Abstract
The accessory protein, Vpr, is a virion-associated protein that is required for HIV-1 replication in macrophages and regulates viral gene expression in T cells. Vpr causes arrest of cell cycle progression at G2/M, presumably through its effect on cyclin B1⋅Cdc2 activity. Here, we show that the ability of Vpr to activate HIV transcription correlates with its ability to induce G2/M growth arrest, and this effect is mediated by the p300 transcriptional co-activator, which promotes cooperative interactions between the Rel A subunit of NF-κB and cyclin B1⋅Cdc2. Vpr cooperates with p300, which regulates NF-κB and the basal transcriptional machinery, to increase HIV gene expression. Similar effects are seen in the absence of Vpr with a kinase-deficient Cdc2, and overexpression of p300 increases levels of HIV Vpr+ replication. Taken together, these data suggest that p300, through its interactions with NF-κB, basal transcriptional components, and Cdks, is modulated by Vpr and regulates HIV replication. The regulation of p300 by Vpr provides a mechanism to enhance viral replication in proliferating cells after growth arrest by increasing viral transcription.
HIV replication is highly responsive to changes in cellular stimulation, such as cellular activation by cytokines or cell cycle progression by growth factors. The regulation of cell cycle progression by HIV has recently become apparent. Specifically, Vpr represents an HIV-encoded protein that influences cell cycle progression by either inducing differentiation (1) or causing cell cycle arrest at the G2/M checkpoint (2–5). Vpr is a 14-kDa accessory protein of HIV that localizes to the nucleus and associates with the virion by binding to the carboxyl terminus of the viral Gag protein (6–9). In nondividing cells, such as macrophages, Vpr plays a role in the translocation of HIV to the nucleus by a mechanism distinct from the karyopherin pathway, which is used by the viral matrix protein for nuclear localization of HIV (10, 11). Vpr is important for marked stimulation of viral replication in macrophages and causes moderate increases in HIV transcription and virus production in T lymphocytes (12–14).
HIV transcription is regulated by two κB sites in the viral enhancer within the long terminal repeat (LTR) (15). These sites are recognized by the NF-κB family of transcription factors, which are typically induced in response to stress and infection (reviewed in ref. 16). NF-κB transcriptional activity is largely controlled by sequestration in the cytoplasm by a family of inhibitory proteins, IκBs, and inhibition is relieved when IκBs are phosphorylated and degraded in response to various cytokines, including tumor necrosis factor alpha (TNF-α), growth factors, and bacterial and viral products (reviewed in ref. 17). NF-κB function is also regulated in the nucleus by other transcription factors, members of the basal transcriptional machinery (18), and transcriptional co-activators such as the p300 and CREB binding protein (CBP) co-activators (19). Recently, NF-κB transcriptional activation was shown to be regulated by modulators of cell cycle progression. Specifically, growth arrest by expression of the p21 cyclin-dependent kinase inhibitor potentiated NF-κB function (19). This effect was mediated by the p300 transcriptional co-activator, which binds to cyclin E⋅Cdk2 complexes inhibited by p21. Thus, it seemed that G1/S-specific cell cycle regulatory proteins could modulate NF-κB activity and thereby increase transcription of viral genes when cell cycle progression was arrested.
The mechanism of HIV transcriptional activation by Vpr is not well understood. Vpr inhibits cell cycle progression at the G2/M checkpoint. Our previous finding that G1/S growth arrest affects HIV transcription raised the possibility that Vpr-induced growth arrest may also affect its transactivation. We therefore examined whether Vpr could affect HIV transcription by p300/CBP through its ability to induce G2/M cell cycle arrest. We demonstrate that Vpr affects HIV gene expression through its effect on p300 and that this effect correlates with the ability of Vpr to arrest cell cycle progression. We implicate cyclin B1⋅Cdc2 complexes associated with p300 as a mediator of this effect. HIV Vpr+ replication is increased when p300 is overexpressed in infected cells, implicating transcriptional co-activation regulated by Cdks in the control of HIV replication.
METHODS
Plasmids, Cell Culture, and Nuclear Extracts.
The HIV-CAT, RSV-Rel A, CMV-Vpr, CMV-Vpr (ΔATG), CMV-Cdc2 (ΔK), CMV-p300, pBS-3′p300, WT 12S E1A, Δp300 12S E1A, and CMV-Vpr mutant expression plasmids have all been described (15, 19, 20, 21, 22). The Vpr mutant, R80A, which has been characterized (23), was constructed in the context of Hxbru Vpr (20). Rous sarcoma virus (RSV)-β-galactosidase or RSV-β-globin and cytomegalovirus (CMV)-alkaline phosphatase (VCL-1012 CMV (Vical, Inc.)) vectors were used as control filler plasmids for transfections with RSV-Rel A, CMV-p300, and CMV-Cdc2 (ΔK). The p300 and Rel A pBluescript constructs used for in vitro translation with T7 RNA polymerase were also previously reported (15, 22). The CMV-CD2 plasmid was prepared by ligating a SalI–HindIII fragment containing the coding region for human CD2 into the SalI–EcoRV sites in the multiple cloning region of VCL-1012 CMV. The CMV-CD4 vector was prepared by ligating an EcoRI–BamHI fragment containing the CD4 coding sequence into the PstI–BamHI sites in the VCL-1012 CMV multiple cloning region.
Jurkat, UM-316, 293 kidney epithelial, and NIH 3T3 cells were grown (24), and nuclear extracts prepared as described previously (25). For induction of NF-κB for electrophoretic mobility shift assay (EMSA), cells were stimulated for 1 h with 200 units/ml recombinant TNF-α (Genzyme).
