Abstract
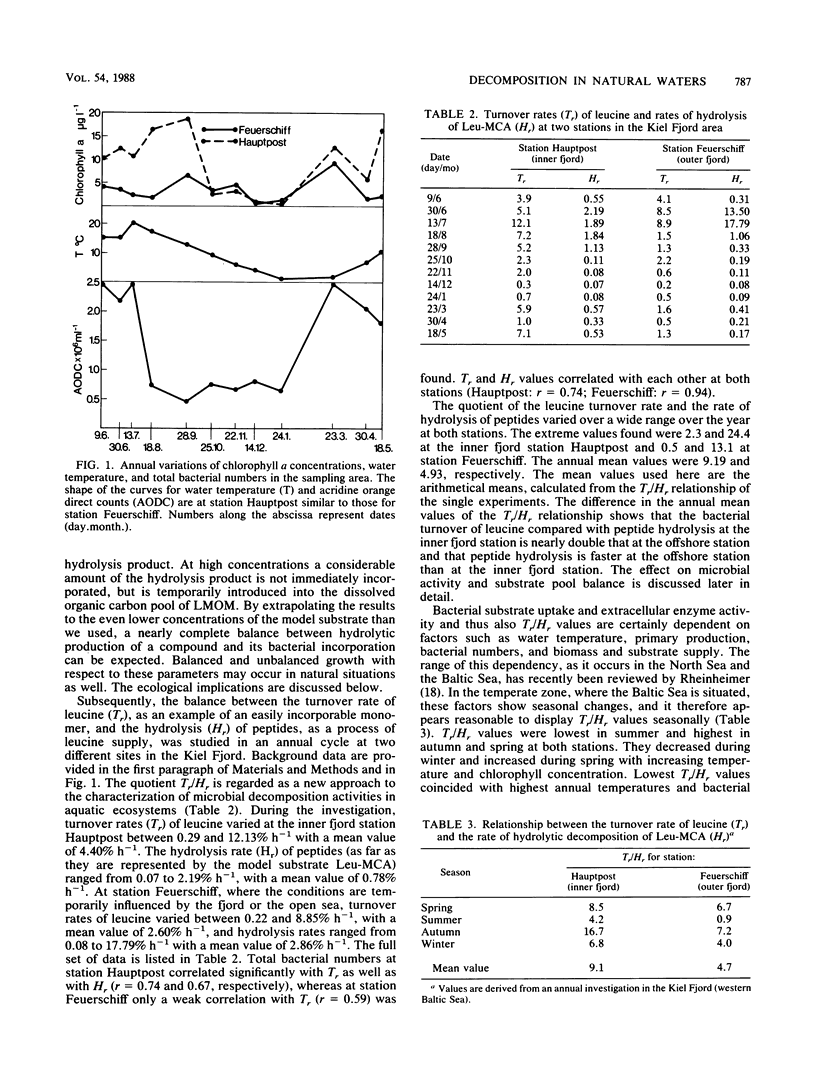
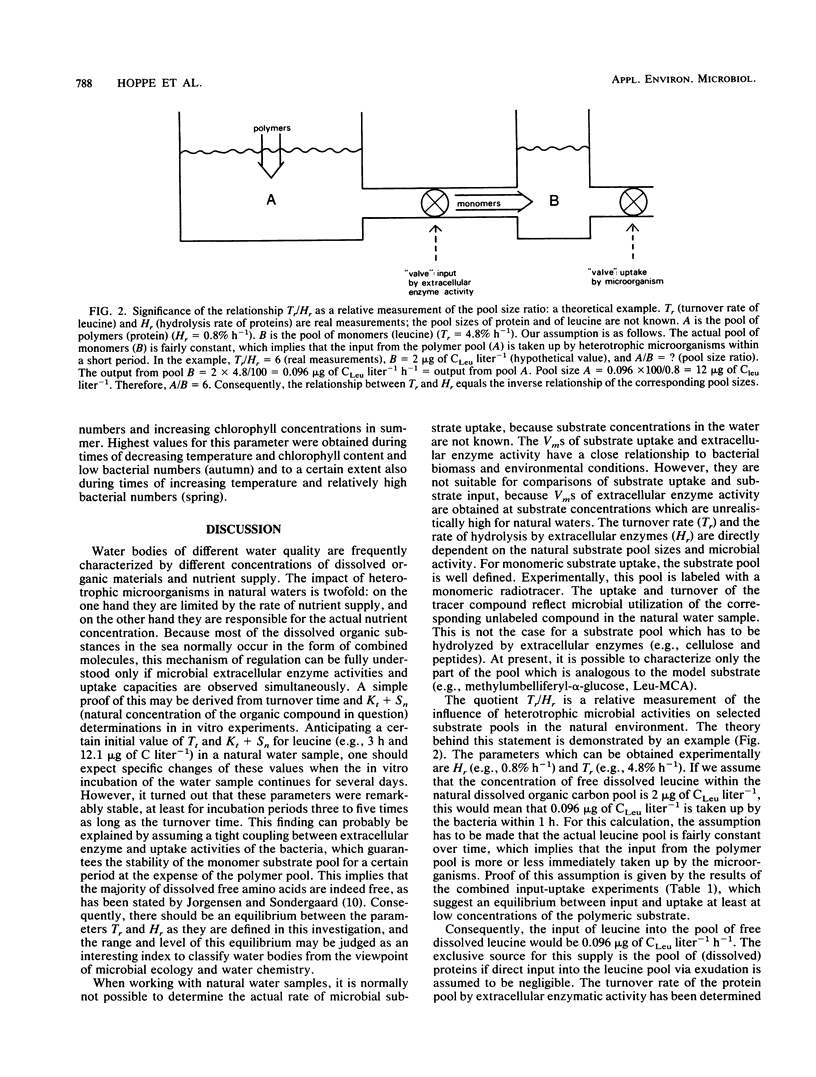
The aim of this study was to define a model for the coupling between extracellular enzyme activity and substrate uptake by bacterial populations in natural waters. The balance between uptake of leucine and extracellular hydrolytic production of leucine from a peptide model substrate was investigated in a combined fluorescence-radiotracer experiment with [3H]leucine as a marker for the leucine pool and l-leucine-4 methyl-7-coumarinylamide (Leu-MCA) as a marker for the pool of dissolved peptide substrates. Results show that at low concentrations of the model substrate the input and uptake processes of leucine are nearly balanced, whereas at high concentrations of the model substrate much more leucine is liberated than taken up. In addition, samples from one polluted and one less polluted station in the Kiel Fjord were investigated for their extracellular enzymatic and uptake properties in an annual cycle. It was found that turnover rates of leucine (Tr, percent per hour) and hydrolysis rates of Leu-MCA (Hr, percent per hour), as well as the quotient Tr/Hr, reflect the impact of environmental conditions on decomposition processes at both sampling sites. The quotient Tr/Hr is interpreted as an indirect measurement of the pool size ratio (polymers/monomers), which may serve as an index of hydrolysis-uptake coupling in bacterial utilization of dissolved protein. Calculated on an annual average basis, turnover rates are ca. nine times higher than hydrolysis rates at the polluted station and ca. five times higher at the less polluted station. From the described model, this would mean that the relative fraction of polymers within the total dissolved organic carbon pool (with regard to the substrate combination dissolved protein-leucine) is about twice that at the polluted than at the less polluted station.