation is responsible for the increase in turbidity during assembly
To confirm that the increase in turbidity is linked to the formation of NLPs, we observed the assembly product after 20 minutes of reaction by electron microscopy (EM). Particles 25 nm ± 7 nm in diameter and uniform in appearance were produced (Fig. 1D) as expected and as previously reported [18]. No particles were seen under EM in grids with core protein only (data not shown), consistent with the previously published observation that recombinant C1-82 is a soluble and monodispersed protein [22].
Effect of salt on in vitro assembly of HCV NLPs
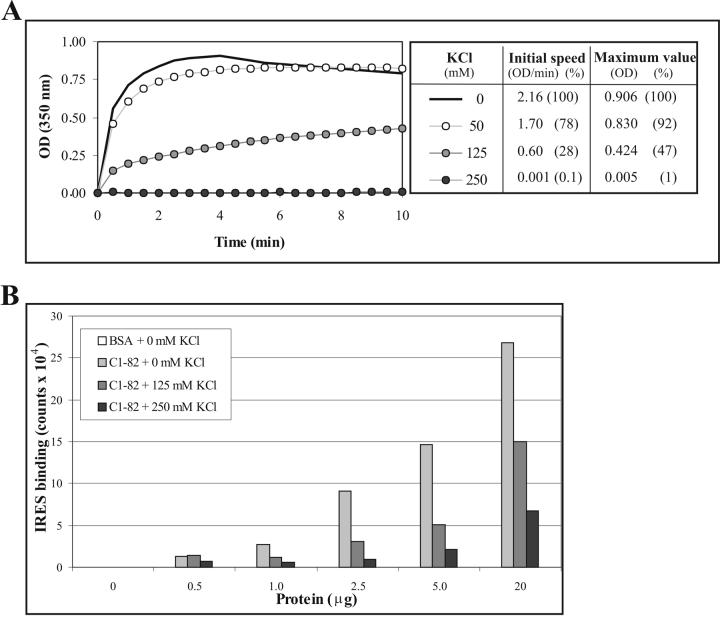
To assess the role of electrostatic forces in NLP assembly, we measured the influence of increasing the concentration of potassium chloride in our assay on the initial speed constant (Fig. 2A). We observed that the increase in absorbance induced by the formation of NLPs decreases proportionally with the increase in potassium chloride concentration, reaching complete inhibition with 250 mM KCl. Other salts, such as potassium nitrate, potassium acetate and sodium chloride, also affected in vitro assembly in a concentration-dependent manner (data not shown). The inhibition of in vitro assembly of NLPs by all these monovalent salts illustrates the fundamental role played by electrostatic forces in the assembly of HCV NLPs.
Figure 2.
Effect of salt on in vitro assembly of C1-82. (A) Kinetics of in vitro assembly of C1-82 (760 pmol) mixed with IRES RNA (38 pmol) in the presence of increasing concentrations of KCl. (B) Different quantities of C1-82 were blotted on a nitrocellulose membrane and incubated with 2.5 μg of radiolabelled IRES in the presence of increasing concentrations of KCl. Radioactivity associated with each dot was analyzed using a Phosphoimager (Typhoon 9200).
It is likely that this inhibition is caused by neutralization of positively charged residues found in C1-82 that are involved in the interaction with nucleic acids. To test this hypothesis, we performed a northwestern dot blot assay using different concentrations of C1-82 proteins incubated with radiolabelled IRES in different salt concentrations. As expected, we observed that increasing the salt concentration efficiently reduced the interaction between the C1-82 protein and the IRES (Fig. 2B). As proposed by van der Schoot and Bruinsma [9], electrostatic and nonspecific attractions between RNA (negatively charged) and protein (positively charged) provide the dynamic driving force for viral assembly.
