Abstract
Virulence genes of Agrobacterium tumefaciens are under the control of positive and negative transcriptional regulators. We found that the transcriptional regulator Ros controls expression of the plant oncogene ipt, which encodes isopentenyl transferase, in A. tumefaciens. This enzyme is involved in biosynthesis of the plant growth hormone cytokinin in the host plant. An ipt promoter∷cat reporter gene fusion showed a 10-fold increase in ipt promoter activity in A. tumefaciens ros mutant strains when compared with wild type. Also, increased levels (10- to 20-fold) of isopentenyl adenosine, the product of the reaction catalyzed by isopentenyl transferase, were detected in ros mutant strains. In vitro studies using purified Ros showed it binds directly to the ipt promoter. Analysis of the deduced Ros amino acid sequence identified a novel type of C2H2 zinc finger. In Ros the peptide loop spacing of the zinc finger is 9 amino acids as opposed to the invariant 12 amino acids in the classical C2H2 motif. Site-directed mutagenesis of Cys-82 and His-92 in this motif showed that these residues are essential for Zn2+ and DNA binding activities of Ros. The existence of such a regulator in Agrobacterium may be due to horizontal interkingdom retrotransfer of the ros gene from plant to bacteria.
Keywords: cytokinin
Tumorigenesis on plants by Agrobacterium tumefaciens is caused by horizontally transferred T-DNA genes that encode products that catalyze the formation of plant growth hormones indoleacetic acid and cytokinin (trans-ribosylzeatin) in the transformed plant cell (1). The presence of elevated local levels of these hormones results in neoplastic transformation at the site of A. tumefaciens infection culminating in the formation of crown gall tumors. The T-DNA is a 25-kb DNA region located on the resident 200-kb Ti plasmid. Situated near the T-DNA is a 29-kb vir regulon (2) containing genes that confer T-DNA processing and interkingdom DNA transfer properties on A. tumefaciens (3–6). Expression of the vir genes is under positive control by a two-component regulatory system VirA/VirG (for review, see ref. 7) and negative control by Ros (8).
Ros is encoded by the chromosomal gene ros (for rough outer surface) and is a repressor protein targeting the operator of the divergent virC and virD promoters (9, 10). The Ros binding site on the virC/D operator was determined by DNase I footprinting and contains a 9-bp inverted repeat designated the “ros box” (11). virC and virD are required for T-DNA processing, an activity without which virulence is not conferred on A. tumefaciens. Mutations in ros result in constitutive expression of virC and virD in the absence of induction by plant phenolic compounds (8). Although Ros does not appear to affect virulence per se, its absence increases the appearance of T-DNA intermediates in A. tumefaciens as a result of the derepression of virC and virD operons (8).
The ros gene was isolated from an A. tumefaciens genomic library and localized to an 825-bp fragment (12). Nucleotide sequencing of this fragment identified a single ORF consisting of 426 bp, coding for a protein of 142 amino acids. From the deduced amino acid sequence Ros is a relatively small protein of 15.5 kDa with a pI of 7.1. The amino terminus of the protein is negatively charged and contains more hydrophobic amino acid residues than the positively charged carboxyl terminus (12).
By using the published Ros sequence, ros homologs have been recently identified in Agrobacterium radiobacter (rosAR), Sinorhizobium meliloti (mucR and ORF2) and Rhizobium etli (rosR) (13–18). rosAR is required for the expression of the exoY glycosyltransferase gene, which is involved in one of the early steps in exopolysaccharide synthesis (14), and thus, is a ros homolog capable of positive transcriptional regulation. rosR contributes to nodulation competitiveness (15). mucR is involved in regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan (16) and binds to a short DNA region located upstream of the mucR coding region (17). ORF2 is located upstream of the syrB coding region. It was hypothesized that the gene product of this ORF2 may interfere with the expression or inhibit the activity of SyrB (18).
Previous studies on the Ros protein in our laboratory showed that the carboxyl-terminal half of Ros contains an amino acid sequence that has some resemblance to C2H2 type zinc fingers (12). The presence of such a sequence in a protein of prokaryotic origin is of great interest because zinc fingers were primarily found in DNA binding proteins of eukaryotic origin (19, 20). However, Ros differs from the classical C2H2 motif in its peptide loop length, 9 amino acids as opposed to the previously invariant 12 amino acids. The Ros peptide loop is properly basic and contains two phenylalanine and leucine residues at positions similar to those found in many typical zinc finger proteins of this type, such as the transcription factor TFIIIA from Xenopus (21–23). Because a shorter peptide loop would affect the structure of the zinc finger, it was important to show that this motif is indeed essential for the DNA binding ability of Ros.
