Abstract
Our three-dimensional (3-D) images showed that paxillin co-localized on actin filaments as fibrous structures, as well as clusters, in endothelial cells (ECs). In living ECs under flow condition, we monitored concurrently the intracellular dynamics of DsRed2-paxillin and GFP-actin by time-lapse video recording and dual-color fluorescence imaging. The results showed that the dynamic motion of paxillin as fibrous structures was associated with actin filaments, but not with microtubules. Our findings suggest that the actin network plays an important role not only in the assembly/disassembly of paxillin at focal adhesions, but also as a track for the intracellular transport of paxillin, which is involved in signaling pathway.
Keywords: Paxillin, Actin filaments, Microtubules, Focal adhesions
Cell spreading and migration, which play integral roles in development, repair and defense, are modulated by extracellular chemical and mechanical stimuli. These stimuli are transduced into well-regulated intracellular events involving membrane receptors, integrins, focal adhesions (FAs), cytoskeleton, and signaling pathways (1-3). Paxillin, which is associated with the cytoplasmic domains of integrins localized at FAs (4), functions as an adaptor protein that interacts with cytoskeletal and signaling proteins to regulate the assembly, disassembly, and signaling of FAs (5). Thus, paxillin serves to integrate and transduce signals from integrins and growth factor receptors to modulate actin filament dynamics and cellular functions (6).
Vascular ECs, which form the inner lining of the blood vessel wall and are subjected to hemodynamic forces, play an important role in the regulation of vascular homeostasis in health and disease (7). During EC migration, paxillin-containing FAs (paxillin-FAs) are formed at the leading edge, remain fixed as the cell migrates over them, and then detach when they are at the cell rear (8). Rho-family small GTPases (Rho, Rac, and Cdc42) regulate the organization of actin filaments and contribute to cell movement (9). Paxillin regulates p190RhoGAP (GTPase-activating protein) activity to inhibit Rho at the leading edge, thus promoting lamellipodial extension (10). Paxillin may also play an important role in regulating Cdc42 activity and hence cell polarity. Tyrosine phosphorylation of paxillin stimulates CrkII binding and may thus activate Rac via a Crk-Cas-Dock180 signaling pathway (11,12). Paxillin is now considered to be at the crossroad of cell adhesion and mechanochemical stimuli in modulating intracellular signal transduction and the consequent changes in actin filament dynamics, cell shape, and cell motility (13). There is a need to elucidate the mechanisms that regulate paxillin-FAs formation and detachment, as well as intracellular transport of paxillin in ECs.
We recently reported the assembly/disassembly and distribution of paxillin-FAs in ECs under static condition by tracking the intracellular dynamics of paxillin at FAs in migrating living ECs following transfection of constructs encoding green fluorescent protein (GFP)-tagged paxillin and by using time-lapse image recording (8). Here we report the dynamics of paxillin-clusters at FAs and in the cytoplasm of living ECs under shear flow. We discovered that paxillin form fibrous structures in the cytoplasm and that they move in close association with and along actin stress fibers (SFs). These findings have significant implications in the molecular and cellular bases of the intracellular transport of paxillin to modulate cellular functions.
Materials and methods
Cell culture
BAECs isolated from the bovine aorta were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, Inc. Gaithersburg, MD) supplemented with 10% fetal calf serum (Invitrogen, Carlsbad, CA), 2 mM L-glutamine, and 1 mM each of penicillin-streptomycin and sodium pyruvate. The cells were grown on glass slides (3 × 1 inch) precoated with 4 μg/cm2 fibronectin (Sigma Chemical Co., St. Louis, MO) in DMEM. The cells were maintained in a humidified 95% air, 5% CO2 incubator at 37°C.
Flow system
A flow system was used to impose shear stress on cultured ECs as described by Frangos et al. (14). In the shear stress experiments, cells were grown on a fibronectin-coated coverglass slide in the flow chamber for microscopic observation. The slide with subconfluent BAECs was mounted in a rectangular flow chamber created by sandwiching a silicon gasket between the slide and an acrylic plate. The chamber, two reservoirs, and a circulation circuit were filled with the cell culture medium. The flow of the medium was driven by a constant pressure head, which was maintained by feedback refilling with a rolled pump. Laminar shear stress was generated by the pressure-driven flow such that the BAECs were subjected to shear stress at 5 dyn/cm2. The medium was kept at a constant temperature of 37°C and equilibrated with a gas mixture of humidified 95% air and 5% CO2. In the control static group, the BAECs were kept for the same duration without flow.