Influence of nucleic acid concentration and species on in vitro assembly of HCV NLPs
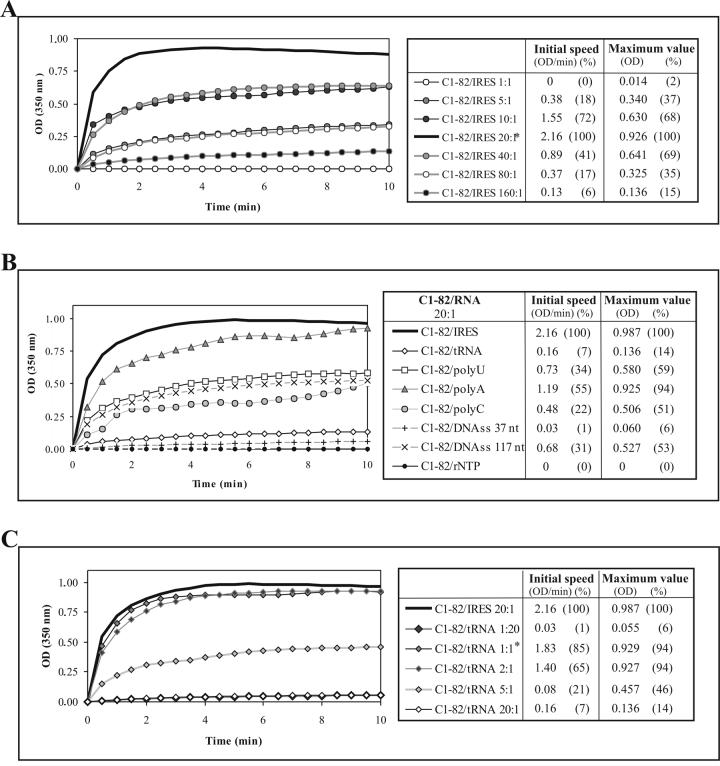
The optimal ratio of C1-82 to IRES for in vitro assembly was evaluated. As shown in Fig. 3A, the kinetics of assembly were optimal when a protein:RNA molar ratio of 20:1 was maintained. With higher amounts of proteins (ratios 40:1, 80:1 and 160:1) or RNA (ratios 1:1, 5:1 and 10:1) assembly was less efficient. These results are in accordance with a previous report [10] in which a similar optimized ratio for in vitro formation of NLPs with truncated core protein (1-124) was observed.
Figure 3.
Influence of nucleic acid species on in vitro assembly of C1-82. (A), (B), (C) Kinetics of in vitro assembly of C1-82 (760 pmol) mixed with different concentrations of IRES RNA (A), different RNAs (38 pmol) to maintain a protein:RNA ratio of 20:1 (B), or different concentrations of tRNA (C). * indicates the optimum ratio for each RNA.
We next examined the capacity of different templates to induce in vitro assembly. The following templates were tested: ribonucleotides (rNTP), structured yeast tRNA, unstructured polynucleotides [200-260mer of polyuridylic acids (polyU), 405-660mer of polyadenylic acid (polyA), 290-434mer of polycytidylic (polyC)], and single-stranded DNA of various sizes (ssDNA: 117 nts, and 37nts) (Fig. 3B). In all cases, we maintained the same protein:RNA ratio (20:1) that had previously shown to be optimal with the IRES. All the RNA types tested, and even ssDNA could initiate NLP assembly. Neither monovalent cations (potassium, sodium) in any concentration (data not shown) nor ribonucleotides can act as nucleation agents, as previously described for viruses of the Bromoviridae family [23]. Electron microscopy analysis of tRNA NLPs and polynucleotide NLPs revealed that they were similar in size and shape to IRES NLPs (data not shown). Kunkel et al. [10] proposed that structured RNA is required for NLP formation. We showed that ssRNA can be a substrate for formation of NLP but structured RNA appeared to be more efficient to trigger the assembly process.
Although tRNA has previously been shown to be efficient in triggering in vitro assembly of HCV core [10, 18], we found it to be less efficient than the IRES in triggering the assembly process in vitro at a ratio of 20:1 (Fig. 3C); we found a ratio of 1:1 to be optimal for this RNA. Therefore, if only 5 IRES (373 nucleotides) molecules are necessary for 100 C1-82 molecules, 100 tRNAs of 75 nucleotides are necessary for optimal assembly for the same amount of C1-82 protein. This result suggests that the size of the RNA is critical in the initiation of assembly. In summary, for a given number of C1-82 subunits, the optimal number of RNA molecules necessary to complete assembly decreases with the length of the RNA. A similar observation was made with alphavirus nucleocapsid protein [21], where the optimal molar ratio of protein/RNA varies with length of RNA used for assembly. The initial speed constant of the optimal protein/tRNA assay was lower than that observed for the IRES RNA. It is possible that the protein has a lower affinity for tRNA than for IRES. Addition of structured or unstructured RNA, but not salt or ribonucleotides, permits electrostatic repulsion between core subunits to be overcome, allowing NLP assembly.