Transfections and Chloramphenicol Acetyltransferase (CAT) Assays.
Jurkat cells were transfected using DEAE-dextran, and CAT assays were performed 48 h after transfection, as previously described (15). 293 cells were transfected by calcium phosphate precipitation, and after 72 h, nuclear extracts were prepared (25).
NIH 3T3 cells were transfected with gAP/DLRIE/DOPE 50/50 (Vical). Specifically, cells were plated to 40% confluency 1 day prior to transfection, washed two times with Opti-MEM, and transfected with lipid–DNA solution made by combining 1.5 ml of Opti-MEM containing 15 μg of DNA plus 1.5 ml of Opti-MEM containing 30 μl of a 2 mg/ml lipid solution. Cells were incubated for 4 h, the lipid solution was removed, fresh medium was added, and cells were lysed and assayed after 48 h. Due to differences in transfection efficiencies observed with these cells, 4 μg of RSV-β-galactosidase was cotransfected in all samples, and extracts were normalized prior to analysis of CAT activity based on β-galactosidase activity (26).
UM-316 cells were plated to 20% confluency in 6-well plates 1 day prior to transfection. Cells were transfected with 5 μg of CMV-p300 or CMV empty control vector and 0.5 μg of CMV-CD4 using Lipofectamine (GIBCO/BRL), according to manufacturer’s instructions.
Cell Cycle Analysis.
293 cells were transfected with 2.5 μg of CMV-CD2 plus 7.5 μg of CMV-Vpr vectors. After 48 h, cells were incubated for 30 min with an antibody to the CD2 cell surface marker (ATCC HB 222 hybridoma supernatant), washed three times with PBS + 2% fetal calf serum, incubated for 30 min with an anti-mouse fluorescein isothiocyanate-conjugated antibody, and washed an additional three times. Cells were resuspended in 875 μl of ice-cold PBS and fixed for 1 h on ice by adding 125 μl of cold PBS containing 2% paraformaldehyde. Cells were washed one time with PBS + 2% fetal calf serum, permeabilized for 15 min at 37°C with 1 ml 2% Tween 20 in PBS, and washed an additional time. Propidium iodide staining was performed by incubating cells for 1 h at 37°C in 960 μl of PBS, 2 units of RNase, and 40 μl of propidium iodide (Boehringer Mannheim). DNA content of CD2 positive cells was measured by fluorescence-activated cell sorting.
Electrophoretic Mobility Shift Assays.
EMSAs were performed with 1.5 μg of nuclear extract from 293 cells and a double-stranded, end-labeled oligomer containing one NF-κB binding site as described previously (25). Complexes were analyzed on 5% polyacrylamide gels in 0.25× TBE buffer.
Protein Binding Assays and Western Blots.
For binding to in vitro translated proteins, immunoprecipitations were performed with anti-cyclin B antibody (SC-245AC, Santa Cruz) or a control mouse IgG1 (M-9269, Sigma) on 50 μg of nuclear extract for 2 h. Immunoprecipitated proteins were washed three times with immunoprecipitation buffer (19), incubated with in vitro translated carboxyl-terminal p300 (T7 TNT rabbit reticulocyte transcription/translation system, Promega), and washed three additional times with immunoprecipitation buffer. Complexes were analyzed by 8% SDS/PAGE.
For detection of associated p300, Rel A, cyclin B1, and Cdc2 by Western blot, immunoprecipitations were performed on 420 μg of nuclear extract, subjected to 8% SDS/PAGE and then to Western analysis with antibodies to p300 (SC-584 and SC-585, Santa Cruz), Rel A (SC-109 and SC-372, Santa Cruz), cyclin B1 (SC-245, Santa Cruz), and Cdc2 (SC-54, Santa Cruz). Antibodies were used at a concentration of 0.5 μg/ml, and the appropriate secondary anti-rabbit antibody or anti-mouse antibody linked to horseradish peroxidase was used at dilutions of 1:8000 and 1:3000, respectively. Proteins were visualized using the enhanced chemiluminescence system (Amersham).
HIV Infections and Reverse Transcriptase Assays.
Transfected CD4 positive 293 cells or UM-316 cells in 6-well tissue culture plates were infected with HIVbru Vpr+ or HIVbru Vpr− by incubating cells with virus for 4 h at 37°C at a multiplicity of infection of 0.05–0.1. Following incubation with virus, cells were washed twice with D-PBS, and fresh DMEM was added to a final volume of 4 ml.
Culture supernatants were assayed for reverse transcriptase (RT) activity as described previously (27). Poly(A)/oligo(dt) was used as template primer, and incorporation of [32P]dTTP was measured after spotting 3 ml of the RT reaction mixture onto DE81 paper, drying, and washing four times in 2 × SSC solution at 22°C. Radioactivity was analyzed on a Betagene (Waltham, MA) betascope, and activity was expressed as cpm/ml of culture supernatants. Triplicate RT assays were performed, and values are expressed as the mean ± standard deviation.
