Abstract
The recently identified PTEN/MMAC1 gene is a candidate tumor suppressor implicated in multiple tumor types based on mutations or homozygous deletions of the gene in certain human cancers. No studies of PTEN/MMAC1 mRNA or protein expression in cancer cells have been reported, primarily because of significant numbers of normal cells contaminating most tumor samples and because of the lack of antibody reagents. We examined PTEN/MMAC1 in advanced prostate cancer for gene mutations or abnormalities in expression by using a series of recently derived xenografts free of normal human cells and a PTEN/MMAC1-specific antibody. Only 1 of 10 tumors contained a homozygous deletion of PTEN/MMAC1, and no mutations were detected in the entire coding region of the remaining nine xenografts. However, five of these showed reduced or absent PTEN/MMAC1 expression by Northern analysis and reverse transcription–PCR of mRNA. PTEN/MMAC1 mRNA expression was restored in nonexpressing prostate cancer cells by in vitro treatment with the demethylating agent 5-azadeoxycytidine. Alterations in PTEN/MMAC1 expression were confirmed at the protein level by immunoblot analysis, and immunohistochemical studies show that the endogenous wild-type PTEN/MMAC1 protein is localized exclusively in the cytoplasm. These results demonstrate that loss of PTEN/MMAC1 expression occurs frequently in advanced prostate cancer.
PTEN/MMAC1 was identified recently as a candidate tumor suppressor phosphatase gene associated with loss of heterozygosity on chromosome 10q23 (1, 2). Germ-line mutations are found in families with Cowden disease, which is characterized by hamartomas and increased susceptibility to breast and thyroid cancers (3). PTEN/MMAC1 is deleted or mutated in sporadic cases of glioblastoma, breast cancer, kidney cancer, melanoma, and endometrial cancer (1, 2, 4–6). A role in prostate cancer was suggested by the observation of homozygous deletions and point mutations in clinical material (7, 8) and in a limited set of prostate cancer cell lines (1, 2). Of importance, all studies of PTEN/MMAC1 in human cancers reported to date have searched exclusively for mutations or deletions of the gene rather than for abnormalities of mRNA or protein expression; therefore, PTEN/MMAC1 abnormalities may be more frequent.
To address this issue in prostate cancer, we have examined the mutational status and the expression of PTEN/MMAC1 at the mRNA and protein levels in a panel of tumor tissues from patients with advanced stage disease. Normally, examination of primary prostate tumor specimens is complicated by their small size and heterogeneity within tumors and by the presence of contaminating normal stromal cells, which confound the detection of tumor-specific mutations or abnormalities in gene expression (9). Advanced disease samples are particularly difficult to obtain because these patients rarely require surgical procedures. Cell lines are not a useful alternative because prostate cancer cells grow poorly in culture, and only one cell line retains the androgen-sensitive, prostate-specific antigen (PSA)-secreting phenotype typical of the clinical disease (10). We have circumvented these issues through the establishment of xenografts from fresh prostate tumor tissue implanted into severe combined immunodeficient mice (11, 12). Because normal cells are eliminated by serial tumor passages, this strategy enriches for homogenous populations of prostate cancer cells that can be used for molecular studies. Consistent with other tumor types, we found that mutations or deletions of PTEN/MMAC1 occur in a small fraction of advanced prostate cancers; however, expression of PTEN/MMAC1 is lost at the mRNA and protein levels in at least 50% of cases. These findings demonstrate that PTEN/MMAC1 abnormalities occur frequently in advanced prostate cancer and that a major mode of gene inactivation is through loss of expression.
MATERIALS AND METHODS
Prostate Cancer Xenografts.
LAPC-3 and -4 and LuCaP-23 have been described (11, 12). LAPC-9, -12, -14, and -15 and LuCaP-35, -41, and -58 were derived from patients with locally advanced or metastatic prostate cancer by using similar methods and will be described in detail elsewhere. All patient materials were obtained with the approval of the local institutional review boards.
Molecular Studies.