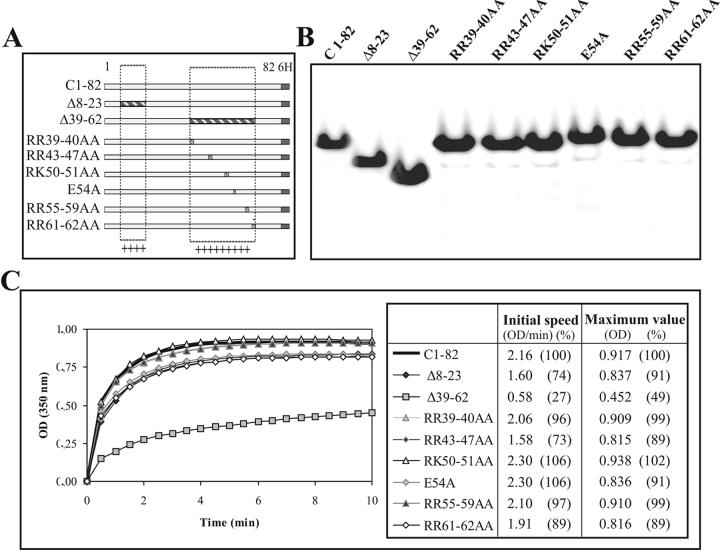
Identification of basic residues critical for in vitro assembly of HCV NLPs
We showed that salt can have a detrimental effect on in vitro assembly, which suggests that charged residues of the protein are important in the recognition of the RNA to trigger the assembly process. To further investigate the importance of charged residues in NLP formation in vitro, we introduced mutations in the two positively charged regions (8-23, 39-62) of C1-82, which are separated by a glycine-rich domain (Fig. 4A). We deleted each of these regions individually and determined the impact on assembly. The recombinant proteins were expressed and purified from E. coli as before (Fig. 4B). As shown in figure 4C, deletion of the 8-23 fragment had little effect on NLP formation if we consider only the maximum value obtained for this mutant. However, the initial speed was affected, showing a decrease of 26% as compared to C1-82. This suggests that deletion in the first charged amino acid cluster can impair the initial rate of particle formation but not the multimerisation process itself. In contrast, deletion of residues 39-62 drastically affected the kinetics of assembly, reducing the maximum value by half and the initial speed by a factor of four (Fig. 4C). This result is consistent with the previously reported observation that particle formation is decreased with a Δ39-64 mutant in a cell-free in vitro assembly system [24]. To investigate if specific regions in the 39-62 fragment are more important for assembly, we mutated several clusters of charged residues. The concentration and purity of the mutant proteins produced were roughly the same for all proteins (Fig. 4B). Most of the mutants were as capable of in vitro assembly as C1-82 (Fig. 4C). However, the mutated proteins RR43-47AA and RR61-62AA showed an 11% decrease in efficiency of NLP formation and decreases of 27% and 11 %, respectively, in the initial speed of assembly (Fig. 4C). None of these specific mutations were as drastic as the Δ39-64 deletion mutant. As no specific region playing a key role in NLP assembly could be identified, it is probably the global charge of the protein that is the driving force for multimerization and the assembly process.
Figure 4.
Role of positively charged residues on in vitro assembly of C1-82. (A) Schematic representation of C1-82 and mutated derivatives. The boxed regions represent the two positively charged regions (8-23, 39-62), which are separated by a glycine-rich domain. The deleted regions in Δ8-23 and Δ39-62 are shaded, and the position of amino acid substitutions in the 39-62 stretch in the other constructs is indicated. (B) SDS-PAGE of purified C1-82 and mutated derivatives. (C) Kinetics of in vitro assembly of the mutants (760 pmol) mixed with IRES RNA (38 pmol) (20:1 ratio).