We have extended our studies to show that Ros indeed contains a bona fide zinc finger that is essential for DNA binding. We also show that Ros represses the expression of the T-DNA-encoded oncogene ipt in A. tumefaciens.
MATERIALS AND METHODS
Site-Directed Mutagenesis.
Site-directed mutagenesis was performed by asymmetric PCR (24). Mutant primers complementary to the zinc finger sequence were synthesized with base mismatches to change His-92 or Cys-82 into alanine. The primers are as follows: 5′ primer RGP5, TACTCATATGACGGAAACTGCATA; 3′ primer RGP3, GCAAAGATCCCAATATGCTGCCAAG; Cys-82 primer C82A, CTTGAACGAGCCACCAGCTTCCAA; His-92 primer H92A, TCGCTCAAACGCCTCCTGACG. Altered nucleotides are underlined. Plasmid pUCD4401 bearing the ros gene under the control of the φ10 promoter of phage T7 in expression vector pET-3a (25) was used as the template. PCR products were verified by sequencing and used to generate full-length ros, with and without the amino acid substitution, and cloned into pET-3a.
Protein Purification.
Proteins were expressed in Escherichia coli BL21 DE3 by induction with isopropyl β-d-thiogalactoside (1 mM, final concentration) for 3 h. Expressed wild-type and mutant Ros proteins were purified from inclusion bodies that were solubilized in 0.1 M NaHCO3/0.1% SDS, at 23°C. Without stirring, the solution was dialyzed overnight against 400 vol of dialysis buffer A (10 mM Tris⋅HCl, pH 8.0/100 mM KCl/100 μM ZnCl2/5% glycerol). Dialysis was performed twice more with stirring against dialysis buffer B (10 mM Tris⋅HCl, pH 8.0/100 mM KCl/5% glycerol/1 mM EDTA). The latter procedure caused precipitation of the protein and the precipitate was collected by centrifugation (13,000 × g, 15 min, 4°C). The precipitated protein was solubilized in 2 M KCl and then gradually diluted by adding storage buffer (20 mM Tris⋅HCl, pH 8.0/25% glycerol) to a final salt concentration of 0.1 M. Protein purity was determined electrophoretically on 12% SDS/PAGE gels.
Electrophoretic Mobility Shift Assay.
A 210-bp DNA fragment containing the divergent virC/virD promoters and a 341-bp ipt promoter fragment were labeled with [α-32P]dATP and used for gel shift assays. Labeled probes (0.1 μg) were incubated with purified Ros protein (5 μg) in binding buffer [10 mM Tris⋅HCl, pH 8.0/BSA (500 ng/ml)/100 mM KCl/5% glycerol/sonicated salmon sperm DNA at a 500-fold excess of the labeled probe] to a total volume of 20 μl for 20 min at 23°C. The protein/DNA reaction products were resolved on a nondenaturing 5% polyacrylamide gel at 5 V/cm, and the bound complex was visualized by autoradiography.
DNase I Footprinting.
The ipt promoter region used in this study is contained in a 341-bp HindIII–PstI T-DNA fragment. The recessed 3′ terminus of the HindIII site was filled by using the Klenow fragment of DNA polymerase I and [α- 32P]dATP. Ros binding and footprinting conditions were as described (11).
Atomic Absorption Spectroscopy.
Atomic absorption spectrometry was performed in the Division of Agriculture and Natural Resources analytical laboratory at University of California, Davis, according to the procedure of Tchou et al. (26) with a Perkin–Elmer spectrometer model 2380 on protein samples containing 0.5–0.9 mg. ZnCl2 solutions determined for zinc content by flame spectroscopy were used as the standard.
Chloramphenicol Acetyltransferase (CAT) Assay.
CAT activity was assayed by the procedure of Shaw (27). A. tumefaciens was grown overnight at 28°C in medium 523 (28) with vigorous shaking (200 revolutions per min), and cells were harvested by centrifugation (7,000 × g, 5 min, 4°C) and lysed by sonication for 1 min at setting 50 using a Microson Ultrasonic cell disrupter (Heat Systems Ultrasonics). Cell debris was removed by centrifugation (9,000 × g, 5 min, 4°C), and the cell-free supernatant was assayed for specific CAT activity. Protein concentrations were determined by BCA protein assay method (Pierce).
Isopentenyl Adenosine (IPA) Assay.
