Abstract
The specificity of T cell-mediated immune responses is primarily determined by the interaction between the T cell receptor (TCR) and the antigenic peptide presented by the major histocompatibility complex (MHC) molecules. To refine our understanding of interactions between the TCR and the antigenic peptide of vesicular stomatitis virus (VSV) presented by the class I MHC molecule H-2Kb, we constructed a TCR α chain transgenic mouse in a TCR α-deficient background to define specific structural features in the TCR β chain that are important for the recognition of the VSV/H-2Kb complex. We found that for a given peptide, a peptide-specific, highly conserved amino acid could always be identified at position 98 of the complementarity-determining region 3 (CDR3) loop of TCR β chains. Further, we demonstrated that substitutions at position 6, but not position 1, of the VSV peptide induced compensatory changes in the TCR in both the amino acid residue at position 98 and the length of the CDR3β loop. We conclude that the amino acid residue at position 98 of the CDR3β loop is a key residue that plays a critical role in determining the specificity of TCR–VSV/H-2Kb interactions and that a specific length of the CDR3β loop is required to facilitate such interactions. Further, these findings suggest that the α and β chains of TCRs interact with amino acid residue(s) toward the N and C termini of the VSV peptide, respectively, providing functional evidence for the orientation of a TCR with its peptide/MHC ligand as observed in the crystal structures of TCR/peptide/MHC complexes.
One of the central events in determining the specificity of T cell-mediated cellular immune responses is the interaction between the αβ T cell receptor (TCR) and the major histocompatibility complex (MHC)/peptide complex. To generate the TCR diversity needed for antigen recognition, sequences encoding TCR α and β chain variable domains are formed through DNA rearrangement, a process that fuses the variable (V), diversity (D, β only), and joining (J) gene segments of TCR α and β loci together (1, 2). The complementarity-determining region 1 (CDR1, 6 amino acids in length) and CDR2 (8–12 amino acids) stretches of TCR α and β chains are encoded by germ-line V gene segments, whereas the CDR3 loops (6–13 amino acids) are encoded by the V(D)J junctions derived from different germ-line V, D, and J segments and N-additions. Due to the absence of somatic hypermutation in TCR genes, such a mechanism limits the diversity of the CDR1 and CDR2 loops to the sequences present in the germ line and makes the CDR3 loops the most diverse regions in TCR chains (1). The striking concentration of TCR diversity in the CDR3 loops has led to the proposal that the CDR3 loops of TCRs interact with antigenic peptides in the T cell recognition of the peptide/MHC complex (1, 3, 4). The crystal structures of TCR/peptide/MHC complexes support such a proposal (5, 6).
A number of structure–function studies have been carried out to test in vivo the proposed CDR3–peptide interaction (7–11). Using an approach analogous to the classical genetic technique of second-site suppression, several groups showed that alterations at the TCR-contacting residues of antigenic peptides presented by class II MHC molecules elicited compensatory changes in the CDR3 loop of the TCR α or β chain (7, 8, 10). These results indicated that there are specific interactions between the CDR3 loops of TCRs and the antigenic peptide presented by class II MHC molecules. On the other hand, similar studies performed in the class I MHC system to date did not find interactions between specific amino acid residue(s) within the CDR3 loops and the peptides presented by class I MHC molecules, although the studies supported the idea that the CDR3 loops of TCRs were the major determinants for recognizing the antigenic peptides presented by class I MHC molecules (9, 11).
Although the solution of the crystal structure of TCR/peptide/MHC complexes has provided a general outline of interactions between TCRs and their cognate ligands, in vivo functional studies are needed to define specific structural features of TCR–peptide interactions important for the recognition of the peptide/MHC complex. We have developed a model system for analyzing the specific features of TCRs of cytotoxic T lymphocytes (CTLs) that recognize the antigenic peptide (amino acid residues 52–59 of the nucleoprotein, RGYVYQGL) of vesicular stomatitis virus (VSV) presented by the class I MHC molecule H-2Kb. From the crystal structure of the VSV peptide/H-2Kb complex (12, 13) as well as functional studies (14), we identified amino acid residues at positions 1, 4, and 6 of the VSV peptide as potential TCR-contacting residues. Further, in a previous study (15), we found that the Vβ13 gene segment was used by 8 of 12 VSV-specific T cell clones, suggesting that Vβ13 plays an important role in the T cell recognition of the VSV peptide/H-2Kb complex. However, as different Vα gene segments were employed in these Vβ13+ VSV-specific CTL clones and there was a high level of diversity in the CDR3 loops of TCR α and β chains (15), it was difficult to rigorously establish a relationship between the structure of the CDR3 loops of TCRs and the VSV peptide bound to the H-2Kb molecule. These results impelled us to take a different approach to analyze the TCR–VSV peptide interaction.