Inhibition of in vitro assembly of HCV NLPs
The core protein is one of the most conserved proteins of all HCV viral genotypes sequenced so far. For this reason, it is an excellent target for the development of viral inhibitors. Recently, based on the same rationale, peptides inhibiting the in vitro assembly of the human immunodeficiency virus (HIV-1) have been developed [25]. Although the efficacy of such peptides has not yet been validated in vivo, they were shown to be a good template for the development of assembly inhibitors.
We selected 11 peptides of 18 amino acids in length, corresponding to fragments of the HCV core protein, and tested each independently for the ability to inhibit NLP assembly in vitro. The peptide was added to the IRES prior to the addition of the protein, to give a final molar ratio of C1-82/peptide/IRES of 20:1000:1. The influence of the different peptides on assembly varied (Fig. 5A). In fact, 4 of the 11 peptides tested, 15-32, 43-60, 64-81, and 71-88, had no affect on in vitro assembly. Five peptides, 1-18, 8-25, 22-39, 29-46, 36-53, partially inhibited assembly by less than 50 %, and two peptides, 50-67 and 57-74, reduced assembly by a factor of 4.5- and 3.3-fold, respectively. Interestingly, the two most efficient inhibitory peptides include amino acids R61-R62 and are rich in basic residues, again highlighting the important role of electrostatic forces in HCV NLP assembly. The peptides most likely inhibit assembly by interacting with the IRES, thus interfering with C1-82 binding to RNA. To test this hypothesis, we performed a gel retardation assay with IRES RNA and the peptides used in the assembly assay (Fig. 5B). As expected, peptides 50-67 and 57-74 bound the IRES very efficiently. This result suggests that the positively charged residues of these peptides play an important role in inhibition of assembly. However, the number of basic residues in peptides 50-67 and 57-74 are not the sole determinant for the inhibition since other basic peptides such as 1-18, 8-25 and 36-53 failed to inhibit the assembly process despite the fact that they could interact efficiently with the IRES and induce a shift of the RNA in the gel retardation assay (Fig. 5B).
Peptide 22-39 was the most efficient in affecting the initial speed of assembly. This peptide was derived from the hydrophobic glycine-rich cluster (25-38) located between the two basic clusters. This peptide does not interact with nucleic acids (Fig. 5C), but can efficiently inhibit in vitro assembly by a factor of 2. Peptides 15-32 and 29-46, each comprising only one-half of the hydrophobic domain, did not affect in vitro NLP assembly. A β-sheet structure was predicted in the region 29 to 38, which overlaps with the sequence of the inhibitory peptide (22-39) [22]. Also, β-sheet in the HCV core 1-120 were recently showed to be important for the interaction between the subunits [26]. Therefore, the peptide (22-39) probably interferes with the subunit interactions by binding on the predicted β-sheet structure.
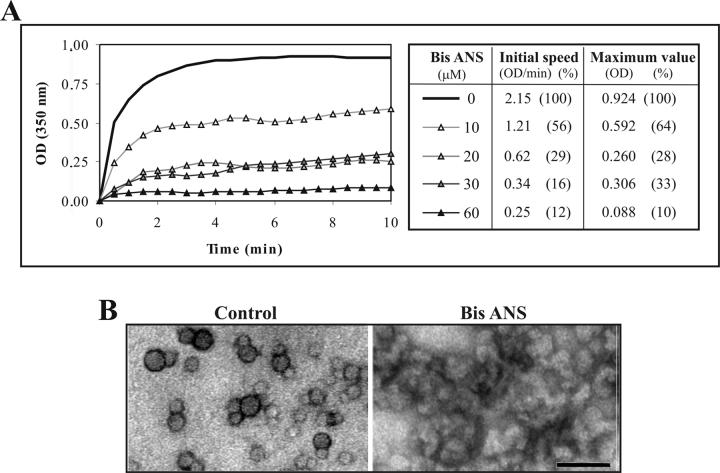
Since the hydrophobic region of core appears to be the most promising target for inhibition of assembly, we next tested chemicals known to interact with hydrophobic clusters. Bis-ANS, a fluorescent probe that binds hydrophobic residues on protein, has been shown to inhibit in vitro HBV capsid assembly [27]. We showed that this compound also inhibits C1-82 assembly in vitro in a dose-dependent manner (Fig. 6A). As little as 20 μM of Bis-ANS reduced both the initial speed and global in vitro assembly by more than 3 fold. Bis-ANS clearly interfered with assembly and led to formation of non-specific aggregates that could be observed under the EM (Fig. 6B).