IPA levels were measured by using the Phytodetek enzyme immunoassay kit (product 8013) from Idetek (Sunnyvale, CA). Strains to be assayed were pregrown in medium 523 broth at 28°C. Cultures were grown to an OD600 of ∼0.7, centrifuged, washed in AB minimal medium, and subcultured at 28°C in fresh AB minimal medium (29). Overnight cultures were lysed as described above, and the cell debris removed by centrifugation (9,000 × g, 5 min, 4°C). The cell-free supernatant was assayed for IPA content by the method outlined in the kit. Protein concentration was determined as described above.
RESULTS
Cys-82 and His-92 Are Essential for DNA Binding.
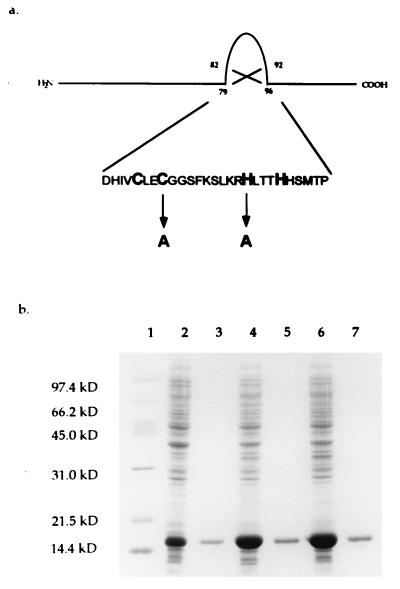
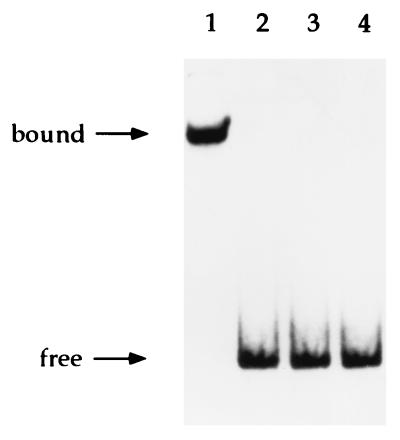
Our previous studies on the amino acid sequence of Ros revealed the presence of a zinc finger motif similar to the C2H2 class (12). However, we still lacked evidence showing that this motif is essential for DNA binding. Because cysteine and histidine residues of known eukaryotic C2H2 zinc fingers have been shown to be essential for binding DNA and Zn2+ (19, 20, 30), we replaced Cys-82 with Ala (RosC82A) and His-92 with Ala (RosH92A) in Ros by site-directed mutagenesis (Fig. 1a). The wild-type and mutant proteins were overexpressed individually in E. coli and purified to apparent homogeneity (Fig. 1b). The purified proteins were analyzed for DNA binding activity. A change in either Cys-82 or His-92 caused the mutant proteins to lose their DNA binding activity at the virC/D operator (Fig. 2), indicating that these residues are essential for DNA binding.
Figure 1.
Location and deduced primary sequence of the zinc finger motif of the Ros protein and sites of amino acid substitution. The nucleotide sequence published previously (12) has been deposited in GenBank (accession no. M65201). (a) The mutations placed at positions 82 and 92 with alanine substitutions are indicated below the wild-type amino acid sequence. (b) Expression and purification of wild-type and mutant Ros proteins. Proteins from total cellular extracts and purification were analyzed in a SDS/12% polyacrylamide gel and stained with Coomassie blue. Lanes: 1, molecular weight standards; 2, 4, and 6, total cellular extracts (∼20 μg per lane) of E. coli BL21(DE3) containing pUCD4401, pUCD4401C82A, and pUCD4401H92A, respectively [these plasmids are the respective wild-type and mutant ros genes cloned into pET-3a (25)]; 3, 5, and 7, approximately 2 μg of purified Ros, RosC82A, and RosH92A, respectively.
Figure 2.
Gel mobility shift analysis of the binding of Ros and Ros mutant proteins to the promoters of virC and virD operons. Lanes: 1–3, wild-type Ros protein and C82A and H92A mutant Ros proteins, respectively; 4, target DNA fragment only. A 210-bp PvirC/virD fragment labeled with [α-32P]dATP was used as a probe.
Cys-82 and His-92 Are Involved in Binding a Single Zinc Ion.
