Abstract
The expression of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II in Escherichia coli converted this organism from the production of organic acids to the production of ethanol. Ethanol was produced during both anaerobic and aerobic growth. The extent to which these ethanologenic enzymes were expressed correlated with the extent of ethanol production. The replacement of organic acids with ethanol as a metabolic product during aerobic and anaerobic growth resulted in dramatic increases in final cell density, indicating that these acids (and the associated decline in pH) are more damaging than the production of ethanol. Of the plasmids examined, the best plasmid for growth and ethanol production expressed pyruvate decarboxylase and alcohol dehydrogenase II at levels of 6.5 and 2.5 IU/mg of total cell protein, respectively.
Full text
PDF

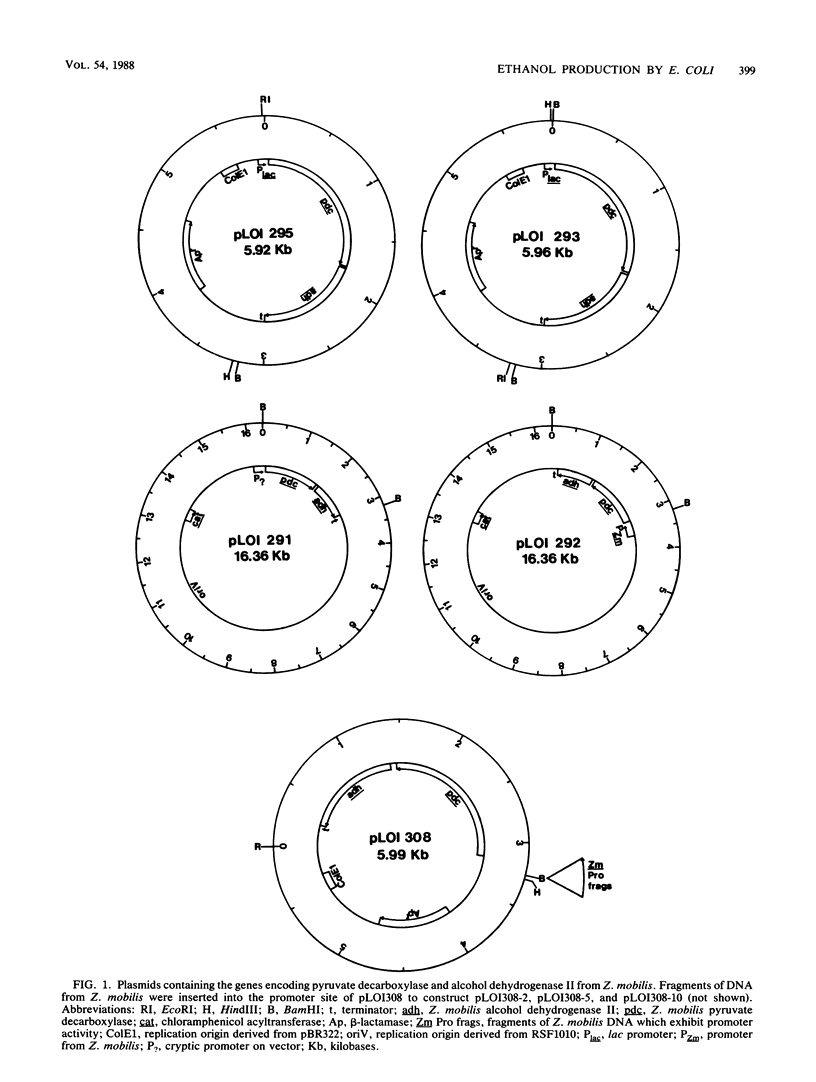
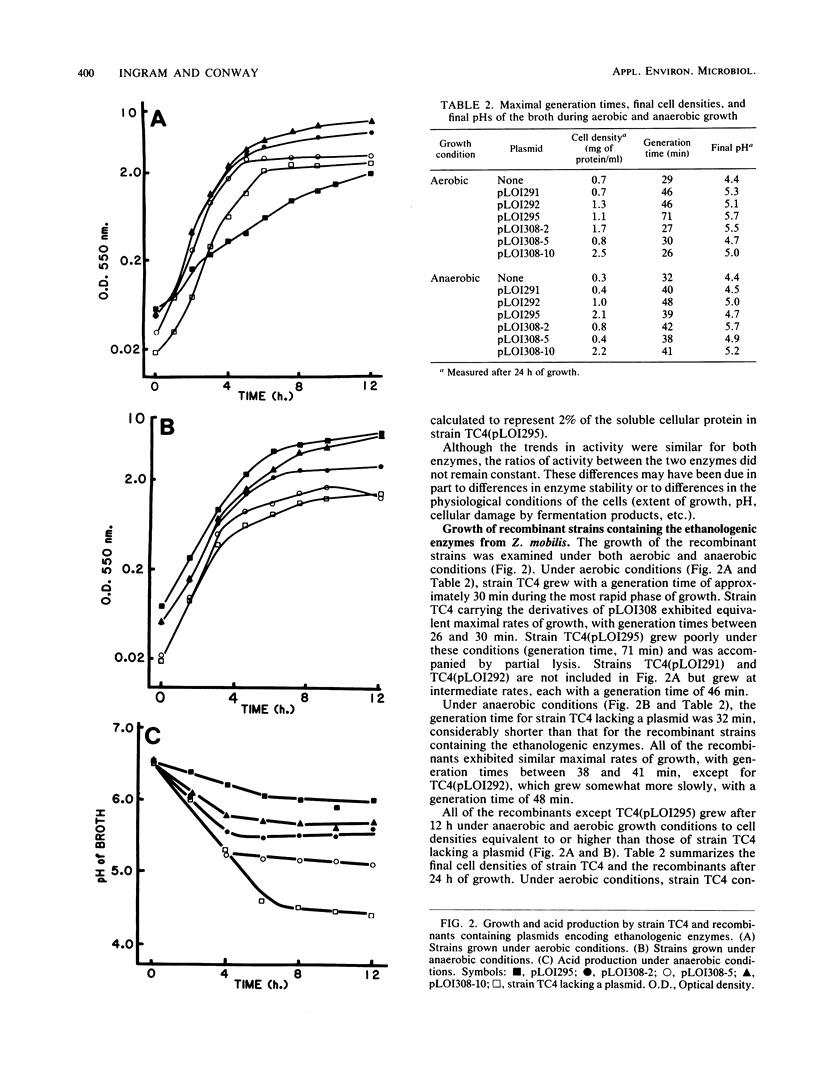
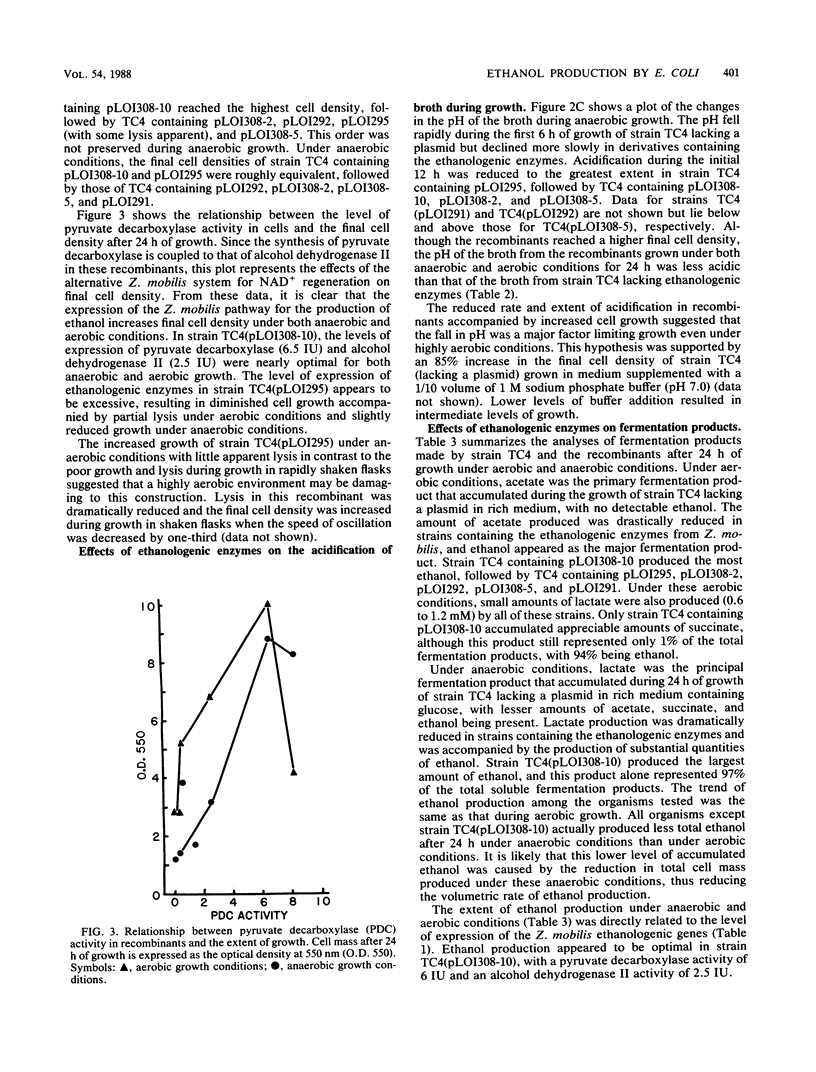



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama S. K., Hammes G. G. Elementary steps in the reaction mechanism of the pyruvate dehydrogenase multienzyme complex from Escherichia coli: kinetics of acetylation and deacetylation. Biochemistry. 1980 Sep 2;19(18):4208–4213. doi: 10.1021/bi00559a011. [DOI] [PubMed] [Google Scholar]