Microsatellite markers D10S215, AFMA086WG9, D10S541, AFM280WE1, and WI-10275 were amplified by PCR from genomic DNA (13). Primers 5′-TTCTGAGGTTATCTTTTTACCACA-3′ (E5A) and 5′-GAAGAGGAAAGGAAAAACATCAA-3′ (E5B) were used to amplify a 300-bp fragment from exon 5 of PTEN/MMAC1. For Southern analysis, 10 μg of genomic DNA isolated from cell lines and xenografts was digested with HindIII, was separated by agarose gel electrophoresis, was transferred to a nylon membrane, and was probed with the 300-bp exon 5 fragment. Single-strand conformation polymorphism was performed on exons 1 through 9 of PTEN/MMAC1 by using intron-based primers as described (8). Controls included wild-type PTEN/MMAC1 and mutant alleles detected previously. Each sample was examined under three different gel electrophoresis conditions.
For Northern analysis, total RNA was isolated from xenografts by using TRIzol (Life Technologies, Gaithersburg, MD) according to the manufacturer’s directions. The Northern filter was probed with a 1.2-kb fragment representing the entire coding region of PTEN/MMAC1. For reverse transcription (RT)–PCR studies, first-strand cDNA was synthesized from total RNA with an oligo(dT) primer and M-MLV reverse transcriptase (Life Technologies). For PCR of human specific PTEN/MMAC1 transcripts, primers 5′-GGACGAACTGGTGTAATGATATG-3′ (RT 5′) and 5′-TCTACTGTTTTTGTGAAGTACAGC-3′ (RT 3′) were used to amplify a 671-bp fragment. Thermal cycling was performed by 35 cycles at 94°C for 1 min, 57.5°C for 1 min, and 72°C for 3 min, followed by a final extension at 72°C for 10 min. PCR for PSA and β-actin was performed as described (11, 14).
For treatment with 5-azadeoxycytidine, LuCaP-35 xenograft cells were placed in tissue culture and grown in RPMI 1640 medium supplemented with 15% fetal calf serum, 5 ng/ml epidermal growth factor, and 1 nM R1881 (NEN). 5-azadeoxycytidine (Sigma) was added to a final concentration of 1 or 2 μM on day 0 and was replenished on day 2. Cells were harvested for RNA isolation on day 4.
Protein Analysis.
Rabbit polyclonal antiserum was generated against a glutathione S-transferase fusion protein containing the carboxyl terminus of PTEN/MMAC1 (amino acids 321–403). Immunoblot analysis was performed by SDS/PAGE using whole cell lysates of minced tumor with PTEN-specific antiserum diluted 1:5,000. Immunohistochemical staining was performed by using modifications of a described immunoperoxidase technique (15). Tissue from xenograft tumors LAPC-4 and -9 was snap frozen in liquid nitrogen and embedded in OCT compound (Sakura Finetek, Torrance, CA). Serial cryostat sections were fixed in acetone and incubated with anti-PTEN antiserum diluted 1:100 or 1:200 in PBS. Antibody localization was performed by using the diaminobenzidene reaction. Controls consisted of substitution of primary antiserum with preimmune serum.
RESULTS
Low Frequency of Mutations or Homozygous Deletions of PTEN/MMAC1 in Advanced Prostate Cancer.
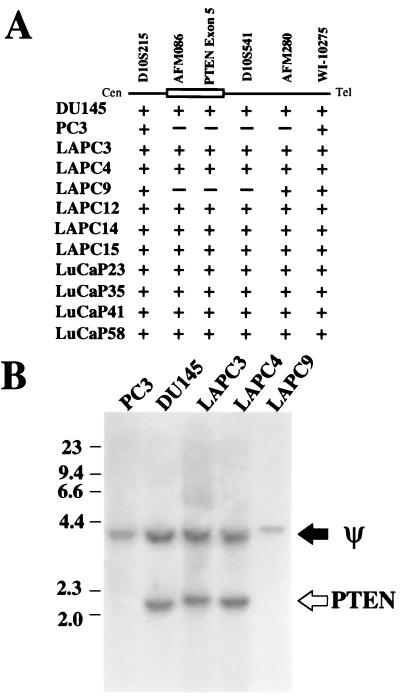
To determine the frequency of homozygous deletion of the PTEN/MMAC1 locus in advanced prostate cancer, genomic DNA extracted from two prostate cancer cell lines and 10 xenografts derived from patients with locally advanced or metastatic disease were examined by using PCR with a series of markers within and adjacent to the PTEN/MMAC1 locus on chromosome 10q23 (Fig. 1A). In agreement with a previous report (1), the PC3 cell line contained a homozygous deletion extending from AFMA086WG9 to AFM280WEI. Only one xenograft, LAPC-9, contained a homozygous deletion in the PTEN/MMAC1 locus. The LAPC-9 deletion is smaller than that found in PC3 because it retains the AFM280 marker. Southern blot analysis with a probe derived from exon 5 of PTEN/MMAC1 confirmed the PCR results. A 2.2-kb band present in DU145, LAPC-3, and LAPC-4 was absent from PC3 and LAPC-9 (Fig. 1B). The 4.0-kb band detected in all lanes represents a pseudogene (Y.E.W., X.W., and C.L.S., unpublished data).