In this study, we generated TCR α chain transgenic mice in a TCR α-deficient background and used these mice to further analyze the interaction between the TCR β chain and the VSV peptide. The use of a TCR α-deficient background ensured that all mature T cells from our transgenic mice express identical TCR α chains (the introduced TCR α chain transgene) on their surfaces. When these mice were immunized with VSV peptide variants altered at potential TCR-contacting residues identified in our previous studies (13–15), the sequences of the TCR β chains of T cells proliferating in response to the altered peptides could then be analyzed to reveal changes that compensate for the altered peptide residue. It was found that substitution at position 6 of the VSV peptide induced compensatory changes in the CDR3β loops of TCRs from responding T cells. These included peptide-specific, reciprocal charge changes in the amino acid residue at position 98 of the CDR3β loops and alterations in the length (i.e., the number of amino acids) of the CDR3β loops. These findings allow us to conclude that the amino acid residue at position 98 of the TCR CDR3β loop is a key residue that plays a critical role in determining the specificity of the interaction between the TCR β chain and the antigenic peptides presented by the class I MHC molecule, and that a specific CDR3β length is required to facilitate the interaction between a TCR and its ligand.
MATERIALS AND METHODS
Preparation of mRNA and cDNA.
mRNA was extracted from 106 cells using the Oligotex Direct mRNA Kit (Qiagen). Single-strand cDNA was made by reverse transcription of mRNA using Moloney murine leukemia virus reverse transcriptase and oligo(dT)15 as primer. Double-strand cDNA was obtained by PCR amplification of single-strand cDNA using Pfu DNA polymerase (Stratagene) and the specific PCR primers (see below).
Construction of TCR α Chain Transgene.
The TCR α chain transgene was constructed by inserting the double-strand cDNA of the VJ segment of the TCR α chain (Vα2) of the VSV peptide-specific CTL clone N30.7 (15) into the TCR α chain shuttle vector (16), kindly provided to us by Mark Davis (Stanford University). Two specific PCR primers for generating the double-strand cDNA of the VJ segment were 5′ primer, aaactcgagacctgtgtggataaaaacctctctgattctggtttgcttttctgtttccaagcagtacaggagaaacgtgaccagcg; and 3′ primer, aaaattgcggccgcttgggcccaagaaactgtcatcaaaacgtactggggctgactgataccgtggt. The underlined nucleotides in the 5′ and 3′ primers represent restriction sites for XhoI and NotI, respectively.
Generation of TCR α Chain Transgenic Mice.
TCR α chain (Vα2) transgenic mice were generated by injecting purified TCR α chain transgene DNA into F2 embryos of CBA and C57BL/6 mice. Mice carrying the Vα2 transgene were identified by Southern analysis of mouse tail DNA. To generate TCR α chain transgenic mice in a TCR α-deficient background, the Vα2 transgenic mice were crossed for two generations with TCR α-deficient mice (17) purchased from the Jackson Laboratory. Southern analysis of mouse tail DNA was used to identify mice that not only carried the introduced Vα2 transgene but also were homozygous for the disrupted TCR Cα allele. These mice are referred to as TCRα−/−TgVα2+ mice.
Southern Analysis of Mouse Tail DNA.
Mouse tail DNA was digested with BamHI, electrophoresed through 0.7% agarose gels, and transferred to Hybond-N membranes (Amersham). The hybridization probe was prepared by PCR using the TCR α transgene construct as template and the following primers: 5′ primer, ctggctagtccagagagttcc; 3′ primer, ggccccattgctcttggaatc, and labeled with 32P using Rediprime DNA labeling system (Amersham). Rapid-hyb buffer (Amersham) was used in all hybridizations, and the most stringent wash was performed in 0.3× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7)/0.1% SDS at 65°C.
Generation of Peptide-Specific CTLs.
Peptides were synthesized and purified to >98% homogeneity by HPLC before being used for immunization. TCRα−/−TgVα2+ mice were immunized in their hind footpads with 15 μg of peptide emulsified in complete Freund’s adjuvant and boosted 1 week later with 15 μg of peptide emulsified in incomplete Freund’s adjuvant. One week after boost, spleen cells were cultured at 5 × 106/ml with 1 μM peptide in 10 ml of Iscove’s MDM supplemented with 10% heat-inactivated fetal bovine serum (HyClone) at 37°C under 9% CO2 in air. Cytolytic assay, flow cytometry, and cell sorting were performed after 5–6 days of culture.
Cytolytic Assay.
CTL activity was measured in a 4-h 51Cr release assay as described (18). The percentage of specific lysis was calculated as [(experimental release-spontaneous release)/(total release-spontaneous release)] × 100.
Fluorescence-Activated Cell Sorter (FACS) Analysis and Cell Sorting.
To detect surface expression of the Vα2 transgene, lymph node cells of 6- to 8-week-old mice were stained with anti-Vα2 (B20.1) and anti-pan TCR (H57–597) antibodies for 30 min at 4°C. To determine the usage of different Vβ families in peptide-specific CTLs, CTLs were stained with anti-CD8 (53–6.7) and a specific anti-Vβ antibody. To obtain pure populations of CD8+Vα2+ T cells, peptide-specific CTLs were stained with antibodies against Vα2 and CD8. All antibodies were purchased from PharMingen. FACS analysis and cell sorting of stained cells were performed on a FACScan and a FACStar (Becton Dickinson), respectively.
Analysis of TCR CDR3β Sequences.
