Abstract
Molecular beacon detection of equilibrium cyclization (MBEC) is a novel, high sensitivity technique that can allow DNA-protein complex formation to be studied under diverse conditions in a cost effective and rapid manner that can be adapted to high throughput screening. To demonstrate the ease and utility of applying MBEC to the investigation of the KD values of protein-DNA complexes, the sequence-specific Escherichia coli integration host factor (IHF) protein has been used as a test system. Competition between a labeled MBEC DNA construct and unlabeled duplex DNA for IHF binding allows the determination of KD values as a function of the DNA duplex sequence. This allows sequence specificity to be monitored while using only a single molecular beacon-labeled DNA. The robustness of MBEC for monitoring protein-DNA complex formation has been further demonstrated by determining the KD values as a function of salt concentration to investigate the net number of salt bridges formed in sequence-specific and -nonspecific IHF-DNA complexes. These MBEC results have been compared with those from other approaches.
INTRODUCTION
Understanding the elements of sequence-specific recognition and stability in DNA-protein complex formation relies on the determination of the association constants under a wide range of experimental conditions (1). For example, the determination of the net number of salt bridges formed in a complex requires that complex formation be studied over a range of salt concentrations (2–5). In addition to ionic strength, pH, temperature, water activity, and accessory factors can have biologically relevant effects on the formation of DNA-protein complexes (3,6).
Currently, measurement of DNA-protein binding in solution with high sensitivity over this range of experimental conditions has been challenging. The available methods include gel mobility shift assays, surface plasmon resonance, centrifugation, Raman spectroscopy, calorimetry, and NMR as well as fluorescence intensity, fluorescence lifetimes, and anisotropy. Each of these methods has distinct advantages and limitations. In this report we present a technique that monitors the binding equilibria of DNA-protein complexes in solution with high sensitivity under diverse conditions that is rapid and cost effective and can be adapted to high throughput screening.
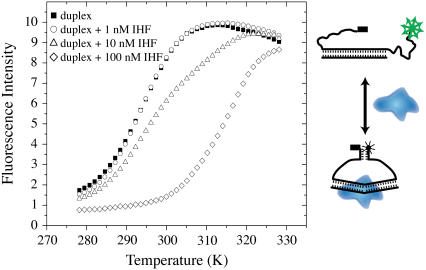
We have previously shown that equilibrium cyclization can be used to determine the KD values of DNA-protein complexes (7). The original approach utilized the fluorescence of 2-aminopurine incorporated into the DNA (7). In this work, the sensitivity has been enhanced by three orders of magnitude through the incorporation of molecular beacon detection (8–11). To illustrate molecular beacon detection of equilibrium cyclization (MBEC), a construct was prepared consisting of a 29-nt duplex flanked by dT19 linkers that are terminated in complementary 5-nt sequences, as depicted in Fig. 1. In the open state the fluorophore, on the 3′ end, and the quencher, on the 5′ end, are distant from one another. In the closed, cyclized state the 5-nt-long reporter duplex is present, and the fluorescence is quenched since the fluorophore and quencher are spatially close to one another. The use of Oregon Green 514 as the fluorophore and Iowa Black as the quencher allows nanomolar sensitivity to be attained, and these dyes have high photostability and are commercially available. When molecular beacons are used to detect hybridization to form the 29-nt-long duplex, the fluorescence increases. IHF binding to the MBEC-labeled DNA leads to a decrease in observed fluorescence, whereas the addition of a competing DNA leads to an increase in fluorescence as in a hybridization experiment.
FIGURE 1.
The MBEC experiment is depicted. The binding of the protein, shown in blue, shifts the equilibrium between the open state and the closed state. In the open state the fluorophore and the quencher are distant from one another. In the closed state the reporter duplex is present, the fluorophore and quencher are close to one another, and the fluorescence is quenched. The binding of the protein IHF shifts the equilibrium toward the closed state. The fluorescence of 4 nM MBEC-H1 DNA in the presence of 1, 10, and 100 nM IHF as a function of temperature is shown. The KD value for this complex is 2.8 nM.
To examine the complexes of a protein with a number of duplex DNAs, only a single MBEC-labeled DNA needs to be prepared. A sample of the MBEC-labeled DNA and the protein of interest are titrated with an unlabeled duplex, or single-stranded, DNA and the change in fluorescence used to monitor the competition. The KD value of the unlabeled duplex DNA can be determined from the fluorescence data and the KD value of the MBEC DNA and the concentrations of the DNAs and the protein.
To demonstrate the utility of MBEC for monitoring protein-DNA interactions, studies were conducted with the multifunctional Escherichia coli integration host factor protein (IHF) as a test system. Since IHF can act as a transcription factor as well as in DNA packaging, it can serve as a model system for studying the interactions of many types of DNA-binding proteins. IHF is a small heterodimeric E. coli protein with two homologous subunits, α and β, which are expressed by unlinked genes (12–14). IHF bends DNA ∼160°, and sequence-specific recognition appears to follow a probabilistic code due to the “wrapping” of the DNA about IHF (2,15–18). The binding of DNA to IHF is thought to be primarily by indirect readout (19–21) and the sequence specificity arises from a combination of direct interactions of Arg residues with the DNA consensus sequence, intercalation of Pro residues to induce DNA kinking, and electrostatic interactions of IHF residues with the DNA backbone (20,22,23).