Figure 1.
Homozygous deletion of PTEN/MMAC1. (A) Map of homozygous deletions. The approximate location of the PTEN/MMAC1 gene is shown as an open box. The ordered set of microsatellite markers on chromosome 10q23 are shown above the line. +, the presence of at least one allele as indicated by amplification of the correct size fragment by PCR of genomic DNA; −, the deletion of both alleles as indicated by the absence of the amplified fragment after PCR of genomic DNA. (B) Southern blot analysis. The filter containing HindIII-digested genomic DNA separated by electrophoresis was probed with the PTEN exon 5 probe. The band representing the pseudogene is indicated by ψ. The band representing the functional PTEN/MMAC1 gene is indicated by PTEN.
In those xenografts with an intact PTEN/MMAC1 locus (all except LAPC-9), we searched for mutations in the coding region of the gene by using single-strand conformation polymorphism analysis. Genomic DNA from exons 1 through 9 was amplified by PCR using intron specific primers that flank each exon and have been shown to detect efficiently mutations in the coding region and in splice donor/acceptor sites (8). Control templates from other tumors containing these mutations were included in the analysis to validate the sensitivity of the assay (8). Exons 1 through 9 from all xenografts studied demonstrated a migration pattern in single-strand conformation polymorphism analysis consistent with wild-type sequences. Within the limits of detection by single-strand conformation polymorphism, these data failed to demonstrate mutations in the coding region of PTEN/MMAC1 in all xenografts with an intact PTEN/MMAC1 gene. Taken together with the Southern analysis, homozygous deletion or mutation in exons 1 through 9 of PTEN/MMAC1 occurs infrequently (1 of 10 cases) in advanced prostate cancer.
Abnormal PTEN/MMAC1 mRNA Expression in Prostate Cancer Xenografts.
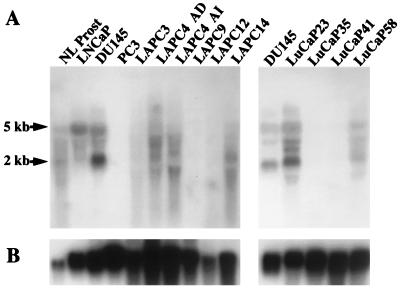
In many cancers, tumor suppressor gene function can be impaired through loss of expression rather than gene deletion (16–20). To determine whether PTEN/MMAC1 expression is down-regulated in prostate cancer xenografts, Northern blot analysis was performed by using a probe spanning the entire coding region of PTEN/MMAC1 (Fig. 2A). Controls for RNA integrity demonstrated equivalent intact ribosomal bands in all lanes (not shown) and expression of glyceraldehyde-3-phosphate dehydrogenase (Fig. 2B). In normal prostate tissue (Fig. 2B, lane 1) and in the human prostate cancer cell lines LNCaP and DU145 (lanes 2–3), two major species migrating at 5 and 2 kb were observed, as were several minor bands, consistent with previous reports (2, 21). As expected, no PTEN/MMAC1 mRNA was detected in PC3 cells or in the LAPC-9 xenograft, both of which contain homozygous deletions of the gene. Surprising to us, the LAPC-3, LAPC-12, LuCaP-35, and LuCaP-41 xenografts all showed reduced or absent PTEN/MMAC1 mRNA expression even though the coding region of each was intact. PTEN/MMAC1 mRNA was expressed in androgen-dependent and androgen-independent sublines of the LAPC-4 xenograft and in LAPC-14, LuCaP-23, and LuCaP-58. The relative abundance of the various PTEN/MMAC1 mRNA isoforms in LAPC-4 and LAPC-14 appeared different from controls. Specifically, the level of the 5-kb transcript was reduced whereas transcripts ranging from ≈2 to 4 kb were increased in intensity, including a 1.5-kb transcript in the androgen-independent subline. The significance of these alternate size transcripts will require detailed characterization of the structure of the various PTEN/MMAC1 mRNA isoforms. At the level of sensitivity of Northern analysis, 5 of 9 xenografts show reduced or absent PTEN/MMAC1 mRNA expression.
