Abstract
The BRCA2 gene was identified based on its involvement in familial breast cancer. The analysis of its sequence predicts that the gene encodes a protein with 3,418 amino acids but provides very few clues pointing to its biological function. In an attempt to address this question, specific antibodies were prepared that identified the gene product of BRCA2 as a 390-kDa nuclear protein. Furthermore, direct binding of human RAD51 to each of the four single 30-amino acid BRC repeats located at the 5′ portion of exon 11 of BRCA2 was demonstrated. Such an interaction is significant, as BRCA2 and RAD51 can be reciprocally coimmunoprecipitated by each of the individual, specific antibodies and form complexes in vivo. Inferring from the function of RAD51 in DNA repair, human pancreatic cancer cells, Capan-1, expressing truncated BRCA2 were shown to be hypersensitive to methyl methanesulfonate (MMS) treatment. Exogenous expression of wild-type BRCA2, but not BRC-deleted mutants, in Capan-1 cells confers resistance to MMS treatment. These results suggest that the interaction between the BRC repeats of BRCA2 and RAD51 is critical for cellular response to DNA damage caused by MMS.
Keywords: breast cancer, DNA repair
BRCA2 was identified (1, 2) based on its initial mapping to chromosome 13q12–13 by linkage analysis of families with inherited breast cancer not attributed to mutations in BRCA1 (3). Germ-line mutations in BRCA2 account for the same percentage of familial breast cancers as BRCA1 (1). Together, these two breast cancer susceptibility genes are responsible for a large percentage of familial cases. In addition to breast cancer, BRCA2 mutations are also linked to other cancers including ovarian (4, 5), hepatocellular (6), pancreatic (5, 7), and prostate (4–6) tumors. However, mutations in BRCA2, like BRCA1, are mainly found in familial breast cancer but seldom occur in sporadic cases (8, 9). There have been over 100 distinct mutations spanning the sequence of this large gene (Breast Cancer Information Core). The majority of these mutations lead to truncation of the gene product.
BRCA2 has 27 exons and expresses an mRNA 11 kb in size (1). The expression pattern of BRCA2 mRNA is similar to that of BRCA1, with highest levels in the testis, thymus, and ovaries (10). During mouse development, Brca2 mRNA is first detected on embryonic day 7.5, a time of rapid proliferation (11). At the cellular level, expression is regulated in a cell cycle-dependent manner with peak expression of BRCA2 mRNA in S phase (12). These results suggest BRCA2 may have a role in proliferating cells. Using a gene knockout method to create mice with BRCA2 mutations, homozygous mutant mice with BRCA2 truncated from the 5′ half of exon 11 cannot survive embryogenesis (refs. 11, 13, and 14, and our unpublished results), suggesting that Brca2, like Brca1, plays an essential role in early embryonic development. Similar to Brca1, Brca2 heterozygotes are phenotypically normal and fertile. Although they are predicted to be genetically predisposed to cancer, they show no evidence to date of increased tumor formation.
The identification of the BRCA2 gene was accomplished, quickly giving rise to the hope that the function(s) of the gene product would soon become clear. However, BRCA2 presents dilemmas similar to BRCA1, as very little insight in its function has been determined. To address this question systematically, we have identified the cellular BRCA2 protein as a nuclear protein and determined the domain responsible for interactions with human RAD51. Furthermore, BRCA2 apparently has a critical role in response to DNA damage. These results provide a molecular basis that begins to explain how mutations of BRCA2 contribute to carcinogenesis.
MATERIALS AND METHODS
Cell Lines and Tissue Culture.
Human pancreatic cancer lines, Capan-1, bladder cancer line, T24, breast epithelial line, MCF10A, and all breast cancer lines were obtained from American Type Culture Collection and cultured as recommended.
Cloning of the BRCA2 cDNA.
Based on the published sequence, a pair of primers from exon 11 (forward primer, 5′-TCAGACAAGCTCAAAGGTAACAATTATGAATC-3′; reverse primer, 5′-CTTAAGAGCTTAGGTGGCACCACAGTCTCAAT-3′) was used in a PCR using normal human genomic DNA as the template. The predicted 2.5-kb fragment obtained from PCR was used as a probe to isolate additional cDNAs from a human fibroblast library as described (15). cDNA fragments were assembled into full-length BRCA2 cDNA. The cDNA clone was completely sequenced, and mutations were corrected by site-specific mutagenesis using Quickchange kit (Stratagene). Full-length BRCA2 cDNA is capable of expressing a 390-kDa BRCA2 protein when transfected into human cells (data not shown).
Antibody Production and Protein Identification.
To identify and characterize cellular BRCA2 protein, polyclonal antibodies (anti-BRCA2) were obtained by using two bacterially expressed and purified glutathione S-transferase (GST)-BRCA2 fusion proteins, BBA and p400, containing amino acids 188–563 and 2336–2478 as antigens, respectively, to generate polyclonal antisera according to standard procedures (15). Immunoprecipitations for identification of cellular BRCA2 were done as previously described (15).
Localization of BRCA2 Protein.