RESULTS
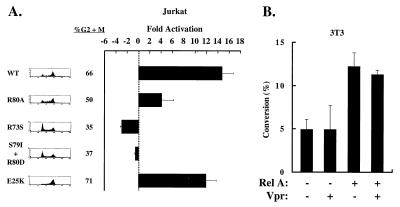
To examine the mechanism of HIV transcriptional activation by Vpr, we first determined whether the ability of Vpr to cause growth arrest correlated with its activation of HIV transcription. Jurkat T leukemia cells were transfected with a vector encoding the chloramphenicol acetyltransferase gene controlled by the HIV enhancer (HIV-CAT) and expression vectors encoding either wild-type or mutant forms of Vpr. The R80A mutant, which fails to arrest cells but retains the ability to translocate to the nucleus (23), and two additional mutants, R73S and S79I/R80D, which are similar to previously characterized cell cycle mutants (23), were tested in these experiments. The E25K mutation, which impairs nuclear localization and virion incorporation of Vpr but allows effective viral replication (20), was also examined. In 293 cells, E25K inhibited cell cycle progression after 48 h, whereas R80A, R73S, and S79I/R80D were impaired in their ability to arrest cells (Fig. 1A). 293 cells were used to document growth arrest because of the high transfection efficiencies achieved, and it was more difficult to assess these effects in Jurkat cells, which are poorly transfected; however, previous studies showed similar cell cycle arrest phenotypes by Vpr and several mutants in Jurkat cells (3, 28). Infection of Jurkat cells with HIV containing the R80A or E25K mutation was also used to confirm that Vpr R80A does not cause cell cycle arrest and that Vpr E25K does arrest cells (R.A.S. and E.A.C., unpublished data), similar to the findings in 293 cells shown here.
Figure 1.
Vpr activation of HIV transcription with Rel A correlates with its ability to regulate cell cycle progression. (A) Vpr mutants defective in G2/M growth arrest are impaired in their ability to activate HIV transcription. For analysis of cell cycle distribution, 293 cells were cotransfected with pCMV-CD2, an expression vector encoding the CD2 antigen, wild-type CMV-Vpr, and the indicated Vpr mutant forms. CD2 positive cells were analyzed by fluorescence-activated cell sorting. The ratios of G2/M to G1 cell populations are designated. For analysis of Vpr transactivation, Jurkat cells were transfected with 5 μg of HIV-CAT, 0.2 μg of RSV Rel A, and 1 μg of CMV-Vpr (WT), CMV-Vpr (R80A), CMV-Vpr (R73S), CMV-Vpr (S79I/R80D), or CMV-Vpr (E25K). Control plasmids were used, such that each transfection received 0.2 μg of RSV and 1 μg of CMV plasmids. Values are represented as -fold activation over basal HIV-CAT activity, and error bars represent SEM. (B) Vpr cannot activate the HIV enhancer in NIH 3T3 cells, which cannot be arrested by Vpr. NIH 3T3 cells were cotransfected with 5 μg of HIV-CAT, 0.5 μg of RSV-Rel A, and 1 μg of CMV-Vpr where indicated. Control plasmids were used such that each sample received 0.2 μg of RSV and 1 μg of CMV plasmids.
To analyze transcriptional enhancement, Jurkat cells were cotransfected with each Vpr expression vector and Rel A to ensure constant NF-κB expression and HIV transcriptional activation (Fig. 1A). Both wild-type Vpr and the E25K mutant stimulated HIV-CAT activity (Fig. 1A; p = 0.001 for Vpr and p = 0.044 for E25K compared with the untranslated negative control, Vpr (ΔATG), by paired t test), suggesting that nuclear localization and virion incorporation of Vpr were not required for activation of HIV transcription by Vpr; however, R80A, R73S, and S79I/R80D were impaired in their ability to transactivate, suggesting a correlation between the transactivation and the cell cycle arrest functions of Vpr. The expression of Vpr was confirmed by Western blot analysis (data not shown).
To examine further whether cell cycle arrest by Vpr correlated with transactivation of the HIV enhancer, we analyzed the effect of Vpr on HIV-CAT activity in NIH 3T3 cells, which are not growth arrested by Vpr (3). NIH 3T3 cells were transfected with HIV-CAT and CMV-Vpr in the presence or absence of Rel A. In contrast to the effects of Vpr in Jurkat cells, no activation of the HIV-LTR in NIH 3T3 cells was observed in the presence of Vpr (Fig. 1B). Furthermore, when Rel A was cotransfected to provide constant amounts of nuclear NF-κB activity, no transactivation by Vpr was observed in these cells. Thus, the inability of Vpr to arrest NIH 3T3 cells in G2/M correlated with a lack of HIV transcriptional activation, further suggesting that the G2/M growth arrest activity of Vpr is required for its transactivation function.
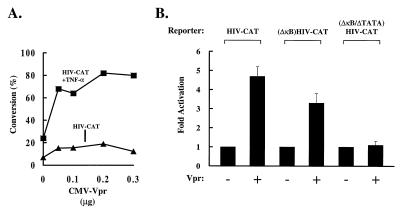
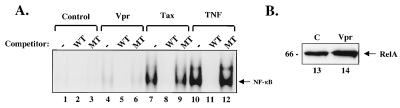
To analyze the specific cis-acting elements of the HIV promoter involved in Vpr transactivation, we next examined whether Vpr could affect basal and TNF-induced HIV transcription. Jurkat cells were transfected with HIV-CAT and increasing amounts of a Vpr expression vector. Cells were then either left unstimulated or stimulated with TNF-α. Vpr induced a dose-responsive increase in CAT activity in the presence or absence of TNF-α (Fig. 2A). To determine the effect of Vpr on specific enhancer regions, Jurkat cells were cotransfected with Vpr and HIV-CAT containing different mutant cis-acting regulatory elements. Though decreased in the absence of the κB sites, the mutant κB enhancer remained Vpr responsive (Fig. 2B), suggesting that NF-κB contributes to transactivation by Vpr but is not completely responsible for this effect. In contrast, no induction was observed using a ΔκB/ΔTATA mutant reporter, suggesting that the basal transcriptional machinery is also required for transactivation by Vpr. Because the ΔκB/ΔTATA mutant reporter is responsive to other viral transactivators (29), it was evident that both the κB sites and the TATA box were necessary for responsiveness to Vpr. The effect of Vpr on NF-κB induction was analyzed biochemically. Nuclear extracts from 293 cells transfected with control plasmids or those expressing either Vpr or Tax were analyzed using EMSA. Expression of Vpr caused no significant change in κB DNA binding activity, in contrast to the known activator, HTLV-1 Tax (21, 30, 31), which increased NF-κB complexes (Fig. 3A). Western blot analysis confirmed that Vpr did not significantly change in the levels of nuclear Rel A (1.3-fold over the control) (Fig. 3B), and no direct interaction with Rel A was found by immunoprecipitation (data not shown).