Cell treatment
Cytoskeleton modulating agents (colchicine and cytochalasin D) were purchased from Sigma Chemical Co. BAECs in culture medium were treated with these agents at a final concentration of 1 μM.
DNA plasmids and transfection
DNA plasmids encoding green fluorescent protein (GFP)-tagged paxillin and red fluorescent protein (DsRed2)-tagged paxillin were provided by Dr. Donna J. Webb (Virginia University, Charlottesville, VA) (15). DNA plasmids encoding GFP-actin and GFP-tubulin were from Clontech Laboratories, Inc. (Mountain Veiw, CA). DNA plasmids were transfected into BAECs at 80% confluence using the GenePORTER™ Transfection Reagent (Gene Therapy Systems, Inc., San Diego, CA). After incubation for 6 h, the cells were washed with DMEM and incubated with fresh DMEM containing 10% serum for another 24-48 h. The transfected cells were then seeded onto fibronectin-coated coverglass slides for the experiments.
Fluorescence staining and confocal microscopy
For fluorescent staining, the BAECs were fixed with 4% paraformaldehyde for 15 min at 37°C in phosphate buffered saline (PBS). The fixed cells were permeabilized with 0.3% Triton X-100, and nonspecific binding was blocked by using 1% normal goat serum. To visualize actin microfilaments, the cells were stained with rhodamineconjugated phalloidin (Molecular Probes, Inc., Eugene, OR). The slides were examined by using a Nikon Diaphot microscope equipped with a PekinElmer Kypton-Argon laser scanning confocal imaging system (PerkinElmer Life Science, Boston, MA). Green fluorescence was excited at a wavelength of 488 nm and detected at 525 nm. Rhodamine was excited at 568 nm and detected at 600 nm. By stepping the objective through the depth of the specimen, a z-series collection of optical sections was obtained and projected to re-construct the 3-dimensional (3-D) or 2-dimensional (2-D) image. After rotating the image, the side view of a 3-D cell was observed by SimplePCI image analysis software of C-Imaging Systems (Compix Inc., Cranberry Township, PA). The images were transferred to a computer for further analysis; Adobe Photoshop (Adobe System, Mountain View, CA) was used to generate RGB images to depict the F-actin in red and the expressed paxillin in green.
Multi-mode time-lapse microscope
Digital images of cell motility were obtained by using an Olympus live-cell confocal microscope system IX81 with a disk scan unit (DSU) (Olympus America Inc., Melville, NY). During microscopy with 40x/60x oil objectives (Olympus, NA 1.10−1.35/1.45), fluorescently labeled cells were revealed by illumination with a 300w xenon lamp. Images were acquired from a cooled charge-coupled device camera (Hamamatsu Orca-AG) (Hamanatsu, Bridgewater, NJ). The two-color fluorescence images were taken alternatively by switching the excitation light paths. Fluorescence images were collected at 1-min intervals. Image acquisition was controlled with MetaMorph® Imaging System (Molecular Devices Corporation, Sunnyvale, CA) to capture and store the images in computer for performing operations such as time lapse, multi-dimensional acquisition, and 3-D reconstruction, as well as making measurements such as morphometry, co-localization, and brightness. To reveal GFP, GFP filter (excitation 460-480HQ, emission 495-540HQ, DM485 dichroic mirror) (Olympus) was used. For two-color fluorescence live-cell imaging, GFP and DsRed2 were revealed with filters for GFP (excitation 460-480HQ, emission 495-540HQ, DM485 dichroic mirror) and DsRed2 (excitation 535-555HQ, emission 570-620HQ, DM560 dichroic mirror) (Olympus).