IHF is a member of the DNABII structural family, and all prokaryotic genomes examined to date have at least one member of this family (24,25). Other members of the DNABII family include TF1 as well as the HU proteins from prokaryotes (20,22,23). IHF participates in chromosome packaging, the regulation of at least 120 genes, the integration of the virus λ, the initiation of DNA replication, and the stabilizing of repressor binding (2,15–17,25,26).
IHF presents an excellent test case for MBEC since there are a number of related DNA sequences that bind to IHF (2,15–17,25,26). IHF binds to ihf sites of the general sequence WATCAANNNNTTR with W being either A or T, R either A or G, and N any nucleotide. The presence of a dA tract downstream from this consensus site can enhance binding, as observed for the H′ site (27). The DNA sequences, with the consensus sequence in bold, studied here are
Hl: d(AGTCACTATGAATCAACTACTTAGATGGT);
H′: d(AAGCATTGCTTATCAATTTGTTGCAACGA);
H2: d(ATGATATAAATATCAATATATTAAATTAG).
The binding affinity and the specificity of IHF depend on the salt concentration (2,16). At potassium concentrations below 100 mM, the specificity of IHF is low and the occluded site for nonspecific binding is much smaller than that for specific binding (2,16). The specificity and the size of the occluded site increase as the salt concentration increases (2,16). The investigation of the association constant as a function of salt concentration has been used to investigate the net number of salt bridges formed in the DNA-IHF complex (2,16).
The algebraic sign of the change in fluorescence depends on the type of experiment being performed. Since IHF bends duplex DNA, the binding of IHF to MBEC-labeled duplex DNA decreases the observed fluorescence. The addition of a competing DNA increases the observed fluorescence. When molecular beacons are used to detect hybridization, the fluorescence increases.
In this study we show that MBEC can be used to investigate both the sequence specificity and the salt dependence of IHF-DNA complex formation. The MBEC results are in agreement with those previously obtained by gel mobility shift and calorimetry experiments. Given the multifunctional nature of IHF and the different modes of interaction with DNA, these results are strongly suggestive that MBEC can be broadly applied to the study of protein-DNA interactions.
MATERIALS AND METHODS
DNA samples
The DNA samples used in this study are listed below with the consensus sequence region shown in bold type. The extinction coefficients, ɛ, at 260 nm, L/(mol-cm), of the DNAs are also listed.
H′: 5′-AAGCATTGCTTATCAATTTGTTGCAACGA-3′ ɛ = 282,900
3′-TTCGTAACGAATAGTTAAACAACGTTGCT-5′ ɛ = 284,000
H1: 5′-AGTCACTATGAATCAACTACTTAGATGGT-3′ ɛ = 291,500
3′-TCAGTGATACTTAGTTGATGAATCTACCA-5′ ɛ = 289,200
H2: 5′-ATGATATAAATATCAATATATTAAATTAG-3′ ɛ = 316,000
3′-TACTATATTTATAGTTATATAATTTAATC-5′ ɛ = 294,500
Scrambled: 5′-CCGGCGCATATATGGCGTATATAGCCCGG-3′ ɛ = 278,800
3′-GGCCGCGTATATACCGCATATATCGGGCC-5′ ɛ = 274,300
ssDNA: 5′-CCGGGCGCGCGATATATGCGCCGCGCCGG-3′ ɛ = 262,400
MBEC-H1, with IB the position of attachment of Iowa Black (Integrated Technologies, Coralville, IA) and OG the point of attachment of Oregon Green (Molecular Probes, Eugene, OR), has ɛ = 668,100 before labeling. The sequence of the labeled strand is 5′-(IB)GCCCA(T19)AGTCACTATGAATCAACTACTTAGATGGT(T19)TGGGC(T-OG)T-3′. The duplex formed between this DNA and the complementary 29mer is referred to as MBEC-H1. The DNA samples were obtained from Integrated DNA Technologies (Coralville, IA).
Molecular beacon donor and acceptor
The Oregon Green 514 (Molecular Probes) was obtained in the form of succinimidyl ester of the carboxylic acid. The Oregon Green was covalently attached to the DNA as described in the Supplementary Material.
IHF preparation
IHF was prepared as previously described (28,29). The activity of the IHF used here had the same activity in gel mobility shift assays as that from previous preparations and reported by others (30). The KD values determined below also indicate that the activity of the IHF was at least comparable to that previously used.
Fluorescence experiments
Fluorescence data were collected with a Fluoromax-2 (Jobin Yvon-Spex, Longjumeau, France) fluorimeter using excitation centered at 505 nm and an emission wavelength of 530 nm with the samples at 310 K. Between titrations, solutions were allowed to sit for 15 min to equilibrate at the specified temperature. The fluorimeter has a four-position cell holder and the temperature was monitored at the sample holder. The incoming light intensity was used to correct for fluctuations in excitation intensity. Additional information on the experimental parameters is contained in the Supplementary Material.