Figure 2.
Definition of cis-acting sites that mediate Vpr stimulation of HIV transcription. (A) Vpr stimulates HIV enhancer activity. Jurkat cells were cotransfected with 5 μg of HIV-CAT and indicated amounts of CMV-Vpr. A control vector, Vpr (ΔATG), was used such that each sample received an equivalent amount of CMV expression plasmid. Forty-two hours post-transfection, cells were stimulated for 6 h with TNF-α (+TNF-α) or left nonstimulated. (B) The κB and TATA sequences are required for Vpr transactivation. Jurkat cells were cotransfected with 5 μg of HIV-CAT, (ΔκB) HIV-CAT, or (ΔκB/ΔTATA) HIV-CAT, and 1 μg of CMV-Vpr where indicated. Control plasmids were used, such that each sample received 1 μg of CMV vectors. Values are represented as -fold activation by Vpr over the basal level of each reporter in the absence of Vpr.
Figure 3.
Vpr does not affect DNA binding or nuclear translocation of NF-κB. (A) Vpr does not induce NF-κB DNA binding. Nuclear extracts from 293 cells transfected with 7.5 μg of a negative control, ATG-deleted CMV-Vpr (control) (lanes 1–3), CMV-Vpr (lanes 4–6), LTR-Tax (lanes 7–9), or stimulated with TNF-α were analyzed by EMSA, using a 32P end-labeled, double-stranded κB oligonucleotide probe. Unlabeled mutant and wild-type κB competitor DNAs (100 ng) were included as indicated. (B) Vpr does not induce Rel A nuclear translocation. Nuclear extracts from 293 cells transfected with 7.5 μg of a negative control CMV-ATG mutant Vpr (control) (lane 1) or CMV-Vpr (lane 2) were analyzed for nuclear localization of Rel A by Western blot. Densitometric analysis revealed that RelA was 1.3-fold higher in Vpr transfected cells.
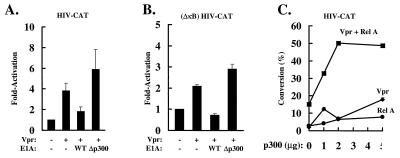
The finding that Vpr transactivation occurred through both the κB sites and the TATA box suggested that Vpr may regulate the function of a transcriptional co-activator, such as p300, which binds to both NF-κB and to the TFIIB and TBP components of the basal transcriptional machinery (19). To investigate the role of p300 in activation of the HIV enhancer by Vpr, the adenovirus E1A gene product was used to inhibit p300-dependent gene activation. The 12S form of adenovirus E1A protein binds to and inactivates p300 (32–36), inhibiting basal and TNF-α-induced NF-κB (37). Wild-type 12S E1A and the 12S E1A p300 binding mutant were cotransfected with HIV-CAT and Vpr in Jurkat T cells. Wild-type 12S E1A repressed activation by Vpr; however, a 12S E1A mutant that fails to bind p300 was unable to exert this repressive effect (Fig. 4A), suggesting that p300 is required for activation of HIV transcription by Vpr. To determine whether p300 is necessary for the activation of the basal transcriptional machinery by Vpr, (ΔκB)HIV-CAT, Vpr, and wild-type 12S E1A or the 12S E1A p300 binding mutant were cotransfected. As previously observed, activation of (ΔκB)HIV-CAT by Vpr was lower than that observed with HIV-CAT; however, the effects by E1A on (ΔκB)HIV-CAT were similar to those observed with HIV-CAT. Wild type but not the 12S E1A p300 binding mutant inhibited Vpr-mediated (ΔκB)HIV-CAT activation (Fig. 4B), suggesting that p300 is necessary for activation of HIV transcription through the basal transcriptional components.
Figure 4.
p300 stimulates HIV transcription. (A) Wild-type 12S E1A but not a 12S E1A p300 binding mutant inhibits activation of the HIV enhancer by Vpr. Jurkat cells were cotransfected with 2 μg of HIV-CAT, 0.5 μg of CMV-Vpr, and 2 μg of wild-type 12S E1A or a 12S E1A p300 binding mutant, as indicated. Relevant control plasmids were used where necessary. Values are represented as -fold activation over the basal level of HIV-CAT. (B) Wild-type 12S E1A but not 12S E1A p300 binding mutant inhibits (ΔκB)HIV-CAT activity. Jurkat cells were cotransfected with 2 μg of (ΔκB)HIV-CAT, 0.5 μg of CMV-Vpr, and either 2 μg of wild-type 12S E1A or a 12S E1A p300 binding mutant, as indicated. Control plasmids were included where necessary. Values are represented as -fold activation over the basal level of HIV-CAT activity. (C) p300 and Vpr function together to activate the HIV enhancer. Jurkat cells were cotransfected with 5 μg of HIV-CAT, 0.2 μg of RSV-RelA where indicated, 1 μg of CMV-Vpr where indicated, and increasing amounts of CMV-p300. Appropriate control vectors were transfected where necessary. Values represent the percent of chloramphenicol conversion in the CAT assay.