Results
Intracellular dynamics of paxillin as fibrous structures
In a previous report on mechanotransduction, paxillin at FAs was found to increase dramatically in ECs stretched biaxially, particularly at the periphery of the cells (16). Mechanotransduction across the cell surface and through the cytoskeleton can be correlated with FA formation (17). Our previous study on the dynamics of paxillin in ECs and its association with the end of actin SFs showed that actin network played an important role in the assembly/disassembly of paxillin at FAs (8). Upon further investigations, especially in ECs transtected with GFP-paxillin, we have discovered the presence of paxillin-containing filamentous structures (paxillin-fibers) in the cytoplasm in addition to paxillin-FAs. Since shear stress can induce remodeling of FAs and reorganization of actin filaments (18), in the present study we investigated paxillin dynamics in ECs transfected with GFP-paxillin and subjected to shear stress. The application of shear stress (5 dyn/cm2) for 30 min caused the dynamic motion of paxillin-fibers (Fig. S1), which underwent continuous shifting as fibrous structures (Fig. S1B and Movie S1B). Some of the preexisting paxillin-clusters at FAs were disassembled at the periphery of cell and shifted toward the cell central region, where they were re-assembled as paxillin-clusters in dynamic motion (Fig. S1C and Movie S1C). These results show that the dynamic assembly/disassembly of paxillin-clusters and paxillin-fibers exist in living ECs under both static and sheared condition.
Association of paxillin-fibers and actin filaments
In a laser scanning confocal microscopic imaging system, a z-series collection of dual-color sections was obtained, and projected to construct 3-D image. After 3-D reconstruction, the side view of the cell was observed by using the SimplePCI image analysis software of C-Imaging Systems (Fig. 1). Figure 1A shows the fluorescence images of an EC that had its paxillin tagged with GFP (green, “Paxillin” column) and actin labeled with rhodamine-conjugated phalloidin (red, “Actin” column), as well as their merged images (“Merge” column), where yellow color shows the co-localization of paxillin and actin. Both in the whole cell view (Fig. 1A-top row) and in the magnified view of the boxed region (Fig. 1A-bottom row), paxillin (Fig. 1A-Paxillin) can be seen to form some filamentous structures (paxillin-fibers) similar to those of actin filaments (Fig. 1A-Actin); furthermore, there is co-localization of paxillin-fibers and actin filaments (yellow color in Fig. 1A-Merge).
Fig. 1. Dual-fluorescence images showing co-localization (yellow) of paxillin-fibers (green) and actin filaments (red) in ECs.
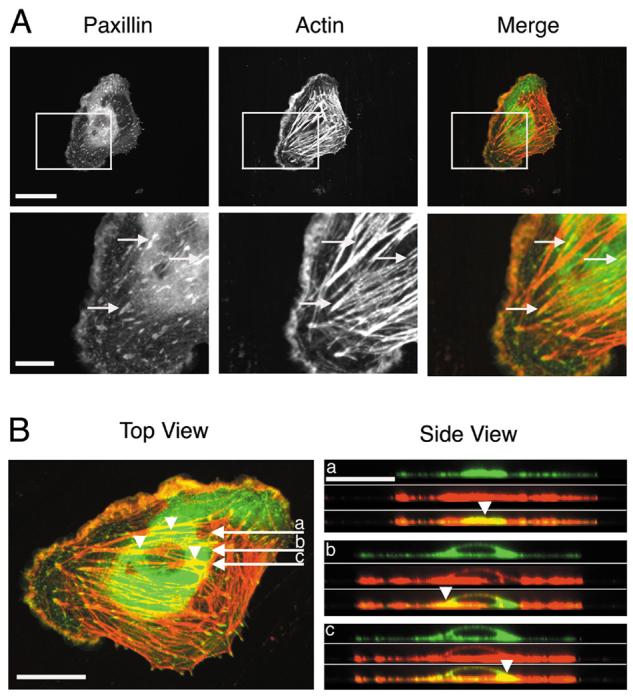
(A) The “Merge” column shows superimposed images from paxillin (“Paxillin” column) and actin (“Actin” column). Arrows show the examples of co-localization of paxillin-fibers and actin filaments. Scale bars: upper row = 30 μm; lower row = 10 μm. (B) In top view (xy-dimension), three arrows (marked a, b and c) indicate the positions of three sections (0.2-μm/section) in side view (xz-dimension) (Movie S2). The three arrowheads in top and side views point to the corresponding positions in these two views. Scale bars = 20 μm.