Figure 2.
Northern blot analysis of PTEN/MMAC1 expression. (A) Total RNA from indicated cell lines and xenografts was analyzed by Northern blot analysis by using a 1.2-kb PTEN/MMAC1 probe. LAPC-4 AD and LAPC-4 AI represent the androgen-dependent and androgen-independent sublines, respectively. (B) The same Northern filter was stripped and reprobed for expression of glyceraldehyde-3-phosphate dehydrogenase.
Loss of PTEN/MMAC1 mRNA Expression in a Majority of Advanced Prostate Cancers.
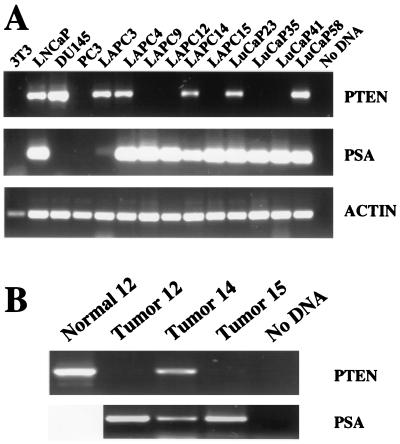
To increase the sensitivity of our analysis of mRNA expression, we developed an RT-PCR assay specific for human PTEN/MMAC1. To avoid detecting the pseudogene or contaminating mouse mRNA, primers (RT 5′ and RT 3′) were designed to amplify only the human PTEN/MMAC1 gene by exploiting mismatches between the human and mouse alleles and the pseudogene. As predicted, a 671-bp product was amplified successfully from LNCaP and DU145 but not murine NIH 3T3 mRNA (Fig. 3A, lanes 1–3). In addition, these primers failed to amplify a product from PC3 mRNA (Fig. 3A, lane 4).
Figure 3.
RT-PCR analysis of PTEN/MMAC1 expression. (A) Expression of PTEN/MMAC1, PSA, and human β-actin in indicated cell lines and xenografts was analyzed by RT-PCR, and ethidium bromide-stained agarose gels are shown. Because the PCR primers for β-actin are directed against the human sequence, the signal in murine NIH 3T3 cells is less pronounced than in other lanes. (B) Expression of PTEN/MMAC1 in peripheral blood leukocytes (normal 12) and PTEN/MMAC1 and PSA in primary prostate tumor tissues before establishment of xenografts was analyzed by RT-PCR. Tumors 12, 14, and 15 were used to establish LAPC-12, 14, and 15, respectively.
Having confirmed the specificity of the RT 5′ and RT 3′ primers, we measured PTEN/MMAC1 expression in the 10 xenografts. The integrity of each RNA sample was confirmed by using primers to the human β-actin gene (Fig. 3A, Bottom). In addition, the prostatic origin of the xenografts was confirmed by expression of PSA (Fig. 3A, Middle). In 5 of 10 xenografts, PTEN/MMAC1 expression was not detected by RT-PCR, consistent with the results from Northern analysis. LAPC-3, which showed no PTEN/MMAC1 expression by Northern blot, was positive by RT-PCR, indicating expression at a reduced level.