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brock F. M., Forsberg C. W., Buchanan-Smith J. G. Proteolytic activity of rumen microorganisms and effects of proteinase inhibitors. Appl Environ Microbiol. 1982 Sep;44(3):561–569. doi: 10.1128/aem.44.3.561-569.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotta M. A., Hespell R. B. Proteolytic activity of the ruminal bacterium Butyrivibrio fibrisolvens. Appl Environ Microbiol. 1986 Jul;52(1):51–58. doi: 10.1128/aem.52.1.51-58.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M. Characterization of beta-Glucosidase Activity in Intertidal Marine Sediments. Appl Environ Microbiol. 1986 Feb;51(2):373–380. doi: 10.1128/aem.51.2.373-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholy S. K., Lynd J. Q. Microbial esterase detection with ultraviolet fluorescence. Appl Microbiol. 1971 Nov;22(5):939–941. doi: 10.1128/am.22.5.939-941.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder A. P., Wang T. T., Greenberg D. B. Pattern recognition analysis of in vivo enzyme-substrate fluorescence velocities in microorganism detection and identification. Appl Environ Microbiol. 1986 May;51(5):969–977. doi: 10.1128/aem.51.5.969-977.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somville M. Measurement and study of substrate specificity of exoglucosidase activity in eutrophic water. Appl Environ Microbiol. 1984 Dec;48(6):1181–1185. doi: 10.1128/aem.48.6.1181-1185.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]