To investigate a role for p300 in transactivation by Vpr further, Jurkat cells were cotransfected with Vpr, Rel A (to provide constant nuclear NF-κB), and increasing amounts of a p300 expression vector. p300 exerted dose-responsive stimulation in combination with Vpr, Rel A, and Vpr plus Rel A, and the Vpr/Rel A/p300 combination was greater than additive (100-fold) over the effect of p300 plus Vpr (36-fold) or p300 plus RelA (16-fold) (Fig. 4C). These data suggested that Vpr, Rel A, and p300 act in concert to activate HIV transcription.
The mechanism of p300 regulation by Vpr was next examined. The p300 co-activator has been shown to stimulate HIV transcription in the presence of the p21 cyclin-dependent kinase inhibitor. Rel A binds to the NH2 terminus of p300, whereas cyclin E/Cdk2 complexes bind to the COOH terminus of p300. Vpr has been shown to cause growth arrest through its ability to inhibit Cdc2 kinase activity. Cyclin B1 had also been detected at low levels in Rel A immunoprecipitations (19), raising the possibility that cyclin B1⋅Cdc2 might also interact with p300. We therefore examined whether p300 could interact with cyclin B1⋅Cdc2 complexes.
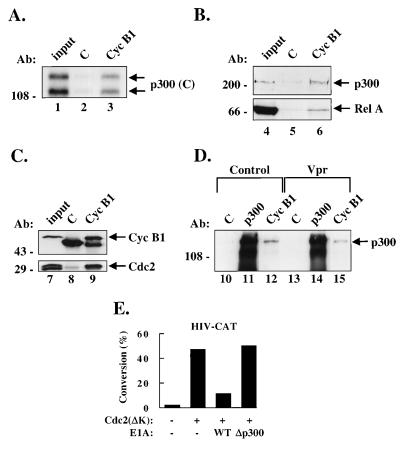
Immunoprecipitations were performed with control antibodies or antibodies to cyclin B1 on nuclear extracts from Jurkat cells, and in vitro translated COOH-terminal p300 were bound to cyclin B1⋅Cdc2 but not to control immunoprecipitations (Fig. 5A). This interaction was confirmed in vivo in TNF-stimulated Jurkat nuclear extracts by immunoprecipitation of cyclin B1 and Western blot analysis for p300 (Fig. 5B). Although p300 was not detected with a control antibody, it was readily detected in the cyclin B1 immunoprecipitates. Western blot analysis for Rel A of the same immunoprecipitated complexes revealed that Rel A was also present in cyclin B1⋅Cdc2⋅p300 complexes and not in control immunoprecipitations (Fig. 5B). The presence of both cyclin B1 and Cdc2 in these complexes was confirmed by a similar cyclin B1 immunoprecipitation, followed by Western blot analysis using cyclin B1 and Cdc2 antibodies (Fig. 5C). Thus, it seemed that cyclin B1⋅Cdc2⋅p300⋅Rel A complexes exist in vivo. Vpr also did not seem to affect the overall level of binding of cyclin B1⋅Cdc2 to p300 (Fig. 5D).
Figure 5.
Cyclin B1⋅Cdc2, p300, and Rel A are associated in the same complex, which is not affected by Vpr overexpression, and effect of kinase-deficient Cdc2 on p300-dependent HIV transcription. (A) Cyclin B1⋅Cdc2 complexes bind p300 in vitro. Immunoprecipitations from Jurkat nuclear extracts using control antibodies (lane 2) or cyclin B1 antibodies (lane 3) were incubated with in vitro translated carboxyl-terminal p300, washed, and analyzed by SDS/PAGE. Lanes 1 and 4 show 10% of the input in vitro translated p300. (B) Cyclin B1⋅Cdc2 complexes bind p300 in vivo. Immunoprecipitations were performed on nuclear extracts from Jurkat cells that had been treated with TNF-α for 1 h with a control antibody (lane 5) or anti-cyclin B1 (lane 6). Complexes were washed and subjected to Western analysis using anti-p300 (Upper) or anti-Rel A (Lower) antibodies. Lane 4 contains 10 μg of Jurkat nuclear extract. (C) Both cyclin B1 and Cdc2 are found in cyclin B1 immunoprecipitations from nuclear extracts. Immunoprecipitations with a control antibody (lane 8) or an antibody to cyclin B1 (lane 9) were performed on Jurkat nuclear extracts and subjected to Western blot analysis using antibodies to cyclin B1 (Upper) or Cdc2 (Lower). Lane 7 contains 10 μg of Jurkat nuclear extract. (D) Expression of Vpr does not affect cyclin B1-p300 binding. 293 cells were transfected with a negative control, ATG deleted CMV-Vpr (control) (lanes 10–12) or CMV-Vpr (lanes 13–15) and immunoprecipitations were performed with control (C) antibodies (lanes 10 and 13), p300 antibodies (lanes 11 and 14), and cyclin B1 antibodies (lanes 12 and 15). Immunoprecipitated complexes were subjected to Western blot analysis for p300. (E) Expression of a kinase-deficient Cdc2 activates HIV transcription through p300 Jurkat cells cotransfected with 5 μg of HIV-CAT, 1 μg of CMV-Cdc 2 (ΔK) where indicated, and either 2 μg wild-type 12S E1A or a 12S E1A p300 binding mutant, as shown. Values are represented as the percent of acetylated chloramphenicol in the CAT assay.