For some tumor suppressors, the frequency of gene inactivation may be higher in cell lines than in clinical material (9, 22). We were able to examine this possibility by measuring PTEN/MMAC1 expression in the original tumor tissue in three cases where clinical material was available. The results were essentially identical to those obtained with the corresponding xenografts (Fig. 3B, lanes 2–4), indicating that loss of PTEN/MMAC1 expression is not a consequence of xenograft passaging. The very faint band in tumor 15 is most likely the result of a small fraction of normal tissue contaminating the tumor biopsy material. It is possible that our failure to detect PTEN/MMAC1 mRNA in some cases could be the result of a polymorphism in the sequence recognized by the RT 5′ or RT 3′ primers. We were able to address this issue for LAPC-12 by examining PTEN/MMAC1 expression in noncancerous tissue (peripheral blood leukocytes) that was available from the patient whose tumor gave rise to this xenograft. PTEN/MMAC1 mRNA was present in normal but not xenograft tissue from patient 12 (Fig. 3B, lane 1), indicating that the RT 5′ or RT 3′ primers accurately predict PTEN/MMAC1 expression in this patient as well as LAPC-15 (Fig. 3B, lane 4) and LuCaP-35 (see below; Fig. 4). The combined results from Northern and RT-PCR analyses demonstrate that expression of PTEN/MMAC1 is down-regulated in more than half (6 of 10 tumors) of advanced prostate cancers examined.
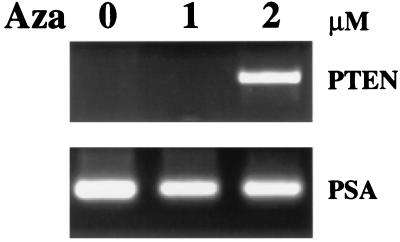
Figure 4.
Restoration of PTEN/MMAC1 expression by 5-azadeoxycytidine. LuCaP-35 xenograft cells cultured in vitro were treated with 5-azadeoxycytidine at a concentration of 0, 1, or 2 μM, and cells were harvested for RNA after 4 days. Expression of PTEN/MMAC1 was detected by RT-PCR and was visualized by ethidium bromide staining. RT-PCR of PSA is shown as a control for the integrity of RNA in all samples.
Restoration of PTEN/MMAC1 mRNA Expression by the Demethylating Agent 5-Azadeoxycytidine.
The failure to detect PTEN/MMAC1 mRNA in a large fraction of prostate cancer xenografts suggests that a major mechanism for loss of function of this tumor suppressor gene is at the level of transcription. One potential explanation is methylation of CG dinucleotides near the region of the promoter or the enhancer, a process recently documented for several tumor suppressors such as the von Hipple Lindau gene and p16/CDKN2 (16–20). Alternative possibilities include mutations in noncoding regions of the mRNA that alter transcript stability or in the genomic locus that render it incompetent for transcription. Until the normal sequence of these regions of the gene is defined, it will be difficult to fully address this issue. However, we were able to examine the possibility of methylation indirectly by using the pharmacological agent 5-azadeoxycytidine, which can demethylate DNA and activate gene transcription. Cells from the LuCaP-35 xenograft can be explanted from the animal and can be propagated short term in tissue culture; therefore, we exposed these cells to 5-azadeoxycytidine by using a protocol developed from studies of other tumor suppressor genes (16–18). In two independent experiments using different cell populations, exposure to 2 μM 5-azadeoxycytidine for 4 days led to reactivation of PTEN/MMAC1 mRNA expression by RT-PCR (Fig. 4). Analogous studies of p16/CDKN2 in prostate cancer cells have shown a similar dose response requiring 2 μM 5-azadeoxycytidine to induce expression of p16/CDKN2 (16). This result demonstrates that the PTEN/MMAC1 locus is intact at the genomic level and is competent for transcription in at least one xenograft and supports the hypothesis that methylation plays a role in transcriptional suppression.
Reduced Levels of PTEN/MMAC1 Protein in Prostate Cancer Xenografts.
