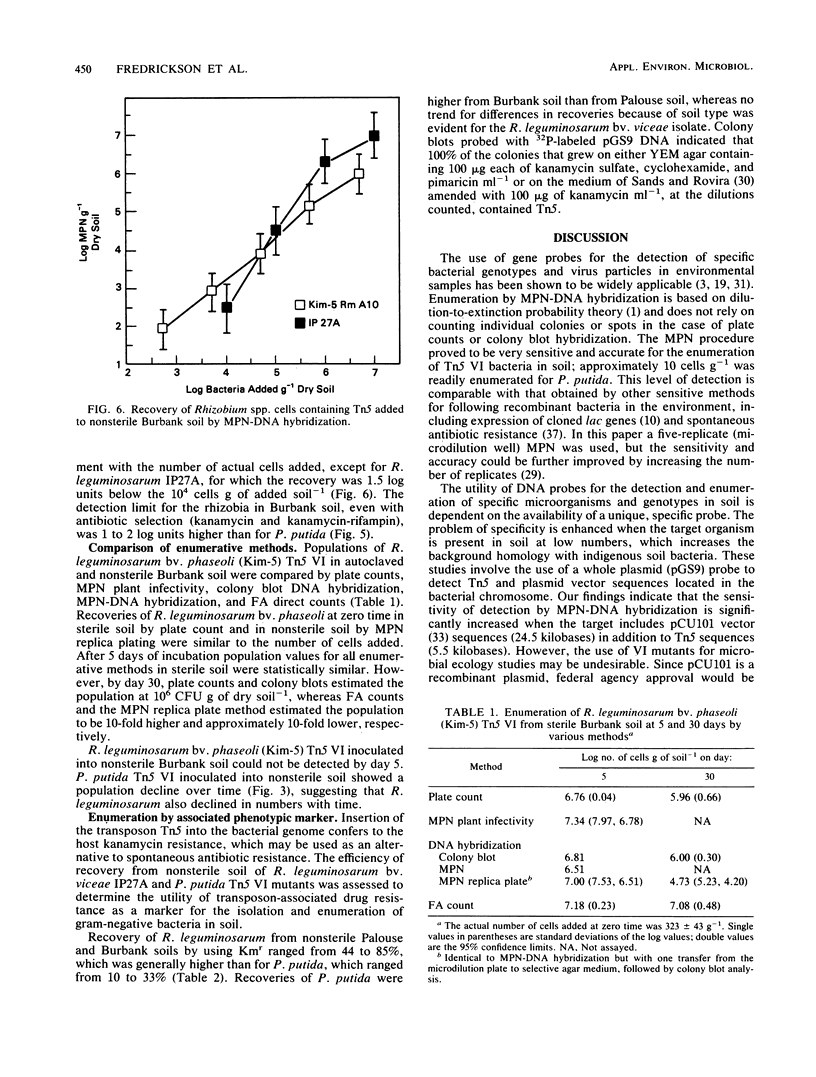
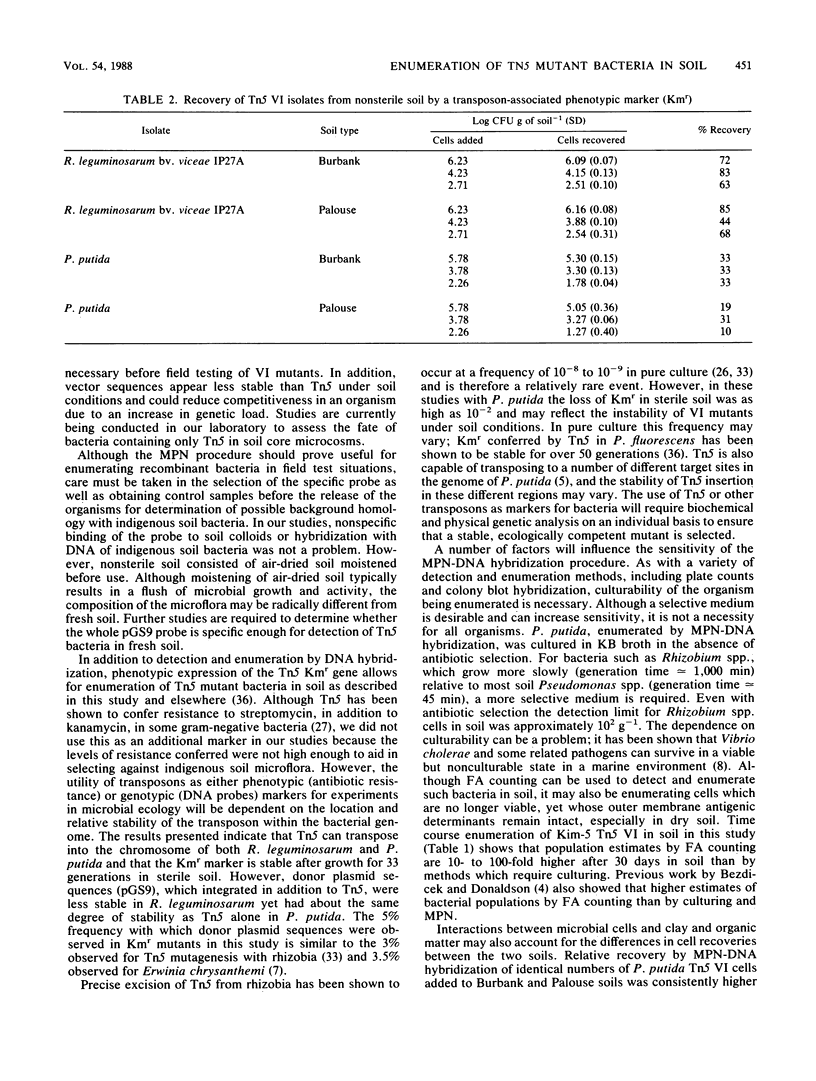
Abstract
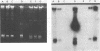
Investigations were made into the utility of DNA hybridization in conjunction with a microdilution most-probable-number procedure for the enumeration of Rhizobium spp. and Pseudomonas putida in soil. Isolates of Rhizobium spp. and P. putida carrying the transposon Tn5 were added to sterile and nonsterile Burbank sandy loam soil and enumerated over time. Soil populations of rhizobia were enumerated by colony hybridization, most-probable-number-DNA hybridization procedure, plate counts, plant infectivity most probable number, and fluorescent antibody counts. Population values compared well for all methods at 5 and 30 days after the addition of cells, although the fluorescent antibody method tended to overestimate the viable population. In nonsterile soil, most-probable-number-DNA hybridization procedure enumerated as few as 10 P. putida Tn5 cells g of soil-1 and 100 R. leguminosarum bv. phaseoli Tn5 cells g of soil-1 and should have utility for following the fate of genetically engineered microorganisms released to the environment. Among the Kmr isolates containing Tn5, approximately 5% gave a dark, more intense autoradiograph when probed with 32P-labeled pGS9 DNA, which facilitated their detection in soil. Hybridization with a pCU101 probe (pGS9 without Tn5) indicated that donor plasmid sequences were being maintained in the bacterial chromosome. Transposon-associated antibiotic resistance was also utilized as a phenotypic marker. Tn5 vector-integrate mutants were successfully enumerated at low populations (10 to 100 cells g of soil-1) in soil by both phenotypic (Kmr) and genotypic (DNA probe) analysis. However, determination of the stability of Tn5 or Tn5 and vector sequences in the bacteria is necessary.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkay T., Fouts D. L., Olson B. H. Preparation of a DNA gene probe for detection of mercury resistance genes in gram-negative bacterial communities. Appl Environ Microbiol. 1985 Mar;49(3):686–692. doi: 10.1128/aem.49.3.686-692.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulnois G. J., Varley J. M., Sharpe G. S., Franklin F. C. Transposon donor plasmids, based on ColIb-P9, for use in Pseudomonas putida and a variety of other gram negative bacteria. Mol Gen Genet. 1985;200(1):65–67. doi: 10.1007/BF00383313. [DOI] [PubMed] [Google Scholar]
- Brill Winston J. Safety concerns and genetic engineering in agriculture. Science. 1985 Jan 25;227(4685):381–384. doi: 10.1126/science.11643810. [DOI] [PubMed] [Google Scholar]