Because Vpr is thought to exert its effect on growth arrest by inhibition of Cdc2 kinase activity and cyclin B1⋅Cdc2 was found in association with p300, we next determined whether a kinase-deficient mutant of Cdc2, Cdc2(ΔK), could affect HIV transcriptional activation by p300. Jurkat cells were cotransfected with HIV-CAT, an expression plasmid encoding Cdc2(ΔK) shown previously to arrest cells at the G2/M phase of the cell cycle (38), and wild-type 12S E1A or the 12S E1A p300 binding mutant. Overexpression of Cdc2(ΔK) stimulated HIV-CAT activity, suggesting that direct inhibition of Cdc2 has the same effect on HIV transcription as does indirect Cdc2 inhibition by Vpr. Cotransfection of optimal amounts of Vpr and Cdc2(ΔK) provided no synergistic activation, and this effect was reduced in the absence of the κB sites, as seen with Vpr (data not shown), suggesting saturation of an activation pathway common to Vpr and Cdc2. In addition, activation of HIV transcription by Cdc2(ΔK) was inhibited by wild-type 12S E1A but not the p300 binding mutant (Fig. 5E), suggesting that transactivation of the HIV-LTR through inhibition of Cdc2 occurs through p300. Thus, the inhibition of p300-associated Cdc2 activity enhances transcriptional activation by Rel A and p300, and it is likely that Vpr functions through this mechanism.
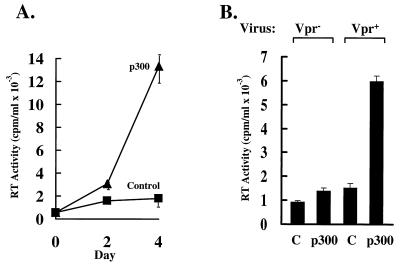
The inhibition of Vpr-mediated HIV enhancer activity by sequestration of p300 by E1A and the involvement of Vpr in HIV replication suggested that p300 may also be involved in HIV replication. To determine whether p300 could regulate HIV replication, CD4–293 cells, a highly transfectable cell line that is highly permissive for HIV, were transfected with CMV-p300 or a control CMV plasmid. These cells were then infected with HIVbru Vpr+, and viral replication was measured at days 0, 2, and 4 by quantification of reverse transcriptase (RT) activity. In the presence of CMV-p300, a gradual increase in p300 compared with control-transfected cells was observed, with a 6- to 7-fold increase 4 days after infection (Fig. 6A, p = .041, paired t test), a significant difference given the high replication of virus in these cells (39). Although CD4–293 cells contain E1A, which inhibits p300 function, the levels of overexpressed p300 were able to overcome to the levels of endogenous E1A, as examined by silver staining and Western blot analysis (data not shown). To confirm this effect in the absence of E1A, a highly transfectable E1A-negative melanoma cell line, UM-316, was used. UM-316 cells were cotransfected with CD4 and p300 or control expression vectors and subsequently infected by HIVbru Vpr+ or HIVbru Vpr−. Reverse transcriptase activity was quantitated 4 days after infection and demonstrated that HIVbru Vpr+ replication was 4.3-fold higher in the presence of p300, whereas HIVbru Vpr− replication was nearly unchanged in the presence of p300 (Fig. 6B). These results demonstrate that Vpr and p300 cooperate to increase HIV replication during acute infection in proliferating cells.
Figure 6.
p300 activates HIVbru Vpr+ replication. (A) Time course for p300 activation of HIV replication stimulated by p300. 293 cells expressing the CD4 molecule were transfected with 6 μg of CMV-p300 or a control CMV plasmid with no insert. Cells were challenged with HIV 24 h later, and replication was measured by RT assays at 0, 2, and 4 days after infection. RT activity is expressed as mean values ± standard deviations of triplicate assays. (B) p300 differentially regulates HIVbru Vpr− and HIVbru Vpr+ replication. UM-316 cells were transfected with 0.5 μg of CMV-CD4 and either 5 μg of a control CMV vector or CMV-p300, as indicated. Cells were infected with HIVbru Vpr− or HIVbru Vpr+ 24 h later, and viral replication was measured on day 4 by RT assays. RT activity is expressed as mean values ± standard deviations of triplicate assays.
DISCUSSION
In this study, we show that activation of HIV transcription by Vpr correlates with its ability to arrest cells in G2/M and is mediated by modulation of transcriptional co-activator function, which is responsive to Cdks. Previous studies had shown that HIV RNA levels increase during the G2/M phase of the cell cycle (40), supporting the notion that Vpr links viral transcription to cell proliferation. Thus, a major role for Vpr in T cells may be to coordinate changes in cell cycle progression with increased viral gene expression through p300/CBP.