To determine whether Ros contains zinc and whether or not the above mutations affect the zinc binding activity, atomic absorption spectroscopy was used to quantify Zn2+ bound by each of the purified wild-type and mutant Ros proteins. The results obtained show that a nearly 1:1 stoichiometric relationship exists between wild-type Ros and Zn2+, whereas negligible amounts of Zn2+ were associated with the mutant proteins (Table 1). These results strongly indicate that Ros contains a single Zn2+ ion presumably sequestered in the C2H2 finger because changing either residue Cys-82 or His-92 results in the loss of Zn2+ sequestration.
Table 1.
Atomic absorption spectrophotometric analysis of Zn2+ bound by wild-type and mutant Ros proteins
| Protein | Amino Acid substitution | Zn2+/protein molar ratio |
|---|---|---|
| Wild-type Ros | None | 0.81 |
| Mutant Ros | C82A | 0.03 |
| Mutant Ros | H92A | 0.01 |
Identification of a Gene Regulated by A. tumefaciens Ros.
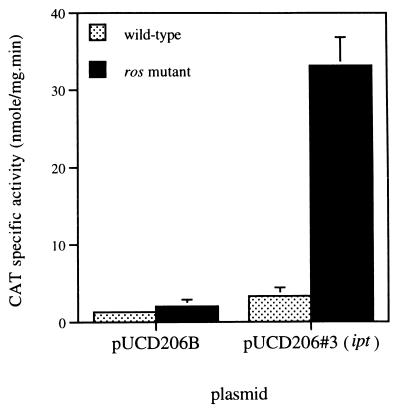
To identify genes that are Ros-regulated and involved in tumorigenesis, we concentrated on genes present on the T-DNA. The T-DNA was isolated, partially digested with SalI, and cloned in front of a promoterless cat gene in the promoter probe vector pUCD206B (9). CAT activities in A. tumefaciens wild-type and ros− mutants were compared. Clones showing elevated CAT activity in ros mutants were selected. The nucleic acid sequences of two cloned fragments were determined and screened in the GenBank database. Both fragments showed a 95% identity to the promoter region of the A. tumefaciens plant oncogene ipt. The ipt gene encodes the enzyme isopentenyl transferase (IPT) that is involved in one of the initial steps in the biosynthesis of cytokinin (31). IPT catalyzes the addition of dimethylallylpyrophosphate to the N6 position of AMP, resulting in the cytokinin ribotide, N6-(Δ2-isopentenyl)AMP. This is subsequently modified to other types of cytokinin ribotides, ribosides, and free bases, including zeatin and dihydrozeatin (1, 32). A 10-fold increase in ipt promoter expression was observed in the ros mutant when compared with wild-type A. tumefaciens (Fig. 3), indicating either a direct or indirect negative regulatory role for Ros on ipt expression.
Figure 3.
ipt promoter expression in wild-type (NT1) and mutant ros (NTR1) strains. pUCD206#3 contains the ipt promoter and pUCD206B is the negative control. Results shown are the average of three determinations.
The Ros Protein Binds Directly to the ipt Promoter.
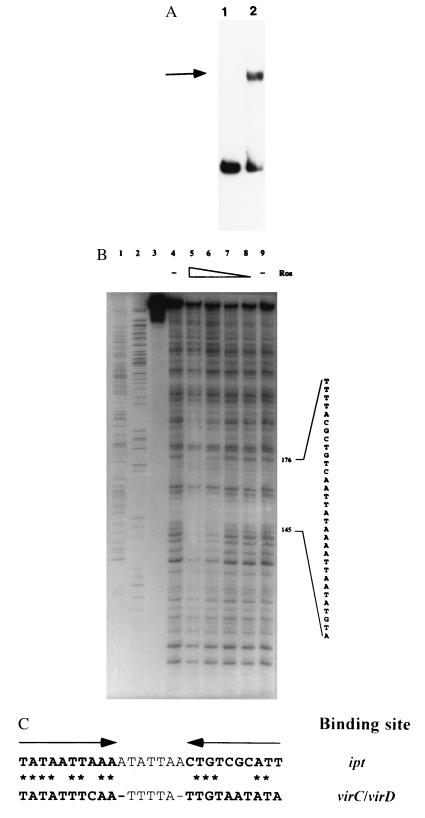
To determine whether Ros acts directly on the ipt promoter, we carried out in vitro gel mobility shift and DNase I footprinting assays. Results from these experiments demonstrate that Ros binds directly to the 5′ nontranscribed region of the ipt gene (Fig. 4 A and B). The region protected by Ros from DNase I digestion covers approximately 40 bp located 485 bp upstream of the ATG start codon and 442 bp relative to the ipt transcriptional start site in plants (33). The ipt transcriptional start site in A. tumefaciens has not been determined. Within the protected region is a sequence similar to the inverted repeat observed in the virC/virD operator (Fig. 4C). However, the sequence on the ipt promoter does not form an inverted repeat. In the ipt left half binding site, 8 of 10 bases match the virC/virD sequence but only 5 of 10 bases match in the right half binding sites. Hence, the specificity of Ros binding to operators appears somewhat flexible.