Biochemical fractionation of cellular proteins was performed as described previously (15). A second approach used was to view a fluorescence-tagged BRCA2 protein in living cells (16). To do so, a mammalian expression plasmid, CHPL, containing either a green fluorescence protein (GFP) alone or a GFP and BRCA2 fusion was constructed and transfected into normal breast epithelial cells, MCF-10A, using a calcium/DNA coprecipitation procedure. Thirty hours after transfection, cells expressing GFP or GFP-BRCA2 were visualized with a Zeiss fluorescence microscope, and the images were captured by using adobe photoshop.
Yeast Two-Hybrid Interaction System.
To determine whether BRCA2 has an intrinsic transactivation domain, full-length (pAS2hFL) or truncated BRCA2 cDNAs [pASNED(927–3418), pASNcoL(1–927), and pASNcoS(1–815)] were fused to the DNA binding domain of Gal4 in the pAS vector and assayed as one hybrid in the yeast Y153 strain. β-Galactosidase units were measured using chlorophenyl-red β-d-galactopyranoside as substrate, and their values were as follows: 232 for the full-length, 406 for the pASNcoL, 91 for pASNcoS, and 0 for pASNED. Therefore, pASNED was used as the bait in two-hybrid screens using a human lymphocyte cDNA library. Detailed screening procedures were described previously (17).
GST Pull-Down Assay and Coimmunoprecipitation.
The construction of bacterial expression plasmids containing GST-BRCA2 fusions and in vitro binding assays were done as previously described (17). [35S]Methionine-labeled proteins were synthesized in vitro by using a TNT kit according to manufacturer’s instructions (Promega). Bacterially expressed and purified RAD51 for the in vitro binding assays was provided by P. Sung (University of Texas Health Science Center at San Antonio). Coimmunoprecipitations were done as described by using cellular lysates labeled with [35S]methionine and antibodies specific for BRCA2 and human RAD51 (kindly provided by S. C. West).
Methyl Methanesulfonate (MMS) Sensitivity Assays.
Human cell lines including Capan-1, T-24, MCF-7, and MCF10A were treated with 0.01% to 0.1% MMS for 40 min. The cells were washed twice with PBS and refed with culture medium. Surviving cells were determined by trypan blue exclusion 48 h after treatment. An expression plasmid, pCNF, which is a pcDNA3 (Invitrogen) derivative with the flag epitope sequences, was used for expressing either full-length or various mutated BRCA2 cDNAs. Transfection of Capan-1 cells with these plasmid DNA was performed by using Lipofectin as provided by GIBCO/BRL. A lacZ reporter gene driven by an SV40 early promoter, pSV2Gal, was included in the transfection as an internal control for transfection efficiency. Forty-eight hours subsequent to transfection, about 1 × 107 cells were harvested for RNA extraction, and 2 × 106 cells were used for measuring β-galactosidase activity as described above. The parallel cultures of transfected or untransfected cells (about 4 × 106) were treated with 0.075% MMS for 40 min, and surviving cells were counted after 8 days.
Detection of mRNA Expression by Reverse Transcription Coupled with PCR.
Total RNA was extracted using the TRI reagent (Molecular Research Center, Cincinnati) and then digested with RNase-free DNase 1 for 2 h at 37°C. An aliquot of RNA was used as template for reverse transcription of cDNA that served as templates for PCR using primers containing Flag sequences and an internal BRCA2 sequence at nucleotide number 464–488. Another aliquot of DNase 1-treated RNA was used directly in PCR for serving as a control for any residual transfected plasmid DNA. The end product of this PCR is a 536-bp DNA fragment.
RESULTS
BRCA2 Is a 390-kDa Nuclear Protein.
To explore potential functions for BRCA2, a full-length cDNA was constructed from four individual cDNAs as shown in Fig. 1A. The predicted 2.5-kb fragment obtained from PCR was used as a probe to isolate additional cDNAs from a human fibroblast library. Two nonoverlapping clones, C49 and C19, were isolated. An additional 1.0-kb DNA fragment encoding the C terminus of BRCA2 was obtained by PCR amplification. Together, these four fragments were completely sequenced and assembled into the full-length, wild-type BRCA2 cDNA, pBSK-BRCA2-FL (Fig. 1A).
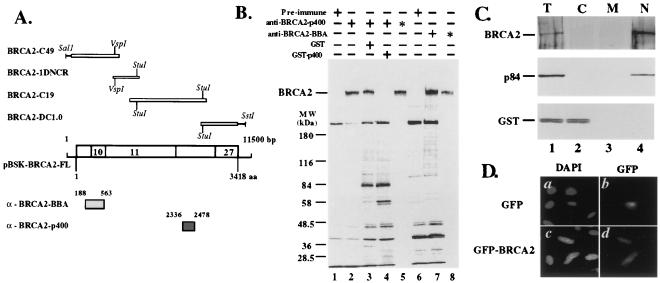
Figure 1.