Double-strand cDNA of TCR CDR3β loops of CD8+Vα2+ cells was prepared as described above. PCR primers used to generate the double-strand cDNA were Vβ7-specific primer, tacagggtctcacggaagaagc; Vβ13-specific primer, aggcctaaaggagctaactccac; Cβ-specific primer, cactgatgttctgtgtgacag. PCR products were cloned into pCR2.1 (Invitrogen), and the TCR CDR3β sequences in the resulting plasmids were determined by DNA sequencing.
RESULTS
Generation of TCR α Chain Transgenic Mice in a TCR α-Deficient Background.
To analyze interactions between the TCR β chain and the VSV peptide presented by the H-2Kb molecule, the TCR α chain (Vα2) from the VSV-specific CTL clone N30.7 (15) was used to generate TCR α chain transgenic mice. This particular TCR α chain was selected for two reasons. First, it is paired with a Vβ13 TCR β chain. It is significant that Vβ13 is used by 75% of VSV-specific CTL clones generated from C57BL/6 mice and is believed to play an important role in the T cell recognition of the H-2Kb/VSV peptide complex (15). Second, anti-Vα2 antibodies are available for detecting the cell surface expression of the Vα2 transgene. To avoid the ambiguity existing in the traditional TCR α chain transgenic mice, where T cells can potentially express either the introduced TCR α transgene or an endogenous TCR α gene or both, we crossed our Vα2 transgenic mice with TCR α chain-deficient mice (17). Due to the targeted disruption of the first exon of the endogenous TCR Cα alleles, T cells from the resulting mice can only express the introduced Vα2 transgene on their surfaces. Accordingly, FACS analysis showed that all peripheral T cells of TCRα−/−TgVα2+ mice expressed the introduced Vα2 transgene on their surfaces (data not shown). Thus, these mice provide us with an animal model for defining the specific features of the interaction between the VSV peptide and the TCR β chain of specifically induced T cells.
TCR β Chains of VSV Peptide-Specific CTLs Have a Highly Conserved G97(V/T)98 Motif in the CDR3 Loop and a Constant CDR3 Length.
To reveal the features of TCRs interacting with the VSV peptide presented by the H-2Kb molecule, TCRα−/−TgVα2+ mice were immunized with the VSV peptide, and the CDR3β sequences of TCR β chains of VSV peptide-specific CTLs from four different mice were analyzed.
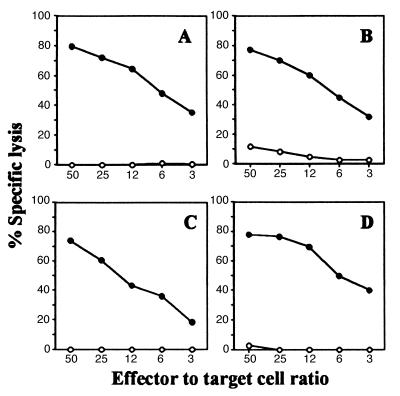
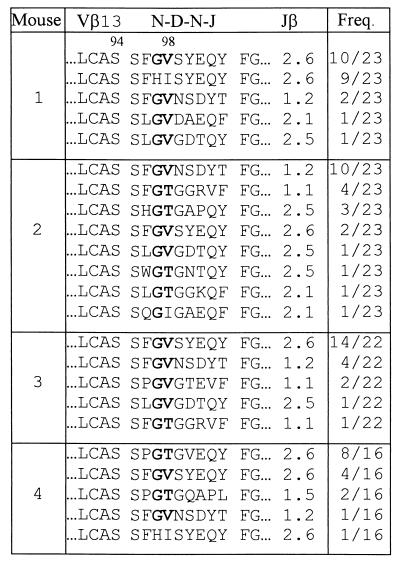
As shown in Fig. 1A, VSV peptide-specific CTLs can be easily induced in TCRα−/−TgVα2+ mice despite the fact that the TCR of every T cell of these mice must use the introduced Vα2 transgenic α chain. This result indicates that endogenous TCR β chains can functionally pair with the introduced Vα2 transgene to recognize and respond to the VSV peptide. FACS analysis using a panel of anti-Vβ antibodies showed that, as in normal C57BL/6 mice (15), Vβ13 is used by more than 80% of VSV peptide-specific CTLs from TCRα−/−TgVα2+ mice (data not shown). Further, analysis of CDR3 loops of TCR β chains revealed two significant structural features (Table 1): 1) almost all the CDR3β loops have a motif of Gly at position 97 and Val or Thr at position 98 [G97(V/T)98], despite the high diversity of amino acid residues at other positions and in the Jβ usage, 2) the length of the CDR3β loop is 9 amino acids in all sequenced TCR β chains when calculated from the amino acid residue following the 94Ser to the amino acid residue preceding the highly conserved Phe-Gly framework (19, 20). Given the fact that these VSV-specific CTLs were generated from four different mice, such data suggest that the VSV peptide presented by the H-2Kb molecule specifically induced T cells that have TCRs with a highly conserved CDR3β loop in terms of the G97(V/T)98 motif and the CDR3 length.
Figure 1.
Induction of peptide-specific CTLs in TCR α−/−Tg Vα2+ mice. TCR α−/−Tg Vα2+ mice were immunized with the VSV peptide (A), the K6 peptide (B), the E6 peptide (C), or the K1 peptide (D). After 1 week in vitro culture, the cytotoxic activity of T cells was measured in a 4-h 51Cr-release assay using RMA/s cells incubated with (•) or without (○) the immunizing peptide as target.