These 3-D images show more clearly the relations between paxillin-fibers and actin filaments (Fig. 1B). Top views (xy-dimensional images) of the EC show that paxillin (green) is present as both clusters and fibers across the whole cell and that actin (red) exists as filaments and filament bundles (SFs). The 3-D images were reconstructed into xz-dimensional sections (each 0.2-μm thick), and three sections (a, b and c) are shown on the right side of Fig. 1B (Side View); these three sections correspond to the three horizontal arrows (a, b and c) in the top view (left side of Fig. 1B). Each of the three xz-sections on the side view is presented as three pictures (top, middle, and bottom) to show paxillin, actin, and merged image, respectively. The merged images demonstrate that the co-localization (yellow) of paxillin and actin extends from top to bottom of the cell; this co-localization exists between the paxillin-fibers and the actin filaments (arrowhead, in section “a”), in addition to the paxillin-clusters at FAs on the ends of actin SFs. In xz-dimensional sections, the co-localization of paxillin-clusters and actin filaments (arrowheads, in sections “b” and “c”) was found in the cytoplasm (perinuclear region). These 3-D reconstructed xz-sections are also presented in Movie S2.
Association of the dynamics of paxillin-fibers with actin filaments, but not microtubules
To investigate the roles of microtubules and actin filaments in the dynamic motion of paxillin-fibers, we co-transfected living ECs with the constructs encoding red fluorescent protein (DsRed2)-tagged paxillin, and GFP-tubulin (Fig. S2A) or GFP-actin (Fig. S2B). The intracellular dynamics of DsRed-paxillin (red) and GFP-tubulin (green) or GFP-actin (green) expressed in ECs were tracked with time-lapse video recording. The boxed regions in Figs. S2A and S2B (double labeled) are enlarged in the pictures below; first in the control state (0′, middle row) and then after cytoskeletal disruption (30′, bottom row) with either colchicine (Fig. S2A) or cytochalasin D (CD) (Fig. S2B). In the middle and bottom rows of both Figs. S2A and S2B, the left column shows paxillin and the right column shows the appropriate cytoskeleton. Paxillin formed paxillin-clusters and paxillin-fibers, and they are co-localized with actin filaments to yield the yellow color (Fig. S2B).
The disruption of microtubules with colchicine (1 μM) for 30 min did not cause the disassembly of paxillin-clusters and paxillin-fibers (Fig. S2A, left column, 30′ vs. 0'), although the microtubule network was almost totally destroyed (Fig. S2A, right column, 30′ vs. 0'; also Movie S3A series). The disruption of actin filaments with CD treatment (1 μM) for 30 min led to not only the disruption of actin filaments (Fig. S2B, right column, 30'vs. 0'), but also the disassembly of paxillin-clusters and paxillin-fibers (Fig. S2B, left column, 30′ vs. 0'). There was neither paxillin-clusters nor paxillin-fibers in the EC, and almost all paxillin became distributed diffusely in the cytoplasm, as the actin network was destroyed (Movie S3B series). These findings indicate that the dynamic motions of paxillin-clusters and paxillin-fibers are dependent on the integrity of actin network, but not microtubules.
The intracellular dynamic motion of paxillin on actin filaments
Using ECs co-transfected with DsRed2-paxillin and GFP-actin, the dynamics of these two types of molecules were observed in the same living cells (Fig. 2). Figures 2AB and Movie 2B show that the DsRed2-labeled paxillin-clusters and paxillin-fibers were co-localized with GFP-labeled actin filaments in static ECs during their dynamic motions. Time-lapse image analysis of the living cells shows that paxillin-clusters at FAs is located mainly at the ends of actin SFs, but paxillin-clusters and paxillin-fibers in the cytoplasm can be seen to undergo dynamic translocation in association with the actin filaments, by sliding either toward the perinuclear regions or the cell periphery (Movie S4B series).
Fig. 2. Dynamic motion of paxillin-fibers and paxillin-clusters in association with actin filaments in living ECs co-transfected with DsRed2-paxillin (red) and GFP-actin (green).