Identification of BRCA2 protein. (A) cDNAs and GST fusion proteins for antibody preparation. Four overlapping cDNAs were used to assemble the full-length cDNA for the BRCA2 coding sequence. Also shown are the two regions of the BRCA2 cDNA used to construct the GST fusion proteins (boxes). These clones were used to express antigens to raise antibodies to BRCA2. The names for each of the antisera are shown in the left margin. (B) Identification of the BRCA2 protein. The indicated GST fusion proteins were used to immunize mice, and sera were used to precipitate BRCA2 protein from 35S-labeled T-24 cells. The position of the size markers and the BRCA2 protein are indicated in the left margin. The antibodies and competitive antigens used in each experiment are indicated above the lanes. Asterisks indicate double immunoprecipitations with the indicated antibodies. (C) Nuclear localization of BRCA2. The letters above each lane denote total extract (T), cytoplasmic fraction (C), membrane fraction (M), and nuclear fraction (N). The BRCA2 protein is indicated. Also shown are the controls, p84 (a known nuclear matrix protein) (36) and GST (a known cytoplasmic protein. Note, in lane 4, the nuclear partitioning of BRCA2 in this assay. (D) Localization of GFP-BRCA2. MCF10A was transiently transfected with expression plasmids, CHPL, containing either GFP or GFP-BRCA2 fusions. Thirty hours after transfection, cells were fixed and stained with DAPI (a and b). GFP fluorescence is shown (b and d). Note the nuclear localization of GFP-BRCA2 (d).
To identify and characterize cellular BRCA2 protein, polyclonal antibodies (anti-BRCA2) were generated by using two bacterially expressed and purified glutathione S-transferase-BRCA2 fusion proteins, BBA and p400, containing amino acids 188–563 and 2336–2478 as antigens, respectively (Fig. 1A). Anti-BRCA2 serum immunoprecipitates a large protein from T24 human bladder carcinoma cells metabolically labeled with [35S]methionine (Fig. 1B, lanes 2 and 7). The immunoprecipitated protein migrates in SDS/PAGE at a position consistent with a molecular mass of 390 kDa, as predicted by the 3,418-aa BRCA2 sequence (1, 2). Preimmune serum did not detect the protein, and preabsorption of the p400 serum with the original antigen, but not GST, eliminated the 390-kDa protein (Fig. 1B, lanes 1, 4, and 6). Because several proteins in addition to the 390-kDa protein are detected by each of the anti-BRCA2 sera, the immunoprecipitates were then denatured and reimmunoprecipitated. In this more stringent protocol, each anti-BRCA2 serum detects only the 390-kDa protein (Fig. 1B, lanes 5 and 8). These results indicate that the product of the BRCA2 gene is a 390-kDa protein.
The subcellular distribution of proteins is an important factor in ascertaining their function. Using biochemical fractionation of T24 human bladder carcinoma cells and immunoblot analysis, BRCA2 protein was mainly distributed in the nuclear fraction (Fig. 1C, lane 4). Consistent with these data, transient transfection of BRCA2 translationally fused to GFP demonstrates nuclear localization in living, human mammary epithelial cells, MCF10A (Fig. 1D, panel d), indicating that BRCA2 is a nuclear protein.
BRC Motif of BRCA2 Directly Interacts with Human RAD51.
To explore the potential function of BRCA2, one approach is to identify proteins with which it interacts. Because the N-terminal region of BRCA2 contains intrinsic transactivation activity, BRCA2 truncated at amino acid 927 was fused to the Gal4 DNA binding domain as a bait for yeast two-hybrid screening (17). The strongest interacting clone contained a cDNA encoding a protein identical to human RAD51 (18). Deletion mapping combined with yeast two-hybrid assays showed that a specific region referred to as NCB (amino acids 927-1596, Fig. 2A) is sufficient for BRCA2 binding to RAD51 in yeast. Furthermore, an in vitro GST pull-down assay was performed by using a series of GST fusion proteins containing contiguous regions of BRCA2 (Fig. 2B) and in vitro translated, radioactively labeled human RAD51 protein. Consistent with the results from the yeast two-hybrid assays, the GST-NCB fusion (Fig. 2B, lane 5), but not other GST-BRCA2 fusions (Fig. 2B, lanes 2–4 and 6–10), binds RAD51. GST-H3A also shows weak binding upon longer exposure of the gels.
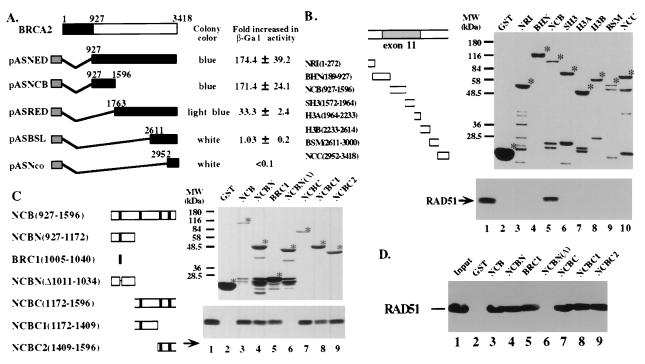
Figure 2.