Table 1.
CDR3β sequences of TCRs of VSV peptide-specific CTLs
TCRα−/−TgVα2+ mice were immunized with the VSV (wild-type) peptide, and TCR CDR3β sequences of CTLs were determined after 1 week in vitro culture. The single-letter amino acid code is used for all listed sequences. The conserved amino acid(s) is shown in boldface. Amino acids preceding “LCAS” of Vβ and following the highly conserved “FG” of Jβ are not shown. Freq. (frequency) column shows the number of times each amino acid sequence was obtained and the total number of sequences analyzed from that mouse.
Substitutions at Position 6 of the VSV Peptide Induce Compensatory Changes in Both the Amino Acid Residue at Position 98 and the Length of the CDR3 Loop of TCR β Chains.
Amino acid residues at positions 1, 4, and 6 of the VSV peptide (RGYVYQGL) have been suggested to make contacts with TCRs (12–14), indicating that alterations at these residues may change or abolish the recognition of the altered peptide by VSV-specific CTLs. Interestingly, the VSV-specific CTL clone N30.7, whose TCR α chain was used to generate the TCRα−/−TgVα2+ mice, recognizes peptides with Ala (A) at position 1 or 4 of the VSV peptide (AGYVYQGL or RGYAYQGL) but fails to recognize the peptide with either Ala (A) or Lys (K) at position 6 of the VSV peptide (RGYVYAGL or RGYVYKGL) (15). This suggests that for this TCR the amino acid residue at position 6 of the VSV peptide could be the key residue determining the specificity for interaction with the corresponding TCR.
To determine whether the amino acid residue at position 6 of the VSV peptide interacts with the β chain of TCRs, the K6 peptide (RGYVYKGL) was used to immunize TCRα−/−TgVα2+ mice. Because the K6 peptide cannot be recognized by N30.7 in vitro and all T cells from TCRα−/−TgVα2+ mice have the same TCR α chain as N30.7, the successful induction of the K6-specific CTLs from TCRα−/−TgVα2+ mice (Fig. 1B) strongly suggests that the amino acid residue at position 6 of the VSV peptide interacts with the β chain of TCRs.
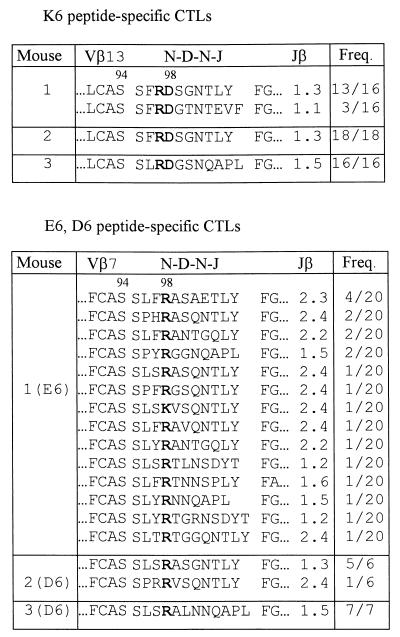
To reveal any changes induced in TCRs of K6 peptide-specific CTLs that can compensate for the alteration (Q → K) at position 6 of the VSV peptide, we analyzed the CDR3β sequences of TCRs of K6 peptide-specific CTLs from three different mice. Because Vβ13 was predominantly used by K6 peptide-specific CTLs from TCRα−/−TgVα2+ mice (data not shown), we focused our analyses on the CDR3β sequences of TCRs of K6-specific, Vβ13+ CTLs. As shown in Table 2, compared with those of VSV peptide-specific CTLs, the CDR3β loops of TCRs of K6 peptide-specific CTLs not only contained a new motif consisting of an Arg (R) at position 97 and an Asp (D) at position 98 (i.e., R97D98 motif), but also had a longer CDR3β loop (10 or 11 amino acids). Taken together with the data obtained from the VSV peptide-specific CTLs (Table 1), these results suggest that the unique motif at positions 97 and 98 of the CDR3β loop as well as the specific CDR3β length of such TCRs are determined by the existence of a particular amino acid at position 6 of the VSV peptide recognized by CTLs.
Table 2.
CDR3β sequences of TCRs of K6, E6, or D6 peptide-specific CTLs
TCRα−/−TgVα2+ mice were immunized with the K6, E6, or D6 peptide, and TCR CDR3β sequences of CTLs were determined after 1 week in vitro culture. The single-letter amino acid code is used for all listed sequences. The conserved amino acid(s) is shown in boldface. Amino acids preceding “LCAS” of Vβ and following the highly conserved “FG” of Jβ are not shown. Freq. (frequency) column shows the number of times each amino acid sequence was obtained and the total number of sequences analyzed from that mouse.