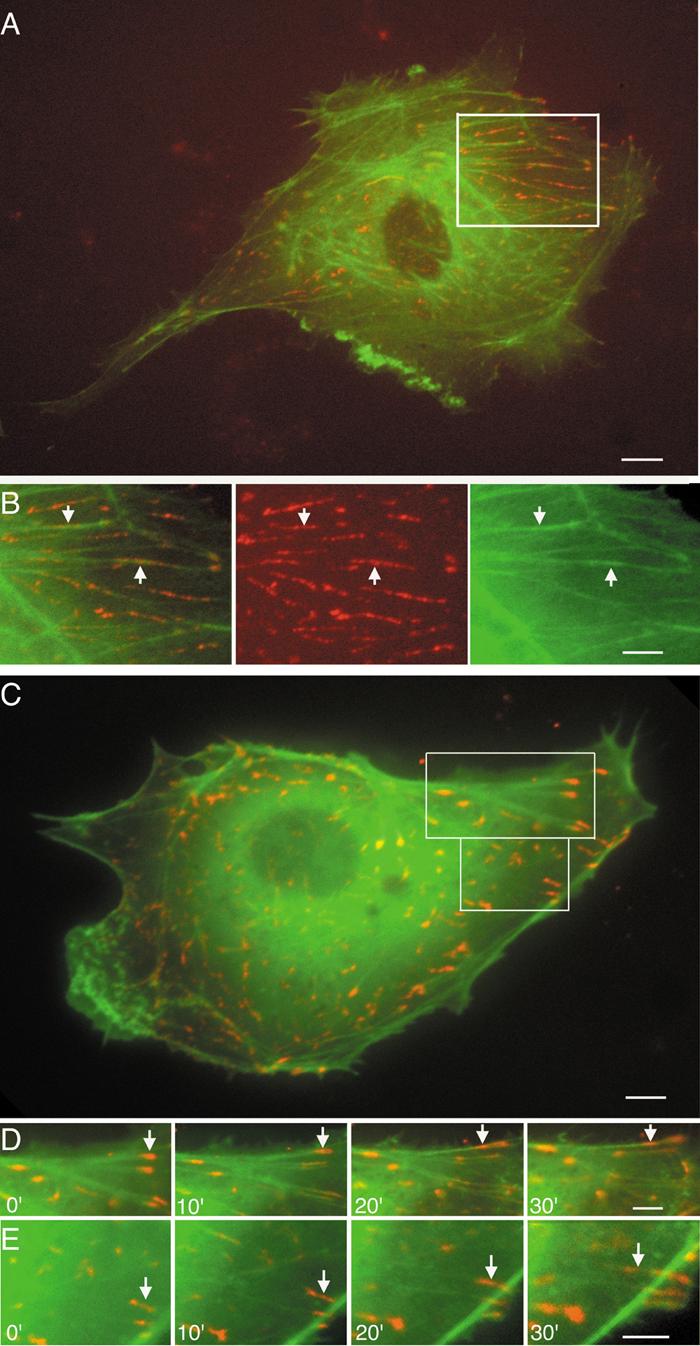
(A) A static EC. (B row) Boxed area in A (Movie S4B series). Arrows point to the dynamic motion of paxillin-fibers on SFs. (C) An EC subjected to shear stress. (D and E rows) Boxed areas in C. Paxillin-clusters (arrows) show disassembling and sliding on actin SFs. Superimposition of DsRed2-paxillin and GFP-actin images appears as yellow (Movie S4DE series). Scale bars: A, C = 10 μm; B, D, E = 8 μm.
The application of shear stress (30 min) caused the disassembly of paxillin-clusters at FAs into smaller ones, which moved along the actin SFs toward the cell center (see figure legends). During these dynamic processes, paxillin-clusters and paxillin-fibers remained associated with actin filaments (Fig. 2CDE and Movie S4DE). Color overlays of sequential images indicate that the majority of paxillin-clusters and paxillin-fibers (red) traversed on the actin filaments (green) to give the yellow color (Fig. S3 and Movie S4D series). The results indicate that the dynamic motions of paxillin-clusters and paxillin-fibers are associated with actin filaments, and suggest that actin filaments may serve as a track for the intracellular transportation of paxillin signaling.
Discussion
Paxillin serves to integrate and transduce signals from integrins and growth factor receptors to modulate actin filament dynamics, assembly/disassembly of FAs, and cell functions. The regulation of actin filament dynamics, cell movement, and signal transduction by paxillin is mediated by its association with a variety of cytoskeletal and signaling proteins (2,4). Paxillin is detected primarily at FAs, but it exists also in the cytoplasm and nucleus (19-21) and can shuttle between the cytoplasmic and nuclear compartments (22-25). Paxillin can affect nuclear transcription, in addition to its functions at FAs (21), and it may constitute a direct signaling pathway from plasma membrane and cytoskeletal network to the nucleus (26,27). Therefore, the mechanism of paxillin transport or shuttling is of considerable importance. The present findings can form a basis to investigate the regulation of paxillin transport by molecules such as Golgi resident ARF (ADP-ribosylation factor), PAK and PIX (28-30).