We find that Vpr exerts its effects on the HIV enhancer through p300, as inhibition of p300 function by 12S E1A, but not a 12S E1A mutant unable to bind p300, eliminates the ability of Vpr to activate NF-κB. In addition, overexpression of p300 enhances the ability of Vpr to activate NF-κB and the HIV enhancer. Vpr, which inhibits the cyclin B1⋅Cdc2 activity (3–5, 28), is not directly associated with p300 but instead regulates cyclin B1⋅Cdc2 activity, which we show interacts with the COOH terminus of this co-activator. Consistent with these observations, a kinase-deficient Cdc2 stimulates a similar increase in HIV transcriptional activation and is inhibited by wild-type 12S E1A but not the 12S E1A p300 binding mutant, providing a link between Cdc2 and NF-κB by p300 in the regulation of HIV transcription. The mechanism by which Vpr inhibits Cdc2 activity is not completely understood; however, the Cdc25 protein that activates Cdc2 is inactive in Vpr-expressing cells (5), suggesting that proteins which normally control Cdc2 are modulated by Vpr, which in turn affects HIV transcription. Our experiments with the Vpr E25K mutant, which is impaired for nuclear localization but retains the ability to arrest cells, suggest that Vpr regulates Cdc2-regulatory proteins that reside in the cytoplasm of the cell and further support an indirect role for Vpr in Cdk modulation, which interacts with p300 in the nucleus to affect NF-κB transcription.
In adherent macrophages, which do not proliferate, the primary role for Vpr seems to be in the localization of the pre-integration complex to the nucleus (10, 11). The ability of Vpr to affect HIV transcription in macrophages has not been tested; however, our results suggest that Vpr may not further induce HIV enhancer function in macrophages, as its ability to arrest the cell cycle would be unnecessary in non-proliferating cells, and NF-κB is already maximally induced in these cells (41). In these cells, it is therefore likely that NF-κB is already regulated by cellular factors that control the cell cycle and are present in resting cells by mechanisms independent of Vpr. However, if macrophages were proliferating, it is likely that Vpr could affect HIV transcription through cell cycle arrest. It has also recently been reported that Vpr represses some NF-κB-regulated cytokine promoters (42). The generalization of those findings, performed in rhabdomyosarcoma cells, is unknown and likely to differ from those reported here. Specifically, those effects involved different target genes in a cell line not normally infected by HIV.
The control of NF-κB activity by E1A has been well studied, and E1A stimulates cell cycle progression by binding to and inactivating cellular proteins, such as pRb, p107, and p300. Conversely, Vpr seems to inhibit cell cycle progression and to increase the activity of proteins (p300/CBP) that are inhibited by E1A. Unlike E1A, Vpr does not seem to bind to p300 but activates the protein through an indirect mechanism. Interestingly, p300 seems to be a molecule that is a point of conversion for these viral proteins as well as for cellular signal transduction pathways (43). In addition, p300 regulates a variety of transcription factors unrelated to NF-κB, including CREB (44), YY1 (45), Myb (46, 47), Jun (48), Fos (49), Sap1a (50), Stat2 (51), myogenic transcription factors (52, 53), nuclear receptors (54), and p53 (55, 56), presumably through its ability to regulate chromatin structure by histone acetylation (57, 58). It also remains likely that p300 may interact with other HIV-encoded proteins that act at different stages of the virus life cycle. For example, Tat is also involved in HIV transcription and interacts with basic components of the transcription complex. p300, through its ability to respond to changes in cell proliferation and cellular activation, thus may play an important role in HIV replication by coordinating changes in cell cycle progression with the transcription of specific viral and cellular genes.
Acknowledgments
We thank Ms. Donna Gschwend and Ms. Nancy Barrett for their assistance with preparation of the manuscript and figures, Drs. Gregory Enders, Jim Koh, and Ed Harlow for the kinase-deficient Cdc2 expression plasmid, Dr. Xavier Danthinne for assistance with cell cycle analysis, and Drs. Neil Perkins, Jonathan Betts, Maria Athanassiou, and other members of the Nabel laboratory for helpful discussions. This work was supported by grants from the Medical Research Council and Fonds pour la Formation de Chercheurs et l’Aide à la Recerche (E.A.C.), Commission for the Advancement of Young Scientist and Scholars of the University of Zurich, Switzerland (M.O.H.), and postdoctoral fellowships from the University of Michigan Immunopathology Training Program and the Cancer Research Institute (L.K.F.).
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: TNF, tumor necrosis factor; CBP, CREB binding protein; LTR, long terminal repeat; EMSA, electrophoretic mobility shift assay; CAT, chloramphenicol acetyltransferase; CMV, cytomegalovirus; RSV, Rous sarcoma virus.