Figure 4.
(A) Gel mobility shift analysis of the binding of Ros protein to the promoter of the T-DNA ipt gene. Lanes: 1, promoter fragment of the ipt gene; 2, same fragment incubated with purified Ros protein. The shifted band is indicated by the arrow. (B) DNase I footprint analysis of the interaction of Ros and the ipt promoter fragment. Lanes: 1 and 2, A and G sequencing reaction products, respectively; 3–9, approximately 4 ng of radiolabeled DNA per lane; 3, free probe; 4 and 9, ipt promoter fragment treated with DNase I only; 5–8, ipt promoter fragment incubated with 10, 5, 2.5, and 1.25 μg of Ros protein, respectively, before treatment with DNase I. Protection against DNase I digestion by Ros was seen between bases 145 and 176 with respect to the ipt sequence (GenBank accession no. X00639). (C) Comparison of virC/D and ipt promoter binding sites for Ros. The arrows above the sequences show the position of the inverted repeat within the virC/D promoter. Asterisks indicate identical base-pair matches between the ipt and virC/D Ros binding sites.
IPA Levels in ros Mutants.
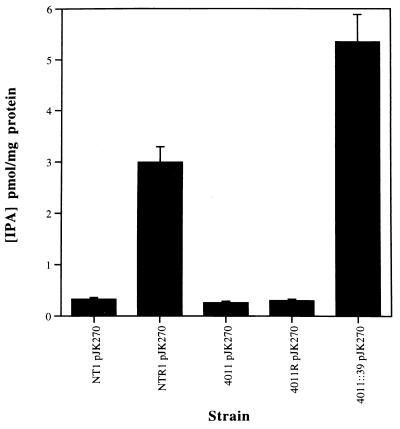
An enzyme immunoassay was used for the quantitative determination of IPA, the product of the reaction catalyzed by IPT. Five A. tumefaciens strains were assayed: the ros mutants NTR1, LBA4011R, and LBA4011∷39 and their respective parents, NT1 and LBA4011. Each strain contained the Ti plasmid pJK270 (34), which carries the ipt gene on its T-DNA. NTR1 and LBA4011R are spontaneous ros mutants described by Steck et al. (35), and LBA4011∷39 is a defined ros mutant described by Cooley et al. (12). The IPA enzyme immunoassay was carried out on cell-free supernatant from each strain. As shown in Fig. 5, a 10-fold increase in IPA production was observed in NTR1 (pJK270) and a 20-fold increase was seen in LBA4011∷39 (pJK270). No significant increase in IPA production was observed for LBA4011R (pJK270). This may be due to the nature of the spontaneous mutation in LBA4011R. IPA was not detected in strains without pJK270 (data not shown). The increase in IPA production in NTR1 (pJK270) and LBA4011∷39 (pJK270) supports the conclusion that Ros negatively regulates expression of the ipt gene.
Figure 5.
IPA production in Agrobacterium parent strains (NT1 pJK270, LBA4011 pJK270) and ros mutants (NTR1 pJK270, LBA4011R pJK270, LBA4011∷39 pJK270). Results are the average of three determinations.
DISCUSSION
A large number of eukaryotic transcriptional regulatory proteins contain zinc fingers composed of a quartet of cysteines and histidines binding a single zinc ion (19, 20, 30). The classical C2H2 zinc finger consists of an invariant 12-amino acid loop connecting the cysteine pair to the histidine pair and thereby forming a DNA binding finger in between. In some proteins this motif is repeated several times resulting in as many as 37 tandem fingers (36). The C2H2 type zinc finger motif was first identified in the Xenopus laevis transcription factor TFIIIA (21–23), and proteins bearing this motif have since been shown to occur in humans (37), plants (38–41), and fungi (23, 42). To date 10 classes of zinc finger proteins have been identified (19, 20). Zinc finger proteins are predominantly associated with controlling transcription in higher eukaryotes; however, it is now clear that they also exist in prokaryotes. Thus, A. tumefaciens Ros and its homologs (RosAR, MucR, and RosR) form a distinct subfamily of the C2H2 type zinc finger (Fig. 6).
Figure 6.