Considering the fact that a negatively charged Asp (D) was used at position 98 of the CDR3β loop for TCRs to interact with the peptide that has a positively charged Lys (K) at position 6, it is likely that there is a specific ionic interaction between these two amino acid residues. To test whether the reciprocal ionic interaction would be found, E6 (RGYVYEGL) and D6 (RGYVYDGL) peptides, which have a negatively charged amino acid at position 6 of the VSV peptide, were used to induce peptide-specific CTLs in TCRα−/−TgVα2+ mice (Fig. 1C), and the CDR3β sequences of peptide-specific CTLs were analyzed. In contrast to the predominant Vβ13 usage (about 80%) in both VSV peptide- and K6 peptide-specific CTLs, we observed that most CD8+ T cells (about 75%) of E6 or D6 peptide-specific CTLs used Vβ7 to pair with the introduced Vα2 (A.M.K., unpublished results). More importantly, it was found that the CDR3β loops of TCRs of E6 or D6 peptide-specific CTLs contained a positively charged Arg (R), or occasionally Lys (K), at position 98 and also contained more amino acids than those of the CDR3β loop of TCRs recognizing the VSV peptide (Table 2). These results indicated that a reciprocal charge change in the amino acid residue at position 98 of the CDR3β loop was made by TCRs of E6 or D6 peptide-specific CTLs to compensate for the alteration (Q → E or Q → D) at position 6 of the VSV peptide. Such results support the suggestion that there is a specific interaction between the amino acid residue at position 6 of the VSV peptide and the amino acid residue at position 98 of the CDR3β loop of TCRs.
Substitutions at Position 1 of the VSV Peptide Do Not Change Either the Conserved G97(V/T)98 Motif or the Length of the CDR3β Loop of TCR β Chains.
To confirm our conclusion that the changes observed in the CDR3β loop of TCRs in response to variant peptides were specifically induced by substitutions at position 6 of the VSV peptide, we next immunized TCRα−/−TgVα2+ mice with peptides altered at a position unlikely to contact the TCR β chain. Several studies have demonstrated that TCRs interact with the peptide/MHC complexes in a diagonal orientation, in which the CDR3 loops of TCR α and β chains map over the N and C termini of the peptide, respectively (5, 6, 10, 21). Accordingly, if the amino acid residue at position 6 of the VSV peptide interacts with the amino acid residue at position 98 of the CDR3β loop of TCRs, as suggested from the above experiments, the amino acid residue at position 1 of the VSV peptide is likely to interact with the CDRα loops of TCRs. Therefore, substitutions at position 1 of the VSV peptide should not induce changes in the CDR3β loop of TCRs. To test this hypothesis, TCRα−/−TgVα2+ mice were immunized with I1 (IGYVYQGL) or K1 (KGYVYQGL) peptides to elicit peptide-specific CTLs. These two particular peptides were selected because they can be recognized by N30.7 in vitro (F.W., unpublished results), thus making it possible for us to obtain I1 or K1 peptide-specific CTLs in TCRα−/−TgVα2+ mice (Fig. 1D) even when the amino acid residue at position 1 of the VSV peptide interacts with the invariable transgenic TCR α chain.
As was the case for the VSV peptide-specific CTLs, Vβ13 was predominantly used by both I1 and K1 peptide-specific CTLs (data not shown). Further, compared with the CDR3β sequences of the VSV peptide-specific CTLs, the CDR3β sequences of TCRs of I1 and K1 peptide-specific CTLs (Table 3) not only contained the same G97(V/T)98 motif but also had the same CDR3 length (9 amino acids), indicating that substitutions at position 1 of the VSV peptide did not induce changes in the CDR3β loop of TCRs. Such results support the argument that the compensatory changes in the CDR3β loop of TCRs induced by substitutions at position 6 of the VSV peptide reflect the specific interaction between the CDR3β loop of TCRs and the amino acid residue at position 6 of the VSV peptide bound to the H-2Kb molecule.
Table 3.
CDR3β sequences of TCRs of I1 or K1 peptide-specific CTLs
TCRα−/−TgVα2+ mice were immunized with the I1 or K1 peptide, and TCR CDR3β sequences of CTLs were determined after 1 week in vitro culture. The single letter amino acid code is used for all listed sequences. The conserved amino acid(s) is shown in boldface. Amino acids preceding “LCAS” of Vβ and following the highly conserved “FG” of Jβ are not shown. Freq. (frequency) column shows the number of times each amino acid sequence was obtained and the total number of sequences analyzed from that mouse.
DISCUSSION
In the present study, we generated TCR α chain transgenic mice in a TCR α-deficient background to define the details of the interaction between the TCR β chain and the VSV peptide presented by the H-2Kb molecule. Two previous findings indicated that such an approach would require the use of mice with a TCR α-deficient background. First, the expression of a functionally rearranged TCR α chain transgene does not prevent the rearrangement and expression of endogenous TCR α chain genes (16, 22–25). Second, two different TCR α chains can be simultaneously expressed on the surface of a T cell (26–28). Thus, T cells from TCR α chain transgenic mice that are not in a TCR α-deficient background can potentially express either the introduced TCR α transgene or an endogenous TCR α gene or both. Such a complicated uncontrolled background of TCR α chain expression makes it very difficult to use such mice to map the peptide-contacting residue(s) in the TCR β chain because the interpretation of the data requires that each TCR β chain be paired with an identical TCR α chain. The use of TCR α chain transgenic mice in a TCR α-deficient background has enabled us to reveal several significant features of the interaction between the TCR β chain and the VSV peptide, summarized in Table 4.
Table 4.