BRC repeats in BRCA2 mediate binding to RAD51. (A) The indicated regions of BRCA2 (solid rectangles) were fused to the DNA binding domain of GAL4 in pAS1. Rad51 was fused to the activation domain of GAL4 in pGAD10. These plasmids, used to cotransform yeast strain Y153, were scored for colony color and assayed for β-galactosidase activity as indicated. (B) GST pull-down assay for the in vitro binding of BRCA2 to RAD51. (Left) A schematic drawing of full-length BRCA2 and the regions fused to GST for bacterial expression. The stippled area denotes exon 11. (Right) SDS/polyacrylamide gel stained with Coomassie blue for the purification of the GST-BRCA2 proteins after binding to glutathione-Sepharose beads. Asterisks indicate the full-length GST fusion proteins. The GST-BRCA2 beads were then used to bind in vitro transcribed and translated 35S-labeled RAD51. The bound proteins were analyzed by SDS/PAGE and autoradiography. Lane 5 demonstrates that the NCB fragment of BRCA2 binds to RAD51. Lane 1 shows the input RAD51. (C) A specific BRC repeat binds to RAD51. (Left) A schematic drawing of the relative positions of the BRC repeats (solid rectangles) in the NCB fragment of BRCA2 and the various truncated and internally deleted constructs fused to GST. (Right) The purification of the GST-NCB proteins after binding to glutathione beads. Asterisks indicate the full-length fusion proteins. The GST beads were used to bind in vitro translated 35S-labeled RAD51 and analyzed as above. Lanes 3–5 and 7–9 demonstrate that the fragments of BRCA2 shown above each lane bind to Rad51. Lane 1 shows the input RAD51 protein. (D) Bacterially expressed and purified human RAD51 was used as the input protein (lane 1) for binding the GST-NCB fusion proteins described in C and indicated above each lane. Note that the same fragments that bind purified RAD51 also bind in vitro translated RAD51.
It was noted that all of the BRCA2 fragments that bind RAD51 contain multiple copies of the recently recognized BRC repeats conserved in human, mouse, monkey, pig, dog, hamster, and rat Brca2 (19, 20). To determine whether these motifs in NCB(927–1596) contribute to RAD51 binding, GST fused to various regions of NCB that contain none or a variable number of the BRC repeats were expressed (Fig. 2C) and assayed for in vitro binding to RAD51. Interestingly, the presence of three (NCBC), two (NCBC2), or one (NCBN, NCBC1) BRC repeat in the GST fusions is sufficient to mediate binding to RAD51 (Fig. 2C, lanes 3–5 and 7–9). A single, isolated BRC1 repeat fused to GST is sufficient for RAD51 binding (Fig. 2C, lane 5), whereas a fusion with a deletion variant of the BRC repeat, NCBN, and GST alone are both negative for binding (Fig. 2C, lanes 6 and 2, respectively). To further test whether such an interaction is direct, both purified recombinant human RAD51 and GST-BRCA2 fusion proteins were used in the in vitro binding assay. Fig. 2D shows that all GST fusions containing one or more BRC repeats (Fig. 2D, lanes 3–5 and 7–9), but not those lacking repeats (Fig. 2D, lanes 2 and 6), bind purified RAD51. These results indicate that BRCA2 directly binds to RAD51 through interactions with the BRC repeats.
Human RAD51 Forms Complexes with BRCA2 in Vivo.
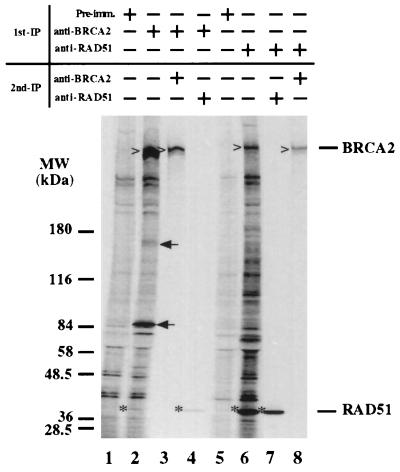
To test whether such interactions between RAD51 and BRCA2 occur in cells, cellular lysates prepared from human bladder carcinoma cell line, T24, labeled with [35S]methionine were used for coimmunoprecipitation by antibodies specific for BRCA2 or RAD51. Fig. 3 shows that, in addition to BRCA2 protein (Fig. 3, carets in lanes 2 and 3), RAD51 is co-immunoprecipitated by the anti-BRCA2 antibodies (Fig. 3). Conversely, RAD51 (Fig. 3) and BRCA2 are also brought down by anti-RAD51 antibody (Fig. 3, carets in lanes 6 and 8). Apparently, not all the molecules of BRCA2 bind to RAD51. Additional specific proteins were also clearly observed in each coimmunoprecipitation (Fig. 3, lanes 2 and 6). It is possible that these proteins are in a complex with BRCA2/RAD51 or associate separately with BRCA2 or RAD51. Combined, these results suggest that BRCA2 and RAD51 form a complex in cells through direct interaction.
Figure 3.