- Clark D., Cronan J. E., Jr Escherichia coli mutants with altered control of alcohol dehydrogenase and nitrate reductase. J Bacteriol. 1980 Jan;141(1):177–183. doi: 10.1128/jb.141.1.177-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Byun M. O., Ingram L. O. Expression Vector for Zymomonas mobilis. Appl Environ Microbiol. 1987 Feb;53(2):235–241. doi: 10.1128/aem.53.2.235-241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney C. L. Bioreactors: design and operation. Science. 1983 Feb 11;219(4585):728–733. doi: 10.1126/science.219.4585.728. [DOI] [PubMed] [Google Scholar]
- Dancey G. F., Shapiro B. M. The NADH dehydrogenase of the respiratory chain of Escherichia coli. II. Kinetics of the purified enzyme and the effects of antibodies elicited against it on membrane-bound and free enzyme. J Biol Chem. 1976 Oct 10;251(19):5921–5928. [PubMed] [Google Scholar]
- Ingram L. O., Conway T., Clark D. P., Sewell G. W., Preston J. F. Genetic engineering of ethanol production in Escherichia coli. Appl Environ Microbiol. 1987 Oct;53(10):2420–2425. doi: 10.1128/aem.53.10.2420-2425.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappe J., Blaschkowski H. P., Gröbner P., Schmitt T. Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur J Biochem. 1974 Dec 16;50(1):253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale A. D., Scopes R. K., Kelly J. M., Wettenhall R. E. The two alcohol dehydrogenases of Zymomonas mobilis. Purification by differential dye ligand chromatography, molecular characterisation and physiological roles. Eur J Biochem. 1986 Jan 2;154(1):119–124. doi: 10.1111/j.1432-1033.1986.tb09366.x. [DOI] [PubMed] [Google Scholar]