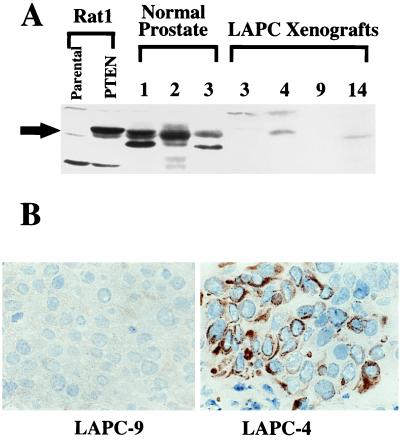
Because mRNA expression of tumor suppressor genes does not reflect necessarily the level of protein expression (23, 24), we examined the level of PTEN/MMAC1 protein in xenografts that expressed the mRNA. First, we prepared polyclonal antisera directed against the carboxyl terminus of the protein by inoculation of rabbits with a glutathione S-transferase PTEN fusion protein. The specificity of the antisera was confirmed by immunoblot analysis of cells engineered to overexpress a cDNA of the coding region of wild-type PTEN/MMAC1. A band migrating at 60 kDa was detected clearly in Rat-1 fibroblasts infected with retrovirus expressing PTEN/MMAC1 (Fig. 5A), which migrated with the same mobility as in vitro-translated protein (data not shown) and was present at levels significantly higher than the endogenous protein in uninfected control cells. Endogenous PTEN/MMAC1 protein expression was detected easily in normal prostate tissue obtained from three individuals. The presence of additional bands migrating faster than full length PTEN/MMAC1 raises the possibility of alternative forms of the protein because of RNA processing, which is consistent with the range of mRNA isoforms observed on Northern analysis (Fig. 2). Other likely explanations include posttranslational modification of the protein or degradation during sample preparation. Further studies using antibodies directed against different PTEN/MMAC1 domains are required to sort out these issues. In the xenografts, expression of PTEN/MMAC1 protein was detected in LAPC-4 and LAPC-14 but not in LAPC-3 in concordance with the relative levels of PTEN/MMAC1 mRNA in these xenografts. As expected, no PTEN/MMAC1 protein was present in LAPC-9 because of homozygous deletion of the gene. These results indicate that the level of PTEN/MMAC1 protein in prostate cancer cells correlates with the level of mRNA expression. The fact that the level of PTEN/MMAC1 protein in normal prostate tissue is higher than in all of the xenografts, despite comparable levels of mRNA, provides further evidence for posttranscriptional or posttranslational regulation.
Figure 5.
PTEN/MMAC1 protein expression. (A) Immunoblot of PTEN/MMAC1 protein. Rabbit polyclonal antibody raised against the C terminus of PTEN/MMAC1 protein was used to probe the whole cell extracts. Rat1-PTEN cells were infected with recombinant retrovirus expressing a cDNA for the PTEN/MMAC1 coding region. Cell extracts from normal prostate tissues from three different individuals (denoted as 1, 2, 3) are shown along with cell extracts from xenografts (LAPC-3, -4, -9, -14). The full length PTEN/MMAC1 protein is indicated by a solid arrow. (B) Immunohistochemical localization of PTEN/MMAC1 in LAPC-4 xenograft tumor (Right) reveals cytoplasmic staining with immunoperoxidase technique (brown). No staining is seen in LAPC-9 (Left) run in parallel on the same slide.
Cytoplasmic Localization of Wild-Type PTEN/MMAC1 Protein.
Knowledge of the cellular localization of signal transduction proteins can provide valuable insight into their functions and substrate preferences. We examined the cellular localization of the endogenous PTEN/MMAC1 phosphatase protein by establishing conditions for immunohistochemical staining of frozen LAPC-4 tumor sections. Consistent with the immunoblot and homozygous deletion data, no staining was observed in LAPC-9 cells (Fig. 5B, Left). In LAPC-4 cells, bright PTEN/MMAC1 staining was observed exclusively in the cytoplasm (Fig. 5B, Right). The amino terminus of PTEN/MMAC1 is homologous to the cytoskeletal protein tensin (1, 2); therefore, this result supports the postulated role of PTEN/MMAC1 as a signal transduction molecule connecting the cytoskeleton to intracellular signaling pathways (1, 2, 21, 25).
DISCUSSION
The initial reports describing the cloning of PTEN/MMAC1 suggested that this phosphatase may play a critical role as a tumor suppressor gene in glioblastoma and breast cancer based on mutations or homozygous deletions of the gene (1, 2). Studies of PTEN/MMAC1 expression at the mRNA or protein level have not been possible because of the presence of normal cells contaminating most tumor samples and a lack of antibody reagents. In this study, we have used xenografts derived from patients with advanced prostate cancer that no longer contained contaminating normal human cells to survey comprehensively the coding region of the PTEN/MMAC1 gene for mutations as well as abnormalities in mRNA expression. We also have developed an antibody to PTEN/MMAC1 to measure protein expression by immunoblot and immunohistochemistry. One tumor contained a homozygous deletion, whereas the remainder had wild-type sequences in all nine coding exons, suggesting that the frequency of PTEN/MMAC1 mutations in advanced prostate cancer is in the range of 10%. Surprisingly, five of the nine xenografts with an intact PTEN/MMAC1 gene showed reduced or absent PTEN/MMAC1 mRNA or protein expression, which was confirmed by analysis of the original patient tumor tissue when available. These results argue that loss of PTEN/MMAC1 expression may occur in as much as half of advanced stage prostate cancers.