Reciprocal coimmunoprecipitation of BRCA2 and RAD51. [35S]Methionine-labeled T-24 cellular lysates were first precipitated with the indicated antibodies or preimmune serum. After dissolution of the immunoprecipitates, the proteins were precipitated a second time with the indicated antibodies (lanes 3, 4, 7, and 8). The precipitated proteins were then analyzed by SDS/PAGE and autoradiographed. Carets (for BRCA2) and asterisks (for RAD51) mark the bands of interest. Note in lanes 2 and 4 that RAD51 (indicated in the right margin) is precipitated by BRCA2, and conversely, in lanes 6 and 8, that BRCA2 (indicated in the right margin) is coprecipitated by RAD51 antibodies. Arrows indicate unknown proteins specifically coimmunoprecipitated with BRCA2.
Human Pancreatic Cancer Cell Line, Capan-1, Contains Truncated BRCA2 and Is Hypersensitive to MMS Treatment.
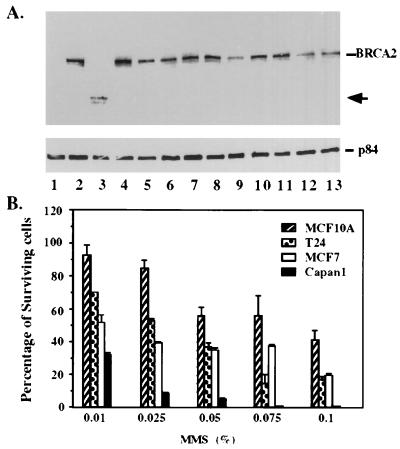
The specific interaction of BRCA2 and RAD51 provides a potential clue that BRCA2 may have a role in response to damage of double-stranded DNA break (21–23). To explore this possibility, cells deficient for BRCA2 were first sought. Among the 13 human cell lines tested, Capan-1, derived from a pancreatic cancer (7), has no detectable full-length BRCA2 protein but, instead, expresses a truncated 224-kDa protein (Fig. 4A, lane 3). This result supports the observation that Capan-1 has a homozygous nonsense mutation at codon 2,003 in the BRCA2 gene (7). To address whether mutation of the BRCA2 gene may impair DNA repair capacity, Capan-1 and three other cell lines including MCF-10A, T24, and MCF-7, which expressed full-length BRCA2 protein, were tested directly for their sensitivity to MMS (24–26). Relative to other cell lines tested, Capan-1 cells seem extremely sensitive to the MMS treatment even at a concentration of 0.075% (Fig. 4B).
Figure 4.
BRCA2-deficient Capan-1 cells are hypersensitive to MMS treatment. (A) Identification of Capan-1 with a truncated BRCA2 protein. Cell lines surveyed for altered BRCA2 protein expression were as follows: lanes 1 and 2, HBL100; lane 3, Capan-1; lane 4, MB361; lane 5, MB175–7; lane 6, MB435; lane 7, MB468; lane 8, MB231; lane 9, BT483; lane 10, SKBR3; lane 11, MCF7; lane 12, ZR75; lane 13, T47D. Full-length BRCA2 is indicated in the right margin. The truncated BRCA2 protein expressed in Capan-1 cells (lane 3) is indicated by the arrow. p84 is a nuclear matrix protein (36) that served as internal control for immunoprecipitations. Cellular lysates were immunoprecipitated by using an equal mixture of anti-p400 and anti-BBA. After separation by SDS/PAGE and blotting, BRCA2 was detected by using rabbit polyclonal anti-BBA antibodies. (B) Hypersensitivity of Capan-1 to MMS treatment. Four different cell lines were treated with various concentrations of MMS (indicated under the x axis), and total surviving cells were counted by trypan blue exclusion assay using hematocytometry. These experiments were repeated three times. Note that, relative to others tested, Capan-1 cells seem to be very sensitive to MMS treatment.
Exogenous Expression of Wild-Type BRCA2 in Capan-1 Confers to MMS Resistance.
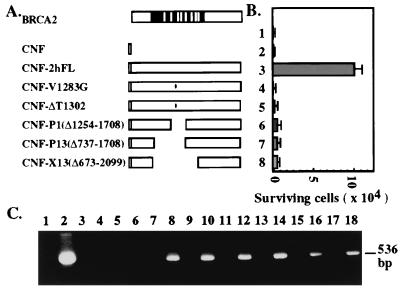
To test whether replacement of the BRCA2 gene in Capan-1 cells rescues its hypersensitivity to MMS, a wild-type, flag-tagged BRCA2 cDNA was exogenously expressed by lipofection. Expression of BRCA2 (Fig. 5 A and B, 2hFL) significantly increased the survival of MMS-treated Capan-1 cells, whereas the vector alone (Fig. 5 A and B) or untransfected cells did not. Because of the limited efficiency of transfections, it is apparent that only a small percentage of the cells can be rescued. The importance of the BRC repeats in response to the DNA repair activity was tested by using a series of BRCA2 mutants deleted for these repeats, including P1, P13, and X13, or familial missense mutations (Breast Cancer Information Core) in region V1283G or ΔT1302 (Fig. 5A). Transfection of these mutant BRCA2 cDNAs into Capan-1 cells have little or no effect on cell survival subsequent to MMS treatment (Fig. 5B). The mRNA products of the transfected BRCA2 cDNAs were expressed at similar levels in these cells (Fig. 5C). These results suggest that the BRCA2–RAD51 interaction by way of the BRC repeats has a role in the cellular response to MMS treatment. It is interesting that the two familial mutations eliminate the ability of BRCA2 to restore resistance to MMS. These two mutations are located between the second (BRC2) and third (BRC3) repeats, not within the repeats themselves. How these mutations abolish its ability to restore resistance to MMS treatment has yet to be determined. It is also noted that the 224-kDa C-terminally truncated BRCA2 protein in Capan-1 cells presumably contains most of the BRC repeats but is apparently unable to confer resistance of cells to MMS treatment. Therefore, it is plausible to suggest that the RAD51-binding BRC repeats are necessary but not sufficient for full function of BRCA2 in its ability to rescue MMS resistance. Other C-terminal regions of BRCA2 may also participate in this process.