In summary, using live-cell fluorescence imaging of paxillin and actin to analyze the dynamics of paxillin-clusters, paxillin-fibers, and actin filaments, we have shown the association of paxillin dynamics with that of actin filaments in living ECs. Our findings suggest that actin network plays an important role not only in the assembly/disassembly of paxillin at FAs, but also as a track for the intracellular transport of paxillin, which plays an important role in cellular signaling cascades.
Supplementary Material
Acknowledgments
The authors wish to express our special thanks to Dr. Donna J. Webb who provided GFP-paxillin and DsRed-paxillin constructs for this study. We also thank Dr. Yi-shuan Li for valuable help and Phu Nguyen, Gerard Norwich, Suli Yuan for their excellent assistance. This work was supported by National Heart, Lung, and Blood Institute Research Grants HL-064382, HL-080518, and HL-085159 (S.C).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Horwitz AR, Parsons JT. Cell migration--movin' on. Science. 1999;286:1102–1103. doi: 10.1126/science.286.5442.1102. [DOI] [PubMed] [Google Scholar]
- 2.Smilenov LB, Mikhailov A, Peham RJ, Jr., Marcantonio EE, Gundersen GG. Focal adhesion motility reveal in stationary fibroblasts. Science. 1999;286:1172–1174. doi: 10.1126/science.286.5442.1172. [DOI] [PubMed] [Google Scholar]
- 3.Small JV, Geiger B, Kaverina I, Bershadsky A. How do microtubules guide migrating cells? Nat. Rev. Mol. Cell Biol. 2002;3:957–964. doi: 10.1038/nrm971. [DOI] [PubMed] [Google Scholar]
- 4.Turner CE, Glenney JR, Jr., Burridge K. Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 1990;111:1059–1068. doi: 10.1083/jcb.111.3.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown MC, Turner CE. Roles for the tubulin- and PTP-PEST-binding paxillin LIM domains in cell adhesion and motility. Int. J. Biochem. Cell Biol. 2002;34:855–863. doi: 10.1016/s1357-2725(01)00154-6. [DOI] [PubMed] [Google Scholar]
- 6.Brown MC, Turner CE. Paxillin: adapting to change. Physiol. Rev. 2004;84:1315–1339. doi: 10.1152/physrev.00002.2004. [DOI] [PubMed] [Google Scholar]
- 7.Davies PF. Flow-mediated endothelial mechanotransduction. Physiol. Rev. 1995;75:519–560. doi: 10.1152/physrev.1995.75.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu YL, Haga JH, Miao H, Wang Y, Li YS, Chien S. Roles of microfilaments and microtubules in paxillin dynamics. Biochem. Biophys. Res. Commun. 2006;348:1463–1471. doi: 10.1016/j.bbrc.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 9.Nobes CD, Hall A. Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 1999;144:1235–1244. doi: 10.1083/jcb.144.6.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kulkarni SV, Gish G, van der Geer P, Henkemeyer M, Pawson T. Role of p120 Ras-GAP in directed cell movement. J. Cell Biol. 2000;149:457–470. doi: 10.1083/jcb.149.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki T, Nakata A, Mukai M, Shinkai K, Yano H, Sabe H, Schaefer E, Tatsuta M, Tsujimura T, Terada N, Kakishita E, Akedo H. Involvement of phosphorylation of Tyr-31 and Tyr-118 of paxillin in MM1 cancer cell migration. Int. J. Cancer. 2002;97:330–335. doi: 10.1002/ijc.1609. [DOI] [PubMed] [Google Scholar]
- 12.Brugnera E, Haney L, Grimsley C, Lu M, Walk SF, Tosello-Trampont AC, Macara IG, Madhani H, Fink GR, Ravichandran KS. Unconventional Rac-GEF activity is mediated through the Dock180-ELMO complex. Nat. Cell Biol. 2002;4:574–582. doi: 10.1038/ncb824. [DOI] [PubMed] [Google Scholar]
- 13.Turner CE. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2000;2:E231–236. doi: 10.1038/35046659. [DOI] [PubMed] [Google Scholar]
- 14.Frangos JA, Eskin SG, McIntire LV, Ives CL. Flow effects on prostacyclin by cultured human endothelial cells. Science. 1985;227:1477–1479. doi: 10.1126/science.3883488. [DOI] [PubMed] [Google Scholar]