Despite the discovery of PTEN/MMAC1 as a tumor suppressor through gene deletion, our data suggest that a primary mode of PTEN/MMAC1 inactivation may be at the level of transcription. There are several potential mechanisms that might explain the loss of mRNA expression. Our ability to restore mRNA expression with the demethylating agent 5-azadeoxycytidine in at least one xenograft argues that the most likely possibility is methylation of CpG islands in the PTEN/MMAC1 genomic locus. Direct proof of this hypothesis will require demonstration of methylated DNA in the PTEN/MMAC1 gene, but interpretation of these experiments is difficult because the promoter and enhancer regions have not yet been defined. The fact that Cairns et al. (7) recently failed to detect methylation of the 5′ region of PTEN/MMAC1 in six clinical prostate cancer samples by using a PCR-based assay (7) demonstrates the inconclusive nature of a negative result. If further studies also fail to show methylation, it is possible that the effect of 5-azadeoxycytidine on PTEN/MMAC1 is mediated indirectly through a second gene, such as a transcription factor that may be the true target of methylation. Whatever the case, it is clear that defining the mechanistic details of transcriptional reactivation of PTEN/MMAC1 by 5-azadeoxycytidine will require more experimentation.
The frequent loss of PTEN/MMAC1 expression suggests that inactivating the function of PTEN/MMAC1 is a critical step in the development or progression of prostate cancer. Because the xenografts and patient material used in this study were from patients with advanced stage disease, it is not yet clear if PTEN/MMAC1 inactivation occurs early or late in disease progression. A recent report examining PTEN/MMAC1 DNA in clinical material suggests that mutations are found in ≈10% of tumors from patients with early or late stage disease and that mutations occur more commonly in metastatic disease (7). Because our results show that PTEN/MMAC1 inactivation occurs frequently at the transcriptional level, early stage clinical material should be reexamined at the level of expression. These studies may be difficult to perform by using RT-PCR because PTEN/MMAC1 is expressed highly by the normal cells that infiltrate these early stage tumors. As indicated by our studies, this problem can be avoided through the use of xenografts, but no early stage prostate cancer xenografts have been propagated successfully to date. It is more likely that antibody reagents such as the one described here that specifically detect the full length protein will be required to define the frequency of gene inactivation in early stage tumors. Because DNA-based surveys of prostate cancer samples fail to detect abnormalities in gene expression, the current frequency of PTEN/MMAC1 gene inactivation is likely to be underestimated.
Having established that PTEN/MMAC1 gene inactivation is a common event in advanced prostate cancer, it will be critical to establish its functional role by analysis of relevant signal transduction pathways and by gene replacement studies. The cytoplasmic location of the protein and its homology to tensin provide an important starting point in the search for physiologically relevant substrates. Our panel of prostate xenografts will be valuable reagents to examine the consequences of gene replacement on prostate cancer growth. Assuming further studies confirm its role as a tumor suppressor, the ability to restore PTEN/MMAC1 expression may have important clinical implications.
Acknowledgments
We thank Arie Belldegrun for assistance in obtaining tissues, Owen Witte for advice, Kent Buhler, Duc Do, Guillermo Rivas, and Peter Shintaku for technical assistance, and Lisa Dove for manuscript preparation. This work was supported by grants from the James S. McDonnell Foundation, the Margaret Early Trust and CaP CURE. C.L.S. is a Scholar of the Leukemia Society of America. Y.E.W. is supported by an American Society of Hematology Scholar Award.