Figure 5.
Ectopic expression of BRCA2 in Capan-1 rescues MMS resistance. (A) Schematic diagrams of the BRCA2 cDNAs used to rescue MMS hypersensitivity of Capan-1 cells in B. The black region denotes exon 11, and white bars are BRC repeats. Constructs named with a “CNF” prefix indicate an N-terminal fusion with the flag epitope (open box). Two of the constructs containing missense mutations, V1283G and ΔT1302, in the vicinity of the BRC repeats are indicated. (B) Graphic summary of cell survival in response to 0.075% MMS treatment. Parallel cultures of transfected or untransfected cells (lane 1) were treated with 0.075% of MMS. Each construct was transfected into 2 × 106 cells. The number of the surviving cells was counted by trypan blue exclusion assay. The experiment was repeated three times, and the transfection efficiency was normalized with β-galactosidase activity from the cotransfected pSV2-Gal. The average number and SD were plotted on the y axis. Note transfection of the cells with wild-type BRCA2 (CNF-2hFL) (lane 3), but none of the mutants (V1283G, ΔT1302, P1, P13, and X13, lanes 4–8, respectively) or the vector CNF alone (lane 2) restored resistance to treatment with MMS. (C) Detection of exogenous BRCA2 mRNA in transfected cells. Total cellular RNA from parallel cell cultures transfected with BRCA2 cDNAs after 48 h was extracted and served as templates for RT-PCR as described in Materials and Methods. The expected PCR product, 536 bp, was analyzed in a 1.5% agarose gel. Lane 1, reaction with all PCR reagents except RNA or cDNA template. Lane 2, reaction with 1 ng of CNF-2hFL DNA as template. RNA was used as templates in the PCR reaction of the following odd lanes and cDNA as templates in even lanes. The templates were prepared from either untransfected cells (lanes 3 and 4) or cells transfected with CNF (lanes 5 and 6), CNF-2hFL (lanes 7 and 8), CNF-V1283G (lanes 9 and 10), CNF-ΔT1302 (lanes 11 and 12), CNF-P1 (lanes 13 and 14), CNF-P13 (lanes 15 and 16), and CNF-X13 (lanes 17 and 18).
DISCUSSION
In an attempt to address the potential biological function of BRCA2, we have determined that the gene product of BRCA2 is a 390-kDa nuclear protein. The BRCA2 390-kDa protein directly binds to human RAD51 through its BRC repeats located at the 5′ portion of exon 11. Such an interaction is significant because BRCA2 and RAD51 apparently form a complex in cells. Using human pancreatic cancer cells, Capan-1, that express a truncated BRCA2, ectopic expression of the wild-type BRCA2, but not BRC-deleted mutants in these cells, restores their resistance to MMS treatment. These data suggest that the BRC repeats are important for the BRCA2 and RAD51 interaction, which seems critical for cellular response to DNA damage.
The above findings underscore the importance of the BRC repeats in BRCA2 because they mediate interactions with RAD51, a critical protein for DNA recombination and double-stranded DNA break repair (21–23). Interestingly, the first four BRC repeats found in the 5′ region of exon 11 are the most conserved in mammals (19, 20) and bind RAD51 strongly. The four repeats at the 3′ end of exon 11 are less conserved and do not or weakly bind to RAD51 (Fig. 2 C and D). These results implicate that the RAD51 binding activity of the BRC repeats is conserved among mammals and may reflect the importance of the BRCA2 function in the RAD51-associated pathway. Although the precise stoichiometry of this complex has to be determined, the eight BRC repeats of BRCA2 apparently did not all bind to RAD51 despite sharing sequence similarity. The four less conserved repeats may have other functions. Recently, the C-terminal region of mouse Brca2 was reported to interact with mouse Rad51 in a two-hybrid system (11, 27). This result is clearly different from our finding that specific BRC repeats instead of the C-terminal region of BRCA2 bind to human RAD51. Although the precise reason for this discrepancy remains unclear, there are several possibilities. First, BRC repeat binding might have been overlooked because there was no systematic examination for RAD51 binding activity in other regions of BRCA2. Alternatively, a weak RAD51 binding activity exists at the C-terminal region of mouse Brca2 that is different from humans.