Amino acid sequence alignment of Ros homologs. Alignment of Ros (ref. 12, GenBank accession no. Q04152), RosAR (ref. 13, P55324), MucR (ref. 16, P55323), RosR (ref. 15, U61146), and RmORF2 (17) was performed with the pileup program (GCG Software, ref. 43). Identical and conserved residues with respect to Ros are shaded. Residues similar to the classical C2H2 zinc finger motif are in boldface type.
Ros is a prokaryotic zinc finger protein with a peptide loop that contains 9 amino acids. The difference in the peptide loop length of Ros and classical C2H2 zinc fingers is an important distinction because it must affect the structure of the finger. Amino acid sequence indicates that the Ros finger has a shorter second β sheet and recognition helix. We investigated the role that this short finger has on DNA and Zn2+ binding ability of Ros. We found that an intact zinc finger is essential for Ros activity, because replacement of either Cys-82 or His-92 with Ala results in the loss of its ability to bind DNA and Zn2+. In either RosC82A or RosH92A, only three ligands are available preventing the required tetrahedral coordination of Zn2+ within the protein (20). This would cause a disruption in the structural integrity of the zinc finger as Zn2+ allows Ros to form a small functional protein domain or “finger” that interacts with the DNA in a sequence specific fashion. In the A. radiobacter Ros homolog, substitution mutagenesis of the two cysteine residues (C79S, C82S) in the zinc finger motif showed them to be essential for complementation of the exopolysaccharide synthesis defect of ros mutant strains (44).
A. tumefaciens may have acquired the ros gene from an eukaryotic source because this organism is well known for its unique ability to transfer and incorporate foreign DNA into plants, and such an event might have taken place in reverse during the course of evolution. All of the ros homologs identified thus far come from organisms that are very closely related to A. tumefaciens and all are involved in plant–microbe interactions, e.g., crown gall formation, symbiosis, and nitrogen fixation. Such a close association between plant and microbe may have favorably contributed to gene retrotransfer from the host to the invading organism. Retrotransfer of genes has been demonstrated between E. coli strains and is thought to occur among distinct organisms (45, 46).
The current model for cytokinin biosynthesis results in the addition of dimethylallylpyrophosphate to the N6 position of AMP by IPT, resulting in the cytokinin ribotide, N6-(Δ2-isopentenyl)AMP, which can be modified to yield various types of cytokinin ribotides, ribosides, and free bases, including zeatin and dihydrozeatin. Despite similarity of function between the endogenous plant IPT and bacterial IPT, there have been no reports of similarity of plant DNA sequences homologous to the bacterial ipt gene (32). Thus Ros may only regulate bacterial ipt expression. This offers an alternative explanation to the origins of Ros in Agrobacterium. The fact that ipt is expressed in a ros− background indicates that the ipt promoter is recognized by the Agrobacterium transcriptional machinery. The presence of eukaryotic promoter elements on the ipt promoter may not necessitate a regulator of eukaryotic origin. Indeed it is not clear whether the same ipt promoter operates in both backgrounds. Ros may simply be a regulator of prokaryotic origin that always had or, during the course of evolution, acquired the ability to regulate ipt expression via the presence of its zinc finger motif.
Whatever the origin of Ros or its zinc finger, it is clear that it binds to the ipt promoter. DNase I footprint experiments showed that Ros protects a region of approximately 40 bp located 485 bp upstream of the ATG start codon. Although the location of this binding site is distant from the Ipt coding region, regulatory elements controlling ipt expression have been found at a similar distance. The existence of an upstream segment between positions −442 and −408 of the Pipt ATG codon that is required for maximal promoter function in roots has been reported (47). Within the protected region is a sequence similar to the virC/D Ros binding site, the “ros box” (11). However, where the virC/D binding site forms a near perfect inverted repeat the ipt binding site does not. Only the left half binding site shows significant homology with 8 of 10 bases matching. The right half binding site has only 5 of 10 bases matching. This may explain why complete protection by Ros is not observed in the DNase I footprint experiment, at least in vitro. However, we have not shown how tightly Ros binds to the ipt operator in vitro. The requirement of the right half binding site for Ros binding has not been investigated for the virC/D promoter. Also the left half binding site may constitute the entire Ros binding site. The DNA binding specificity of Ros is further complicated by the finding that the inside-out ros-box motif upstream of the A. radiobacter ros promoter is not required for autoregulation of ros expression and the remaining upstream sequence, 300 bp long, contains no obvious promoter motifs (44). Therefore, the exact DNA sequence(s) that Ros binds to remains elusive.