- Osman Y. A., Conway T., Bonetti S. J., Ingram L. O. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J Bacteriol. 1987 Aug;169(8):3726–3736. doi: 10.1128/jb.169.8.3726-3736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal M. C., Chippaux M., Abou-Jaoudé A., Blaschkowski H. P., Knappe J. Mutants of Escherichia coli K12 with defects in anaerobic pyruvate metabolism. J Gen Microbiol. 1981 May;124(1):35–42. doi: 10.1099/00221287-124-1-35. [DOI] [PubMed] [Google Scholar]
- Rudolph F. B., Purich D. L., Fromm H. J. Coenzyme A-linked aldehyde dehydrogenase from Escherichia coli. I. Partial purification, properties, and kinetic studies of the enzyme. J Biol Chem. 1968 Nov 10;243(21):5539–5545. [PubMed] [Google Scholar]
- Tarmy E. M., Kaplan N. O. Kinetics of Escherichia coli B D-lactate dehydrogenase and evidence for pyruvate-controlled change in conformation. J Biol Chem. 1968 May 25;243(10):2587–2596. [PubMed] [Google Scholar]
- Thomson J. W., Shapiro B. M. The respiratory chain NADH dehydrogenase of Escherichia coli. Isolation of an NADH:quinone oxidoreductase from membranes and comparison with the membrane-bound NADH:dichlorophenolindophenol oxidoreductase. J Biol Chem. 1981 Mar 25;256(6):3077–3084. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