References
- 1.Levy D N, Fernandes L S, Williams W V, Weiner D B. Cell. 1993;72:541–550. doi: 10.1016/0092-8674(93)90073-y. [DOI] [PubMed] [Google Scholar]
- 2.Rogel M E, Wu L I, Emerman M. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jowett J B M, Planelles V, Poon B, Shah N P, Chen M-L, Chen I S Y. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He J, Chhoe S, Walker R, Di Marzio P, Morgan D O, Landau N R. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Re F, Braaten D, Franke E K, Luban J. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paxton W, Connor R I, Landau N R. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavallee C, Yao X-J, Ladha A, Gottlinger H, Haseltine W A, Cohen E A. J Virol. 1994;68:1926–1934. doi: 10.1128/jvi.68.3.1926-1934.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu Y L, Bennett R P, Wills J W, Gorelick R, Ratner L. J Virol. 1995;69:6873–6879. doi: 10.1128/jvi.69.11.6873-6879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo E, Mammano F, Cohen E A, Gottlinger H G. J Virol. 1995;69:2759–2764. doi: 10.1128/jvi.69.5.2759-2764.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallay P, Stitt V, Mundy C, Oettinger M, Trono D. J Virol. 1996;70:1027–1032. doi: 10.1128/jvi.70.2.1027-1032.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzinger N, Bukrinsky M, Haggerty S, Ragland A, Kervalramani V, Lee M, Gendelman H, Ratno L, Stevenson M, Emerman M. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balliet W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 13.Connor R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 14.Cohen E A, Terwilliger E F, Jalinoos Y, Proulx J, Sodroski J G, Haseltine W A. J AIDS. 1990;3:11–18. [PubMed] [Google Scholar]
- 15.Nabel G, Baltimore D. Nature (London) 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 16.Siebenlist U, Franzoso G, Brown K. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 17.Thanos D, Maniatis T. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 18.Miyamoto S, Verma I M. Adv Cancer Res. 1995;66:255–292. [PubMed] [Google Scholar]
- 19.Perkins N D, Felzien L K, Betts J C, Leung K, Beach D H, Nabel G J. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- 20.Yao X-J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leung K, Nabel G J. Nature (London) 1988;333:776–778. doi: 10.1038/333776a0. [DOI] [PubMed] [Google Scholar]
- 22.Duckett C S, Perkins N D, Kowalik T F, Schmid R M, Huang E S, Baldwin A S, Jr, Nabel G J. Mol Cell Biol. 1993;13:1315–1322. doi: 10.1128/mcb.13.3.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N R. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Z, Perkins N D, Ohno T, Nabel E G, Nabel G J. Nat Med. 1995;1:1052–1056. doi: 10.1038/nm1095-1052. [DOI] [PubMed] [Google Scholar]
- 25.Perkins N D, Agranoff A B, Duckett C S, Nabel G J. J Virol. 1994;68:6820–6823. doi: 10.1128/jvi.68.10.6820-6823.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 27.Potts B J. In: Techniques in HIV Research. Aldovini A, Walker B D, editors. New York: Stockton; 1990. pp. 103–106. [Google Scholar]
- 28.Bartz S R, Rogel M E, Emerman M. J Virol. 1996;70:2324–2331. doi: 10.1128/jvi.70.4.2324-2331.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bielinska A, Krasnow S, Nabel G J. J Virol. 1989;63:4097–4100. doi: 10.1128/jvi.63.9.4097-4100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bohnlein E, Lowenthal J W, Siekevitz M, Ballard D W, Franza B R, Greene W C. Cell. 1988;53:827–836. doi: 10.1016/0092-8674(88)90099-2. [DOI] [PubMed] [Google Scholar]
- 31.Ruben S, Poteat H, Tan T H, Kawakami K, Roeder R, Haseltine W, Rosen C A. Science. 1988;241:89–91. doi: 10.1126/science.2838905. [DOI] [PubMed] [Google Scholar]
- 32.Song C-Z, Loewenstein P M, Green M. J Virol. 1995;69:2907–2911. doi: 10.1128/jvi.69.5.2907-2911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stein R W, Corrigan M, Yaciuk P, Whelan J, Moran E. J Virol. 1990;64:4421–4427. doi: 10.1128/jvi.64.9.4421-4427.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ventura A M, Arens M Q, Srinivasan A, Chinnadurai G. Proc Natl Acad Sci USA. 1990;87:1310–1314. doi: 10.1073/pnas.87.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H-G H, Rikitake Y, Carter M C, Yaciuk P, Abraham S E, Zerler B, Moran E. J Virol. 1993;67:476–488. doi: 10.1128/jvi.67.1.476-488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whyte P, Williamson N M, Harlow E. Cell. 1996;56:67–75. doi: 10.1016/0092-8674(89)90984-7. [DOI] [PubMed] [Google Scholar]
- 37.Parker S F, Felzien L K, Perkins N D, Imperiale M J, Nabel G J. J Virol. 1997;71:2004–2012. doi: 10.1128/jvi.71.3.2004-2012.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 39.Wu B, Woffendin C, Duckett C S, Ohno T, Nabel G J. Proc Natl Acad Sci USA. 1995;92:1480–1484. doi: 10.1073/pnas.92.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emerman M. Curr Biol. 1996;6:1096–1103. doi: 10.1016/s0960-9822(02)00676-0. [DOI] [PubMed] [Google Scholar]
- 41.Griffin G E, Leung K, Folks T M, Kunkel S, Nabel G J. Nature (London) 1989;339:70–73. doi: 10.1038/339070a0. [DOI] [PubMed] [Google Scholar]
- 42.Ayyavoo V, Mahboubi A, Mahalingam S, Ramalingam R, Kudchodkar S, Williams W V, Green D R, Weiner D B. Nat Med. 1997;3:1117–1123. doi: 10.1038/nm1097-1117. [DOI] [PubMed] [Google Scholar]
- 43.Nakajima T, Fukamizu A, Takahashi J, Gage F H, Fisher T, Blenis J, Montminy M. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]
- 44.Kwok R P S, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G E, Green M R, Goodman R H. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 45.Lee J S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 46.Dai P, Akimaru H, Tanaka Y, Hou D, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 47.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Loscher B. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 48.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 49.Bannister A J, Kouzarides T. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janknecht R, Nordheim A. Oncogene. 1996;12:1961–1969. [PubMed] [Google Scholar]
- 51.Bhattacharya S, Eckner R, Grossman S, Oldfield E, Arany Z, D’Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 52.Eckner R, Yao T-P, Oldread E, Livingston D M. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 53.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. J Biol Chem. 1996;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 54.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 55.Gu W, Shi X-L, Roeder R G. Nature (London) 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 56.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 57.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 58.Bannister A J, Kouzarides T. Nature (London) 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]