Increased levels of ipt expression and, hence, IPA production found in various ros mutants supports the regulation of ipt by Ros. Increased IPA levels may be expected to increase tumorigenesis; however, based on tumor size, this is not the case (8). It is likely that once the T-DNA is inside the plant, the function of Ros is superseded by the plants endogenous regulatory machinery.
Ros, therefore, appears essential for negative regulation of genes involved not only in the processing of T-DNA, by repression of vir gene expression in A. tumefaciens in the absence of association with its host plant, but also in ensuring that the cytokinin biosynthetic gene (ipt) is not expressed or at most only expressed at low levels in the bacterial cell. From the pleiotropic nature of the ros mutation, it is clear that regulation by Ros is not confined to the regulation of virC/D and ipt expression (9). The Ros subfamily of C2H2 zinc finger proteins are involved in diverse functions (13–18), demonstrating their global nature as both positive and negative regulators of transcription in prokaryotes.
Acknowledgments
We thank Donald Phillips, Michael Syvanen, and Patricia Zambryski for reviewing the manuscript. This work was supported by National Institutes of Health Grant GM45550 from the National Institute of General Medical Sciences.
ABBREVIATIONS
- IPA
isopentenyl adenosine
- T-DNA
transferred DNA
- CAT
chloramphenicol acetyltransferase
References
- 1.Morris R O. Annu Rev Plant Physiol. 1986;37:509–538. [Google Scholar]
- 2.Rogowsky P M, Powell B S, Shirasu K, Lin T S, Morel P, Zyprian E M, Steck T R, Kado C I. Plasmid. 1990;23:85–106. doi: 10.1016/0147-619x(90)90028-b. [DOI] [PubMed] [Google Scholar]
- 3.Gelvin S B, Filichkin S A. In: Molecular Mechanisms of Bacterial Virulence. Kado C I, Crosa J H, editors. Dordrecht, The Netherlands: Kluwer; 1994. pp. 207–222. [Google Scholar]
- 4.Tinland B, Hohn B. In: Virus Strategies, Molecular Biology and Pathogenesis. Doerfler W, Böhm P, editors. Berlin: VCH; 1995. pp. 349–359. [Google Scholar]
- 5.Hooykaas P J J, Beijersbergen A G M. Annu Rev Phytopathol. 1994;32:157–179. [Google Scholar]
- 6.Zambryski P C. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:465–490. [Google Scholar]
- 7.Winans S C. Microbiol Rev. 1992;56:12–31. doi: 10.1128/mr.56.1.12-31.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Close T J, Rogowsky P M, Kado C I, Winans S C, Yanofsky M F, Nester E W. J Bacteriol. 1987;169:5113–5118. doi: 10.1128/jb.169.11.5113-5118.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Close T J, Tait R C, Kado C I. J Bacteriol. 1985;164:774–781. doi: 10.1128/jb.164.2.774-781.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tait R C, Kado C I. Mol Microbiol. 1988;2:385–392. doi: 10.1111/j.1365-2958.1988.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 11.D’Souza-Ault M R, Cooley M B, Kado C I. J Bacteriol. 1993;175:3486–3490. doi: 10.1128/jb.175.11.3486-3490.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooley M B, D’Souza M R, Kado C I. J Bacteriol. 1991;173:2608–2616. doi: 10.1128/jb.173.8.2608-2616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brightwell G, Hussain H, Tiburtius A, Yoeman K H, Johnston A W B. Mol Plant-Microbe Interact. 1995;8:747–754. doi: 10.1094/mpmi-8-0747. [DOI] [PubMed] [Google Scholar]
- 14.Tiburtius A, DeLuca N G, Hussain H, Johnston A W B. Microbiology. 1996;142:2621–2629. doi: 10.1099/00221287-142-9-2621. [DOI] [PubMed] [Google Scholar]
- 15.Bittinger M A, Milner J L, Seville B J, Handelsman J. Mol Plant-Microbe Interact. 1997;10:180–186. doi: 10.1094/MPMI.1997.10.2.180. [DOI] [PubMed] [Google Scholar]
- 16.Keller M, Roxlau A, Weng W M, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Pühler A. Mol Plant-Microbe Interact. 1995;8:267–277. doi: 10.1094/mpmi-8-0267. [DOI] [PubMed] [Google Scholar]