- Chatterjee A. K., Thurn K. K., Feese D. A. Tn5-Induced Mutations in the Enterobacterial Phytopathogen Erwinia chrysanthemi. Appl Environ Microbiol. 1983 Feb;45(2):644–650. doi: 10.1128/aem.45.2.644-650.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss R., 3rd Genetic manipulation of microorganisms: potential benefits and biohazards. Annu Rev Microbiol. 1976;30:507–533. doi: 10.1146/annurev.mi.30.100176.002451. [DOI] [PubMed] [Google Scholar]
- Fitts R., Diamond M., Hamilton C., Neri M. DNA-DNA hybridization assay for detection of Salmonella spp. in foods. Appl Environ Microbiol. 1983 Nov;46(5):1146–1151. doi: 10.1128/aem.46.5.1146-1151.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasson M. J. Indicator technique for antimetabolic toxin production by phytopathogenic species of pseudomonas. Appl Environ Microbiol. 1980 Jan;39(1):25–29. doi: 10.1128/aem.39.1.25-29.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. E., Payne W. L., Aulisio C. C. Detection and enumeration of virulent Yersinia enterocolitica in food by DNA colony hybridization. Appl Environ Microbiol. 1983 Sep;46(3):636–641. doi: 10.1128/aem.46.3.636-641.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Estes M. K., Metcalf T. G., Melnick J. L. Detection of hepatitis A virus in seeded estuarine samples by hybridization with cDNA probes. Appl Environ Microbiol. 1986 Oct;52(4):711–717. doi: 10.1128/aem.52.4.711-717.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Liang L. N., Sinclair J. L., Mallory L. M., Alexander M. Fate in model ecosystems of microbial species of potential use in genetic engineering. Appl Environ Microbiol. 1982 Sep;44(3):708–714. doi: 10.1128/aem.44.3.708-714.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnoky P., Kiss G. B., Ott I., Kondorosi A. Tn5 carries a streptomycin resistance determinant downstream from the kanamycin resistance gene. Mol Gen Genet. 1983;191(2):288–294. doi: 10.1007/BF00334828. [DOI] [PubMed] [Google Scholar]
- Rissler J. F. Research needs for biotic environmental effects of genetically-engineered microorganisms. Recomb DNA Tech Bull. 1984 Mar;7(1):20–30. [PubMed] [Google Scholar]
- Rowe R., Todd R., Waide J. Microtechnique for most-probable-number analysis. Appl Environ Microbiol. 1977 Mar;33(3):675–680. doi: 10.1128/aem.33.3.675-680.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sands D. C., Rovira A. D. Isolation of fluorescent pseudomonads with a selective medium. Appl Microbiol. 1970 Sep;20(3):513–514. doi: 10.1128/am.20.3.513-514.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayler G. S., Shields M. S., Tedford E. T., Breen A., Hooper S. W., Sirotkin K. M., Davis J. W. Application of DNA-DNA colony hybridization to the detection of catabolic genotypes in environmental samples. Appl Environ Microbiol. 1985 May;49(5):1295–1303. doi: 10.1128/aem.49.5.1295-1303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983 Dec;156(3):1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]