ABBREVIATIONS
- RT
reverse transcription
- PSA
prostate-specific antigen
References
- 1.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 2.Steck P A, Pershouse M A, Jasser S A, Yung W K A, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, et al. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 3.Liaw D, Marsh D J, Li J, Dahia P L M, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, et al. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 4.Guldberg P, thor Straten P, Birck A, Ahrenkiel V, Kirkin A F, Zeuthen J. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 5.Tashiro H, Blazes M S, Wu R, Cho K R, Bose S, Wang S I, Li J, Parsons R, Ellenson L H. Cancer Res. 1997;57:3935–3940. [PubMed] [Google Scholar]
- 6.Kong D, Suzuki A, Zou T, Sakaurada A, Kemp L, Wakatsuki S, Yokoyama T, Yamakawa H, Furukawa T, Sato M, et al. Nat Genet. 1997;17:143–144. doi: 10.1038/ng1097-143. [DOI] [PubMed] [Google Scholar]
- 7.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman J G, Jen J, Isaacs W B, Bova G S, Sidransky D. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 8.Suzuki H, Freije D, Nusskern D R, Okami K, Cairns P, Sidransky D, Isaacs W B, Bova G S. Cancer Res. 1998;58:204–209. [PubMed] [Google Scholar]
- 9.Cairns P, Polascik T J, Eby Y, Tokino K, Califano J, Merlo A, Mao L, Herath J, Jenkins R, Westra W, et al. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 10.Royai R, Lange P H, Vessella R. Semin Oncol. 1996;23:35–40. [PubMed] [Google Scholar]
- 11.Klein K A, Reiter R E, Redula J, Moradi H, Zhu X L, Brothman A R, Lamb D J, Marcelli M, Belldegrun A, Witte O N, et al. Nat Med. 1997;3:402–408. doi: 10.1038/nm0497-402. [DOI] [PubMed] [Google Scholar]
- 12.Ellis W J, Vessella R L, Buhler K R, Bladou F, True L D, Bigler S A, Curtis D, Lange P H. Clin Cancer Res. 1996;2:1039–1048. [PubMed] [Google Scholar]
- 13.Hudson T J, Stein L D, Gerety S S, Ma J, Castle A B, Silva J, Slonim D K, Baptista R, Kruglyak L, Xu S-H, et al. Science. 1995;270:1945–1954. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- 14.Wei Q, Xu X, Cheng L, Legerski R J, Ali–Osman F. Cancer Res. 1995;55:5025–5029. [PubMed] [Google Scholar]
- 15.Said J W, Pinkus J L, Shintaku I P, deVos S, Matsumura F, Yamashiro S, Pinkus G S. Mod Pathol. 1998;11:1–5. [PubMed] [Google Scholar]
- 16.Jarrard D F, Bova G S, Ewing C M, Pin S S, Nguyen S H, Baylin S B, Cairns P, Sidransky D, Herman J G, Isaacs W B. Genes Chromosomes Cancer. 1997;19:90–96. [PubMed] [Google Scholar]
- 17.Herman J G, Latif F, Weng Y, Lerman M I, Zbar B, Liu S, Samid D, Duan D S, Gnarra J R, Linehan W M, et al. Proc Natl Acad Sci USA. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 19.Herman J G, Merlo A, Mao L, Lapidus R G, Issa J P, Davidson N E, Sidransky D, Baylin S B. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 20.Gonzalez–Zulueta M, Bender C M, Yang A S, Nguyen T, Beart R W, Van Tornout J M, Jones P A. Cancer Res. 1995;55:4531–4535. [PubMed] [Google Scholar]
- 21.Li D M, Sun H. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 22.Cairns P, Mao L, Merlo A, Lee D J, Schwab D, Eby Y, Tokino K, van der Riet P, Blaugrund J E, Sidransky D. Science. 1994;265:415–417. doi: 10.1126/science.8023167. [DOI] [PubMed] [Google Scholar]
- 23.Pagano M, Tam S W, Theodoras A M, Beer–Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 24.Loda M, Cukor B, Tam S W, Lavin P, Fiorentino M, Draetta G F, Jessup J M, Pagano M. Nat Med. 1997;3:231–234. doi: 10.1038/nm0297-231. [DOI] [PubMed] [Google Scholar]
- 25.Myers M P, Stolarov J P, Eng C, Li J, Wang S I, Wigler M H, Parsons R, Tonks N K. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]