RAD51 is known to be a component in a complex containing other proteins such as RAD52, RAD55, and RAD57 that is involved in DNA recombination or double-stranded DNA break repair (21–23). Apparently, in addition to BRCA2, several other cellular proteins were specifically co-immunoprecipitated by anti-RAD51. However, some of these proteins were not found in the immunoprecipitates with anti-BRCA2. Two proteins with molecular mass of 150 and 85 kDa, respectively, are particularly noticeable in the complex with BRCA2 (Fig. 3, lane 2). Further identification of these proteins will be of great interest because they will probably provide additional information concerning how BRCA2 functions.
The ability of exogenous expression of BRCA2 to rescue the hypersensitivity of BRCA2-deficient cells to MMS indicates a positive role of BRCA2 in the cellular response to DNA damage. It has been reported that mouse Brca2 may have a role in DNA repair as Brca2 −/− mouse embryos are hypersensitive to γ-irradiation (11). Despite the fact that the mechanism of MMS-induced DNA damage is not completely clear, many reports suggested that MMS induces double-stranded DNA breaks and, therefore, mimics γ-irradiation (24–26, 28). This scenario is plausible because the BRCA2 interacting protein, RAD51, is primarily involved in double-stranded DNA break repair. Results obtained by Holt’s group also suggested that Capan-1 is hypersensitive to γ-irradiation (J. Holt, personal communication). However, some reports suggest that MMS serves as an alkylating agent that causes single point mutations (29). If this is the case, BRCA2 may also have a role in the DNA excision repair pathway possibly mediated by different repair proteins. However, the current results did not provide such evidence. Nevertheless, these initial results will allow us to establish an assay for addressing the BRCA2 structure/function relationship in response to DNA damage.
It was reported that BRCA1 also interacts and colocalizes with RAD51 (30), although no BRC repeats are found in BRCA1. Our preliminary results do not support a direct interaction between RAD51 and BRCA1 by either in vitro or yeast two-hybrid binding assays. Homologs of the BRCT domain in BRCA1 are located in various proteins, including a specific BRCA1-interacting protein, BARD1 (31), and others involved in the repair and recombination process (32, 33), suggesting (though not proving) that BRCA1 may have a role in these activities. This line of thinking is particularly revealing because deletion of Brca1 or Brca2 in mice resulted in failure of early embryogenesis, and loss of function of these genes in humans with hereditary breast cancer resulted in unrestricted growth and tumor formation. Further experimental evidence is needed to substantiate this hypothesis.
Whether mutations in either BRCA1 or BRCA2 are threshold events in breast cancer formation remain to be determined. If both BRCA1 and BRCA2 are only involved in the DNA repair process as mutS is in colon cancer (34, 35), exogenous expression of those wild-type genes in cancer cells may only rescue their repair activity and may not be able to suppress their tumorigenicity. This distinction is important for the future treatment for BRCA-related cancers. In yeast cells, where the basic machinery for repair and recombination activities is well studied, a BRCA2 counterpart is not apparent, suggesting that BRCA2 may serve as a positive regulator for this machinery. How BRCA2 positively participates in these pathways in mammalian cells remains to be resolved. It is also quite possible that BRCA1 and BRCA2 may have important function other than response to DNA damage. The large size of BRCA1 or BRCA2 furnishes ample motifs for interactions with other cellular proteins involved in different cellular function. Nevertheless, our results provide a reasonable rationale of how a loss of BRCA2 function may compromise RAD51-mediated DNA repair systems, thereby destabilizing their genomes and leading to cancerous growth.
Acknowledgments
We thank Drs. S. C. West and P. Sung for their antibodies and purified Rad51 protein, D. Jones for antibody production, H. Chew for initial DNA sequence analysis, and J. Holt for his unpublished observations. This work was supported in part by the Susan Komen Breast Cancer Foundation and National Institutes of Health Grant P50-CA58183.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: MMS, methyl methanesulfonate; GFP, green fluorescence protein; GST, glutathione S-transferase.