Changes in the CDR3β loop of TCRs induced by substitutions at a residue of the VSV peptide
| Peptide | Peptide sequence | Motif in the CDR3β loop | Length of the CDR3β loop |
|---|---|---|---|
| 9798 | |||
| VSV | RGYVYQGL | G V/T | 9 |
| K6 | -----K-- | R D | 10 (11) |
| E6 | -----E-- | R | 11 (12) |
| D6 | -----D-- | R | 11 (12) |
| I1 | I------- | G V/T | 9 |
| K1 | K------- | G V/T | 9 |
The single-letter code for each amino acid is used. For peptide variants, only residues different from those of the VSV (wild-type) peptide are shown. For peptide variants with substitutions at position 6 of the VSV peptide, the most common CDR3β loop length is listed, with the second most common length shown in parentheses.
Analysis of CDR3β sequences of TCRs that interact with the wild-type VSV peptide showed that there was a highly conserved G97(V/T)98 motif in almost all analyzed CDR3β loops (Table 1). The occurrence of Gly at position 97 is interesting, in that Gly provides maximal main chain flexibility. This may be used to allow the CDR3β loop to access a number of different conformations, which lead to productive binding interaction. The choice of Gly, which lacks a side chain, may also allow for a closer approach between the peptide and the CDR3β loop than would other amino acids. The utilization of either Val or Thr at position 98 of the CDR3β loop suggests that this residue may be involved in packing interactions with the wild-type VSV peptide. As Val and Thr are isosteric, they can participate in very similar types of van der Waals interactions. Further, because both residues are β-branched amino acids, they may be responsible for enforcing a particular conformation of the CDR3β loop. This idea is supported by the observation that the only other residue identified at position 98 of the CDR3β loop is Ile, another β-branched amino acid.
As is the case for the wild-type VSV peptide, a peptide-specific, highly conserved motif can also be identified at position 97 and/or position 98 of the CDR3β loop of TCRs that interact with the K6, E6, and D6 peptides (Table 2). Further, we found that a charged amino acid at position 6 of the VSV peptide always selected an oppositely charged amino acid at position 98 of the CDR3β loop (Table 2). Such results, along with the results obtained from immunization with the wild-type VSV peptide, strongly suggest that there is a specific interaction between the amino acid residue at position 6 of the VSV peptide and the amino acid residue at position 98 of the CDR3β loop.
Jorgensen et al. (8) were the first to use single-chain TCR transgenic mice to map TCR-peptide contacts. Working in a class II MHC system, they demonstrated that charge substitutions in the moth cytochrome c peptide elicited reciprocal changes in the CDR3β loop of TCRs. However, in this system, charge substitutions (T → E or T → K) in the moth cytochrome c peptide induced reciprocal charge changes at several different positions within the CDR3β loop of TCRs. Therefore, the data did not reveal a specific position as a peptide-contacting site. In contrast, working in the class I MHC system, we found that reciprocal charge changes in the CDR3β loop induced by substitutions at position 6 of the VSV peptide always occurred at position 98 of the CDR3β loop. Thus, for the VSV/H-2Kb system, a particular amino acid residue within the TCR CDR3β loop can be identified as a key peptide-interacting residue for mediating the specificity of the interaction between the TCR β chain and the antigenic peptides presented by MHC molecules. The differences between our results and the results of Jorgensen et al. (8) may reflect the fundamental differences in modes of peptide binding and presentation found for the class I and class II systems (29).
In contrast to our results, Turner et al. (11) did not find reciprocal charge changes in the CDR3β loop of TCRs when they immunized TCR α chain transgenic mice with charge variants of the ovalbumin peptide, which is presented by H-2Kb. Given that the same MHC molecule is used in our system and theirs, it is not clear why different results were obtained. One reason may be the difference in the antigenic peptides used in the two studies. In particular, the existence of two adjacent charged amino acid residues in the ovalbumin peptide (SIINFEKL) may prevent the appearance of a simple reciprocal charge change in the CDR3β loop of TCRs that recognize the altered peptides. Alternatively, the different results may reflect the importance of using a TCR α-deficient background to provide the necessary precision required for studying the interaction between the TCR β chain and the antigenic peptide.
Our results suggest that the particular length of the CDR3β loop is important for the recognition of the VSV peptide and its variants. Three observations support this suggestion. First, all sequenced CDR3β loops of TCRs recognizing the VSV peptide were 9 amino acids in length (Table 1). Second, substitutions at position 6 of the VSV peptide not only induced compensatory changes in a specific residue within the CDR3β loop but also increased the length of the CDR3β loop from 9 amino acids to 10–12 amino acids (Table 2). Third, substitutions at position 1 of the VSV peptide, which did not induce compensatory sequence changes in the CDR3β loop, also did not alter the 9-amino acid length of the CDR3β loop (Table 3).