- 15.Laukaitis CM, Webb DJ, Donais K, Horwitz AF. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cell. J. Cell Biol. 2001;153:1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swada Y, Sheetz MP. Force transduction by Triton cytoskeletons. J. Cell Biol. 2002;156:609–615. doi: 10.1083/jcb.200110068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:1124–1127. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Butler P, Wang Y, Hu YL, Han DC, Usami S, Guan JL, Chien S. The role of the dynamics of focal adhesion kinase in the mechanotaxis of endothelial cells. Proc. Natl. Acad. Sci. USA. 2002;99:3546–3551. doi: 10.1073/pnas.052018099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nix DA, Beckerle MC. Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Bio. 1997;138:1139–1147. doi: 10.1083/jcb.138.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L, Guerrero J, Hong H, DeFranco DB, Stallcup MR. Interaction of the tau2 transcriptional activation domain of glucocorticoid receptor with a novel steroid receptor coactivator, Hic-5, which localizes to both focal adhesions and the nuclear matrix. Mol. Biol. Cell. 2000;11:2007–2018. doi: 10.1091/mbc.11.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Gilmore TD. Zyxin and paxillin proteins: focal adhesion plaque LIM domain proteins go nuclear. Biochim. Biophys. Acta. 2003;1593:115–120. doi: 10.1016/s0167-4889(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 22.Kanungo J, Pratt SJ, Marie H, Longmore GD. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell. 2000;11:3299–3313. doi: 10.1091/mbc.11.10.3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nix DA, Fradelizi J, Bockholt S, Menichi B, Louvard D, Friederich E, Beckerle MC. Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 2001;276:34759–34767. doi: 10.1074/jbc.M102820200. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Gilmore TD. LIM domain protein Trip6 has a conserved nuclear export signal, nuclear targeting sequences, and multiple transactivation domains. Biochim. Biophys. Acta. 2001;1538:260–272. doi: 10.1016/s0167-4889(01)00077-5. [DOI] [PubMed] [Google Scholar]
- 25.Woods AJ, Roberts MS, Choudhary J, Barry ST, Mazaki Y, Morley SJ, Critchley DR, Norman JC. Paxillin associates with poly(A)-binding protein 1 at the dense endoplasmic reticulum and the leading edge of migrating cells. J. Biol. Chem. 2002;277:6428–6437. doi: 10.1074/jbc.M109446200. [DOI] [PubMed] [Google Scholar]
- 26.Bianchi E, Denti S, Granata A, Bossi G, Geginat J, Villa A, Rogge L, Pardi R. Integrin LFA-1 interacts with the transcriptional co-activator JAB1 to modulate AP-1 activity. Nature. 2000;404:617–621. doi: 10.1038/35007098. [DOI] [PubMed] [Google Scholar]
- 27.Hsueh YP, Wang TF, Yang FC, Sheng M. Nuclear translocation and transcription regulation by the membrane-associated guanylate kinase CASK/LIN-2. Nature. 2000;404:298–302. doi: 10.1038/35005118. [DOI] [PubMed] [Google Scholar]
- 28.Norman JC, Jones D, Barry ST, Holt MR, Cockcroft S, Critchley DR. ARF1 mediates paxillin recruitment to focal adhesions and potentiates Rho-stimulated stress fiber formation in intact and permeabilized Swiss 3T3 fibroblasts. J. Cell Biol. 1998;143:1981–1995. doi: 10.1083/jcb.143.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turner CE, West KA, Brown MC. Paxillin-ARF GAP signaling and the cytoskeleton. Curr Opin. Cell Biol. 2001;13:593–599. doi: 10.1016/s0955-0674(00)00256-8. [DOI] [PubMed] [Google Scholar]
- 30.Kondo A, Hashimoto S, Yano H, Nagayama K, Mazaki Y, Sabe H. A new paxillin-binding protein, PAG3/Papalpha/KIAA0400, bearing an ADP-ribosylation factor GTPase-activating protein activity, is involved in paxillin recruitment to focal adhesions and cell migration. Mol. Biol. Cell. 2000;11:1315–1327. doi: 10.1091/mbc.11.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