References
- 1.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, Collins N, Gregory S, Gumbs C, Micklem G, et al. Nature (London) 1995;378:789–792. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 2.Tavtigian S V, Simard J, Rommens J, Couch F, Shattuckeidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppalyonnet D, et al. Nat Genet. 1996;12:333–337. doi: 10.1038/ng0396-333. [DOI] [PubMed] [Google Scholar]
- 3.Wooster R, Neuhausen S L, Mangion J, Quirk Y, Ford D, Collins N, Nguyen K, Seal S, Tran T, Averill D, et al. Science. 1994;265:2088–2090. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsson J, Johannesdottir G, Arason A, Bergthorsson J T, Ingvarsson S, Egilsson V, Barkardottir R B. Am J Hum Genet. 1996;58:749–756. [PMC free article] [PubMed] [Google Scholar]
- 5.Thorlacius S, Olafsdottir G, Tryggvadottir L, Neuhausen S, Jonasson J G, Tavtigian S V, Tulinius H, Ogmundsdottir H M, Eyfjörd J E. Nat Genet. 1996;13:117–119. doi: 10.1038/ng0596-117. [DOI] [PubMed] [Google Scholar]
- 6.Katagiri T, Nakamura Y, Miki Y. Cancer Res. 1996;56:4575–4577. [PubMed] [Google Scholar]
- 7.Goggins M, Schutte M, Lu J, Moskaluk C A, Weinstein C L, Petersen G M, Yeo C J, Jackson C E, Lynch H T, Hruban R H, Kern S E. Cancer Res. 1996;56:5360–5364. [PubMed] [Google Scholar]
- 8.Futreal P A, Liu Q, Shattuck-Eidens D, Cochran C, Harshman K, Tavtigian S, Bennett L M, Haugen-Strano A, Swensen J, Miki Y, et al. Science. 1994;266:120–122. doi: 10.1126/science.7939630. [DOI] [PubMed] [Google Scholar]
- 9.Miki Y, Katagiri T, Kasumi F, Yoshimoto T, Nakamura Y. Nat Genet. 1996;13:245–247. doi: 10.1038/ng0696-245. [DOI] [PubMed] [Google Scholar]
- 10.Rajan J V, Marquis S T, Gardner H P, Chodosh L A. Dev Biol. 1997;184:385–401. doi: 10.1006/dbio.1997.8526. [DOI] [PubMed] [Google Scholar]
- 11.Sharan S K, Morimatsu M, Albrecht U, Lim D S, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A. Nature (London) 1997;386:804–810. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 12.Vaughn J P, Cirisano F D, Huper G, Berchuck A, Futreal P A, Marks J R, Iglehart J D. Cancer Res. 1996;56:4590–4594. [PubMed] [Google Scholar]
- 13.Ludwig T, Chapman D L, Papaioannou V E, Efstratiadis A. Genes Dev. 1997;11:1226–1241. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki A, de la Pompa J L, Hakem R, Elia A, Yoshida R, Mo R, Nishina H, Chuang T, Wakeham A, Itie A, Koo W, Billia P, Ho A, Fukumoto M, Hui C C, Mak T W. Genes Dev. 1997;11:1242–1252. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Riley D J, Chen P-L, Lee W-H. Mol Cell Biol. 1997;17:6049–6056. doi: 10.1128/mcb.17.10.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Sharp Z D, Lee W-H. J Biol Chem. 1997;272:24081–24087. doi: 10.1074/jbc.272.38.24081. [DOI] [PubMed] [Google Scholar]
- 17.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 18.Shinohara A, Ogawa H, Matsuda Y, Ushio N, Ikeo K, Ogawa T. Nat Genet. 1993;4:239–243. doi: 10.1038/ng0793-239. [DOI] [PubMed] [Google Scholar]
- 19.Bork P, Blomberg N, Nilges M. Nat Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 20.Bignell G, Micklem G, Stratton M R, Ashworth A, Wooster R. Hum Mol Genet. 1997;6:53–58. doi: 10.1093/hmg/6.1.53. [DOI] [PubMed] [Google Scholar]
- 21.Benson F E, Stasiak A, West S C. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donovan J W, Milne G T, Weaver D T. Genes Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- 23.Hays S L, Firmenich A A, Berg P. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haaf T, Golub E I, Reddy G, Radding C M, Ward D C. Proc Natl Acad Sci USA. 1995;92:2298–2302. doi: 10.1073/pnas.92.6.2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinohara A, Ogawa H, Ogawa T. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 26.Morita T, Yoshimura Y, Yamamoto A, Murata K, Mori M, Yamamoto H. Proc Natl Acad Sci USA. 1993;90:6577–6580. doi: 10.1073/pnas.90.14.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuta R, LaSalle J M, Cheng H-L, Shinohara A, Ogawa H, Copeland N, Jenkins N A, Lalande M, Alt F W. Proc Natl Acad Sci USA. 1997;94:6927–6932. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pati D, Keller C, Groudine M, Plon S E. Mol Cell Biol. 1997;17:3037–3046. doi: 10.1128/mcb.17.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bezzubova O, Silbergleit A, Yamaguchi-Iwai Y, Takeda S, Buerstedde J M. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 30.Scully R, Chen J J, Plug A, Xiao Y H, Weaver D, Feunteun J, Ashley T, Livingston D M. Cell. 1997;88:265–275. doi: 10.1016/s0092-8674(00)81847-4. [DOI] [PubMed] [Google Scholar]
- 31.Wu L J C, Wang Z W, Tsan J T, Spillman M A, Phung A, Xu X L, Yang M C W, Hwang L Y, Bowcock A M, Baer R. Nat Genet. 1996;14:430–440. doi: 10.1038/ng1296-430. [DOI] [PubMed] [Google Scholar]
- 32.Koonin E V, Altschul S F, Bork P. Nat Genet. 1996;13:266–267. doi: 10.1038/ng0796-266. [DOI] [PubMed] [Google Scholar]
- 33.Bork P, Hofmann K, Bucher P, Neuwald A F, Altschul S F, Koonin E V. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 34.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 35.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 36.Durfee T, Mancini M, Jones D, Elledge S J, Lee W-H. J Cell Biol. 1994;127:609–622. doi: 10.1083/jcb.127.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]