Figure 6.
Inhibition by Bis-ANS of in vitro assembly of C1-82. (A) Kinetics of in vitro assembly of C1-82 (760 pmol) mixed with IRES RNA (38 pmol) in the presence of increasing amounts of Bis-ANS. (B) Electron micrograph of negatively stained NLPs harvested following in vitro assembly. Left panel, control NLPs; Right panel: In vitro assembly in the presence of 60 μM of Bis-ANS. Bar =50 nm.
It is not clear if inhibitors (Bis-ANS or peptides) will efficiently inhibit assembly using in vivo systems. Bis-ANS was shown to trigger detrimental secondary effects in cells and is therefore not suitable for therapy in its present form [27]. However, the inhibitory effects of this compound confirm that targeting hydrophobic clusters of the core is relevant to the development of assembly inhibitors for HCV NLPs. Peptide inhibitors are small peptides that usually have a short half-life and poor cell permeability. However, peptides could be a valuable starting point for drug design of assembly inhibitors.
In conclusion, we have developed a simple, robust and efficient method with which to perform extensive studies of HCV nucleocapsid assembly in vitro. We showed that electrostatic interactions with the RNA template govern NLP formation in vitro and we have identified a region of the protein (22-39) that could be important for capsid assembly. This method could be expanded and used to perform large-scale screening of assembly inhibitors such as inhibiting peptides or banks of chemicals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Thomson BJ, Finch RG. Hepatitis C virus infection. Clinical Microbiology and Infection. 2005;11:86–94. doi: 10.1111/j.1469-0691.2004.01061.x. [DOI] [PubMed] [Google Scholar]
- [2].Matsuura Y, Miyamura The molecular biology of hepatitis C virus. Semin. Virol. 1993;4:297–304. [Google Scholar]
- [3].Saracco G, Olivero A, Ciancio A, Carenzi S, Rizzetto M. Therapy of chronic hepatitis C: a critical review. Curr Drug Targets Infect Disord. 2003;3:25–32. doi: 10.2174/1568005033342127. [DOI] [PubMed] [Google Scholar]
- [4].McLauchlan J, Lemberg MK, Hope G, Martoglio B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002;21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ogino T, Fukuda H, Imajoh-Ohmi S, Kohara M, Nomoto A. Membrane Binding Properties and Terminal Residues of the Mature Hepatitis C Virus Capsid Protein in Insect Cells. J. Virol. 2004;78:11766–11777. doi: 10.1128/JVI.78.21.11766-11777.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Suzuki R, Tamura K, Li J, Ishii K, Matsuura Y, Miyamura T, Suzuki T. Ubiquitin-Mediated Degradation of Hepatitis C Virus Core Protein Is Regulated by Processing at Its Carboxyl Terminus. Virology. 2001;280:301–309. doi: 10.1006/viro.2000.0785. [DOI] [PubMed] [Google Scholar]
- [7].Shih C, Chen C, Chen S, Lee Y. Modulation of the trans-suppression activity of hepatitis C virus core protein by phosphorylation. J. Virol. 1995;69:1160–1171. doi: 10.1128/jvi.69.2.1160-1171.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Majeau N, Bolduc M, Duvignaud J-B, Fromentin R, Leclerc D.Effect of cAMP-dependent Protein Kinase A (PKA) on HCV Nucleocapsid Assembly and Degradation Biochemistry and Cell Biology In press 2007 [DOI] [PubMed] [Google Scholar]
- [9].van der Schoot P, Bruinsma R. Electrostatics and the assembly of an RNA virus. Physical Review E (Statistical, Nonlinear, and Soft Matter Physics) 2005;71:061928–061912. doi: 10.1103/PhysRevE.71.061928. [DOI] [PubMed] [Google Scholar]