CDR3β loops have a great potential to vary in length due to imprecise V–D and D–J joins and to the random addition of nucleotides during the V–D–J rearrangement. In previous studies, the length of the CDR3β loop was found to vary as a function of the Vβ and Jβ usage (20), and a highly restricted Vβ–Jβ response led to a conserved CDR3β loop length (30). However, these observations cannot be used to explain our results because we found that regardless of the Jβ usage, the length of the CDR3β loops of TCRs that interact with the VSV peptide or its I1 and K1 variants was 9 amino acids in all cases (Tables 1 and 3). Further, we demonstrated that the same CDR3β length can be used by TCRs having different Vβ segments and that TCRs using the same Vβ segment can have different CDR3β lengths. Therefore, it can be concluded that changes in the recombined germline segments are not responsible for the alterations in the CDR3β length observed in our study. Instead, we propose that the CDR3β length alters to properly position the amino acid residue at position 98 of the CDR3β loop so that this key residue can interact with the amino acid residue at position 6 of the VSV peptide. It is interesting that all observed alterations in the CDR3β length were due to the addition of amino acid(s) C-terminal to the conserved residue in the CDR3β loop.
In contrast to substitutions at position 6 of the VSV peptide, substitutions at position 1 of the VSV peptide did not induce changes in the CDR3β loop in terms of the amino acid motif at positions 97 and/or 98 and the length of the CDR3β loop (Table 3). These results provide strong evidence that the amino acid residue at position 1 of the VSV peptide does not interact with the CDR3β loop. Given that position 1 and position 6 of the VSV peptide are at the opposite ends of the VSV peptide, these results suggest that the amino acid residues toward the C terminus of the VSV peptide interact with the CDR3 loop of the TCR β chain, whereas the amino acid residues toward the N terminus of the peptide contact the CDR3 loop of the TCR α chain. Thus, our study provides functional evidence for the current view of the orientation of a TCR with its peptide/MHC ligand.
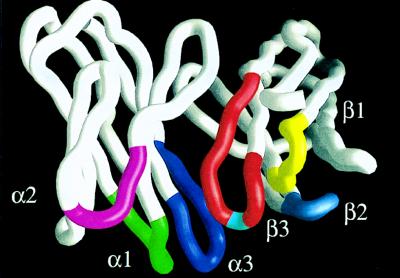
With the solution of the crystal structure of TCR/peptide/MHC complexes, our knowledge of interactions between TCRs and their cognate ligands has been greatly enriched (5, 6). However, in vivo functional studies are needed to reveal the biological importance of the interactions observed in the crystal structures. Here, using TCR α transgenic mice in a TCR α-deficient background as an analytic system, we demonstrated that there was a specific interaction between the amino acid residue at position 98 of the CDR3β loop of TCRs and the amino acid residue at position 6 of the VSV peptide presented by H-2Kb. Consistent with these findings, the crystal structure of a H-2Kb-restricted, VSV peptide-specific TCR (31) showed that the amino acid residue at position 98 of the CDR3β is at the apex of the loop (Fig. 2), indicating that this residue is the most accessible residue in the TCR β chain for contact with the exposed side chain(s) of the peptide presented by an MHC molecule. The crystal structure of two other class I MHC-restricted TCRs (5, 6) showed similar results, even though the length of the CDR3β loop of these TCRs is not the same. Thus, the three-dimensional structures are consistent with the in vivo functional results, suggesting that the amino acid residue at position 98 of the CDR3β loop is a key residue for mediating the specificity of the interaction between the TCR β chain and the antigenic peptides presented by the class I MHC molecule.
Figure 2.
The α carbon trace of Vα and Vβ regions of an H-2Kb-restricted, VSV peptide-specific TCR showing the position of residue 98 in the CDR3β loop. The TCR heterodimer was isolated from the VSV peptide-specific T cell clone N15 (15). CDR loops of the TCR α chain are labeled as α1, α2, and α3. CDR loops of the TCR β chain are labeled as β1, β2, and β3. Within CDR3β loop, the amino acid residue at position 98 is shown in light blue at the apex of the loop; other residues are shown in red. Figure is made with grasp (32).
Acknowledgments
We thank Drs. B. K. Birshtein, M. D. Scharff, S. Almo, A. Davidson, and D. Ostrov for critically reading the manuscript and for their insightful comments. We thank D. Gebhard at the FACS Facility and J. Horner at the Transgenic Mice Facility for their help, C. Thomson for generating computer graphic pictures, and Ms. M. Muranelli for her excellent secretarial work. This work was supported in part by National Institutes of Health Grants R01 AI07289-32, 5T52CA09173-23, and R01AR42533-5. FACS Facility of AECOM Cancer Center is supported by National Cancer Institute Cancer Center Support Grant P30-CA13330.