- 17.Bertram-Drogatz P A, Rüberg S, Becker A, Pühler A. Mol Gen Genet. 1997;254:529–538. doi: 10.1007/s004380050448. [DOI] [PubMed] [Google Scholar]
- 18.Barnett M J, Long S R. Mol Plant-Microbe Interact. 1997;10:550–559. doi: 10.1094/MPMI.1997.10.5.550. [DOI] [PubMed] [Google Scholar]
- 19.Berg J M, Shi Y. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 20.Schwabe W R, Klug A. Nat Struct Biol. 1994;1:345–349. doi: 10.1038/nsb0694-345. [DOI] [PubMed] [Google Scholar]
- 21.Ginsberg A M, King B O, Roeder R G. Cell. 1984;39:479–489. doi: 10.1016/0092-8674(84)90455-0. [DOI] [PubMed] [Google Scholar]
- 22.Miller M, McLachan A D, Klug A. EMBO J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartshorn T A, Blumberg H, Young E T. Nature (London) 1986;320:283–287. doi: 10.1038/320283a0. [DOI] [PubMed] [Google Scholar]
- 24.Perrin G, Gilliland G. Nucleic Acids Res. 1990;18:7433–7438. doi: 10.1093/nar/18.24.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studier W F, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 26.Tchou J, Michaels M L, Miller J H, Grollman A P. J Biol Chem. 1993;268:26738–26744. [PubMed] [Google Scholar]
- 27.Shaw W V. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- 28.Kado C I, Heskett M G, Langley R A. Physiol Plant Pathol. 1971;2:47–57. [Google Scholar]
- 29.Chilton M-D, Currier T C, Farrand S K, Bendich A J, Gordon M P, Nester E W. Proc Natl Acad Sci USA. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klug A, Rhodes D. Trends Biochem Sci. 1987;12:464–469. [Google Scholar]
- 31.Buchmann I, Marner F-J, Schröder G, Waffenschmidt S, Schröder J. EMBO J. 1985;4:853–859. doi: 10.1002/j.1460-2075.1985.tb03710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Binns A N. Annu Rev Plant Physiol Plant Mol Biol. 1994;45:173–196. [Google Scholar]
- 33.Goldberg S B, Flick J S, Rogers S G. Nucleic Acids Res. 1984;12:4665–4677. doi: 10.1093/nar/12.11.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kao J C, Perry K L, Kado C I. Mol Gen Genet. 1982;188:425–432. doi: 10.1007/BF00330044. [DOI] [PubMed] [Google Scholar]
- 35.Steck T R, Lin T-S, Kado C I. Proc Natl Acad Sci USA. 1989;86:2133–2137. doi: 10.1073/pnas.86.7.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhodes D, Klug A. Sci Am. 1993;268:56–65. doi: 10.1038/scientificamerican0293-56. [DOI] [PubMed] [Google Scholar]
- 37.Kadonga J T, Carner K R, Masiarz F R, Tjian R. Cell. 1987;51:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 38.Takatsuji H, Mori M, Benfy P N, Ren L, Chua N-H. EMBO J. 1992;11:241–249. doi: 10.1002/j.1460-2075.1992.tb05047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takatsuji H, Nakamura N, Katsumoto Y. Plant Cell. 1994;6:947–958. doi: 10.1105/tpc.6.7.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakamoto A, Minami M, Huh G H, Iwabuchi M. Eur J Biochem. 1993;217:1049–1056. doi: 10.1111/j.1432-1033.1993.tb18336.x. [DOI] [PubMed] [Google Scholar]
- 41.Tague B W, Goodman H M. Plant Mol Biol. 1995;28:267–279. doi: 10.1007/BF00020246. [DOI] [PubMed] [Google Scholar]
- 42.Lints R, Davis M A, Hynes M J. Mol Microbiol. 1995;15:965–975. doi: 10.1111/j.1365-2958.1995.tb02365.x. [DOI] [PubMed] [Google Scholar]
- 43.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hussain H, Johnston A W B. Mol Plant-Microbe Interact. 1997;10:1087–1093. doi: 10.1094/MPMI.1997.10.9.1087. [DOI] [PubMed] [Google Scholar]
- 45.Heinemann J A. Trends Genet. 1991;7:181–185. doi: 10.1016/0168-9525(91)90433-q. [DOI] [PubMed] [Google Scholar]
- 46.Heinemann J A, Ankenbauer R G. J Bacteriol. 1993;175:583–588. doi: 10.1128/jb.175.3.583-588.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strabala T J, Crowell D N, Amasino R M. Plant Mol Biol. 1993;21:1011–1021. doi: 10.1007/BF00023599. [DOI] [PubMed] [Google Scholar]