- [10].Kunkel M, Lorinczi M, Rijnbrand R, Lemon SM, Watowich SJ. Self-Assembly of Nucleocapsid-Like Particles from Recombinant Hepatitis C Virus Core Protein. J. Virol. 2001;75:2119–2129. doi: 10.1128/JVI.75.5.2119-2129.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shimoike T, Mimori S, Tani H, Matsuura Y, Miyamura T. Interaction of Hepatitis C Virus Core Protein with Viral Sense RNA and Suppression of Its Translation. J. Virol. 1999;73:9718–9725. doi: 10.1128/jvi.73.12.9718-9725.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Tanaka Y, Shimoike T, Ishii K, Suzuki R, Suzuki T, Ushijima H, Matsuura Y, Miyamura T. Selective Binding of Hepatitis C Virus Core Protein to Synthetic Oligonucleotides Corresponding to the 5′ Untranslated Region of the Viral Genome. Virology. 2000;270:229–236. doi: 10.1006/viro.2000.0252. [DOI] [PubMed] [Google Scholar]
- [13].Bukh J, Purcell RH, Miller RH. Sequence Analysis of the 5′ Noncoding Region of Hepatitis C Virus. Proceedings of the National Academy of Sciences. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Honda M, Beard MR, Ping L-H, Lemon SM. A Phylogenetically Conserved Stem-Loop Structure at the 5′ Border of the Internal Ribosome Entry Site of Hepatitis C Virus Is Required for Cap-Independent Viral Translation. The Journal of Virology. 1999;73:1165–1174. doi: 10.1128/jvi.73.2.1165-1174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Beales LP, Rowlands DJ, Holzenburg A. The internal ribosome entry site (IRES) of hepatitis C virus visualized by electron microscopy. RNA. 2001;7:661–670. doi: 10.1017/s1355838201001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Piron M, Beguiristain N, Nadal A, Martinez-Salas E, Gomez J. Characterizing the function and structural organization of the 5′ tRNA-like motif within the hepatitis C virus quasispecies. Nucleic Acids Research. 2005;33:1487–1502. doi: 10.1093/nar/gki290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Majeau N, Gagne V, Boivin A, Bolduc M, Majeau J-A, Ouellet D, Leclerc D. The N-terminal half of the core protein of hepatitis C virus is sufficient for nucleocapsid formation. J Gen Virol. 2004;85:971–981. doi: 10.1099/vir.0.79775-0. [DOI] [PubMed] [Google Scholar]
- [19].Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. Journal of Molecular Biology. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- [20].Rijnbrand R, Bredenbeek PJ, Haasnoot PC, Kieft JS, Spaan WJ, Lemon SM. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA. 2001;7:585–597. doi: 10.1017/s1355838201000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Tellinghuisen TL, Hamburger AE, Fisher BR, Ostendorp R, Kuhn RJ. In Vitro Assembly of Alphavirus Cores by Using Nucleocapsid Protein Expressed in Escherichia coli. J. Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Duvignaud J-B, Savard C, Fromentin R, Majeau N, Leclerc D, Gagné SM. Structural studies of the N-terminal half of Hepatitis C virus core protein : HCV-C82, an intrinsically unstructured protein? Personnal communication. 2007 doi: 10.1016/j.bbrc.2008.10.141. [DOI] [PubMed] [Google Scholar]
- [23].Bancroft JB, Hiebert E, Bracker CE. The effects of various polyanions on shell formation of some spherical viruses. Virology. 1969;39:924–930. doi: 10.1016/0042-6822(69)90029-4. [DOI] [PubMed] [Google Scholar]
- [24].Klein K, Dellos S, Lingappa J. Identification of Residues in the Hepatitis C Virus Core Protein That Are Critical for Capsid Assembly in a Cell-Free System. J. Virol. 2005;79:6814–6826. doi: 10.1128/JVI.79.11.6814-6826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sticht J, Humbert M, Findlow S, Bodem J, Muller B, Dietrich U, Werner J, Krausslich H-G. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 2005;12:671–677. doi: 10.1038/nsmb964. [DOI] [PubMed] [Google Scholar]
- [26].Rodriguez-Casado MMPCA. Conformational features of truncated hepatitis C virus core protein in virus-like particles. Biopolymers. 2006;82:334–338. doi: 10.1002/bip.20474. [DOI] [PubMed] [Google Scholar]
- [27].Zlotnick A, Ceres P, Singh S, Johnson JM. A Small Molecule Inhibits and Misdirects Assembly of Hepatitis B Virus Capsids. The Journal of Virology. 2002;76:4848–4854. doi: 10.1128/JVI.76.10.4848-4854.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]