ABBREVIATIONS
- TCR
T cell receptor
- MHC
major histocompatibility complex
- CDR
complementarity-determining region
- CTL
cytotoxic T lymphocyte
- VSV
vesicular stomatitis virus
- Tg
transgene
- FACS
fluorescence-activated cell sorter
References
- 1.Davis M M, Bjorkman P J. Nature (London) 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Davis M M. Annu Rev Biochem. 1990;59:475–496. doi: 10.1146/annurev.bi.59.070190.002355. [DOI] [PubMed] [Google Scholar]
- 3.Chothia C, Boswell D R, Lesk A M. EMBO J. 1988;7:3745–3755. doi: 10.1002/j.1460-2075.1988.tb03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claverie J M, Prochnicka-Chalufour A, Bougueleret L. Immunol Today. 1989;10:10–14. doi: 10.1016/0167-5699(89)90058-3. [DOI] [PubMed] [Google Scholar]
- 5.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 6.Garcia K C, Degano M, Stanfield R L, Brunmark A, Jackson M R, Peterson P A, Teyton L, Wilson I A. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 7.Hsu B L, Donermeyer D L, Allen P M. J Immunol. 1996;157:2291–2298. [PubMed] [Google Scholar]
- 8.Jorgensen J L, Esser U, de St. Groth B F, Reay P A, Davis M M. Nature (London) 1992;355:224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 9.Kelly J M, Sterry S J, Cose S, Turner S J, Fecondo J, Rodda S, Fink P J, Carbone F R. Eur J Immunol. 1993;23:3318–3326. doi: 10.1002/eji.1830231239. [DOI] [PubMed] [Google Scholar]
- 10.Sant’Angelo D B, Waterbury G, Preston-Hurlburt P, Yoon S T, Medzhitov R, Hong S C, Janeway C A., Jr Immunity. 1996;4:367–376. doi: 10.1016/s1074-7613(00)80250-2. [DOI] [PubMed] [Google Scholar]
- 11.Turner S J, Jameson S C, Carbone F R. J Immunol. 1997;159:2312–2317. [PubMed] [Google Scholar]
- 12.Fremont D H, Matsumura M, Stura E A, Peterson P A, Wilson I A. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 13.Zhang W, Young A C, Imarai M, Nathenson S G, Sacchettini J C. Proc Natl Acad Sci USA. 1992;89:8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata K, Imarai M, van Bleek G M, Joyce S, Nathenson S G. Proc Natl Acad Sci USA. 1992;89:3135–3139. doi: 10.1073/pnas.89.7.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imarai M, Goyarts E C, van Bleek G M, Nathenson S G. Cell Immunol. 1995;160:33–42. doi: 10.1016/0008-8749(95)80006-5. [DOI] [PubMed] [Google Scholar]
- 16.Berg L J, de St. Groth B F, Ivars F, Goodnow C C, Gilfillan S, Garchon H J, Davis M M. Mol Cell Biol. 1988;8:5459–5469. doi: 10.1128/mcb.8.12.5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mombaerts P, Clarke A R, Rudnicki M A, Iacomini J, Itohara S, Lafaille J J, Wang L, Ichikawa Y, Jaenisch R, Hooper M L, et al. Nature (London) 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 18.van Bleek G M, Nathenson S G. Nature (London) 1990;348:213–216. doi: 10.1038/348213a0. [DOI] [PubMed] [Google Scholar]
- 19.Candeias S, Waltzinger C, Benoist C, Mathis D. J Exp Med. 1991;174:989–1000. doi: 10.1084/jem.174.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. Proc Natl Acad Sci USA. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun R, Shepard S E, Geier S S, Thomson C T, Sheil J M, Nathenson S G. Immunity. 1995;3:573–592. doi: 10.1016/1074-7613(95)90128-0. [DOI] [PubMed] [Google Scholar]
- 22.Bluthmann H, Kisielow P, Uematsu Y, Malissen M, Krimpenfort P, Berns A, von Boehmer H, Steinmetz M. Nature (London) 1988;334:156–159. doi: 10.1038/334156a0. [DOI] [PubMed] [Google Scholar]
- 23.Borgulya P, Kishi H, Uematsu Y, von Boehmer H. Cell. 1992;69:529–537. doi: 10.1016/0092-8674(92)90453-j. [DOI] [PubMed] [Google Scholar]
- 24.Brandle D, Muller C, Rulicke T, Hengartner H, Pircher H. Proc Natl Acad Sci USA. 1992;89:9529–9533. doi: 10.1073/pnas.89.20.9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Boehmer H. Annu Rev Immunol. 1990;8:531–556. doi: 10.1146/annurev.iy.08.040190.002531. [DOI] [PubMed] [Google Scholar]
- 26.Heath W R, Carbone F R, Bertolino P, Kelly J, Cose S, Miller J F. Eur J Immunol. 1995;25:1617–1623. doi: 10.1002/eji.1830250622. [DOI] [PubMed] [Google Scholar]
- 27.Malissen M, Trucy J, Letourneur F, Rebai N, Dunn D E, Fitch F W, Hood L, Malissen B. Cell. 1988;55:49–59. doi: 10.1016/0092-8674(88)90008-6. [DOI] [PubMed] [Google Scholar]
- 28.Padovan E, Casorati G, Dellabona P, Meyer S, Brockhaus M, Lanzavecchia A. Science. 1993;262:422–424. doi: 10.1126/science.8211163. [DOI] [PubMed] [Google Scholar]
- 29.Engelhard V H. Annu Rev Immunol. 1994;12:181–207. doi: 10.1146/annurev.iy.12.040194.001145. [DOI] [PubMed] [Google Scholar]
- 30.McHeyzer-Williams M G, Davis M M. Science. 1995;268:106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- 31.Wang J-H, Lim K, Smolyar A, Teng M-K, Liu J-H, Tse A G D, Liu J, Hussey R E, Chishti Y, Thomson C T, Sweet R M, Nathenson S G, Chang H-C, Sacchettini J C, Reinherz E L. EMBO J. 1998;17:10–26. doi: 10.1093/emboj/17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]