The DNAs were annealed at 363 K for 10 min and then allowed to sit overnight in a water bath, and 4 nM of MBEC-H1 was used in each of the titration experiments. An initial volume of 330 μL of MBEC-H1 was used for each experiment. Siliconized tips were used to prevent “sticking” of the Oregon Green 514 labeled DNA. The stock solutions of oligonucleotides used for the competition titration experiments were 0.2, 0.5, and 1 μM. Dilution was less than 15% in all of the titrations and was corrected for. All buffers used contained 10 mM HEPES, 0.5 mM EDTA, 60 mM KCl, 5% glycerol at pH 8.0.
Temperature dependence of MBEC
The temperature dependence of the fluorescence of the MBEC-H1 DNA in the presence of 1, 10, and 100 nM IHF as well as in the absence of IHF was determined and shown in the Supplementary Material. The results indicate that 310 K and 10 nM IHF are appropriate for the competition experiments. As a control the fluorescence of the single-stranded MBEC-H1 DNA was also determined as a function of temperature as shown in the Supplementary Material.
Fitting of fluorescence data
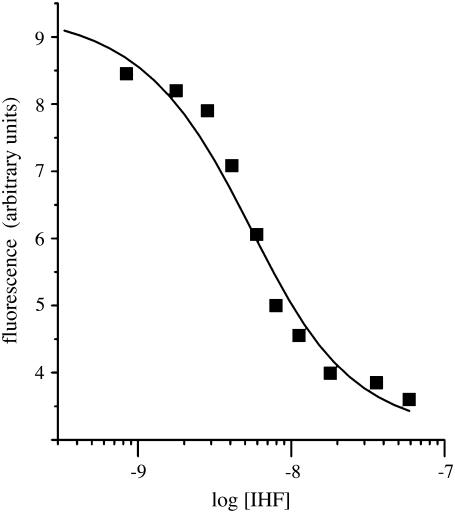
The IHF titration shown in Fig. 2 was fit using the Origin 6.0 program (OriginLab, Northampton, MA). Data were corrected for dilution and fit assuming a 1:1 binding interaction using the following equation:
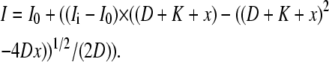 |
Where I0 is the initial intensity, Ii is the intensity at saturation, D is the total DNA concentration, K is the KD, and x is the total concentration of IHF. The competition data were fit using the DynaFit program (BioKin, Pullman, WA) and the script used is given in the Supplementary Material. To verify the fitting procedures, fits were carried out on simulated data. Simulated data points were generated using Microsoft Excel (Microsoft, Redmond, WA). The equilibrium competition was modeled using
 |
FIGURE 2.
The fluorescence of MBEC-H1 DNA as a function of IHF concentration is shown. The KD value for this complex from this single data set was determined to be 2.8 nM.
On average, 11 iterations were needed to establish less than 1% change in free IHF concentration from the previous iteration, as detailed in the Supplementary Material. Simulations were carried out for the competitor concentrations of 0, 1.44, 3.01, 6.16, 9.97, 14.7, 20.8, 29.0, 41.1, 52.7, 66.2, and 99 nM. The simulations give the fraction bound, fb, for calculating the total fluorescence intensity using
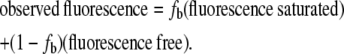 |
Fits of simulated two-equilibrium competition data
The simulated data were fit using the DynaFit program described above, and the fits are shown in the Supplementary Material. The KD values determined by the fitting procedure were within 1.2% of the input values.
Reproducibility of MBEC competition data
The reproducibility of the experimental competition data was determined by repeating the H1 competition experiment four times with independent samples. The KD values from the separate runs are 6.4, 10.9, and 10.8 nM each with a 10% error. The KD value of the average data is 8.9 ± 0.5 nM. The experimental data and analysis of the reproducibility is presented in the Supplementary Material.
Gel electrophoresis experiments
Polyacrylamide gels (6.5%) were prepared from 9.75 mL of 40% acrylamide, 1.5 mL 10× TBE (890 mM Tris, 890 mM boric acid, 0.5 M EDTA with pH 8.0), 300 μL of ammonium persulfate, and 48.42 mL of doubly distilled H2O. This solution was degassed for 10 min before the 7.75 × 7.75 inch and 7.75 × 6.75 inch gels were cast after initiating polymerization with 30 μL of TEMED (N,N,N′,N′-tetramethylethylenediamine). The polymerized gels had 20 lanes, and 1 L of 0.25× TBE was used as running buffer. The gel was prerun for 20 min at 180 V, and the samples were then added to the wells and the gel was run at 180 V for 2–3 h at 5°C.
The IHF concentrations were 0, 1.44, 3.01, 6.16, 9.97, 14.7, 20.8, 29, 41.1, 52.7, 66.2, 99, 120, 160, 190, 500, and 6000 nM. Each sample also contained 4 nM of MBEC-H1 construct and 10 nM of one of the unlabeled constructs as well as 2% Ficoll (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK) in a total volume of 20 μL. The DNA marker lane contained 4 μL of DNA markers, 2 μL of gel loading solution dye, and 2 μL of 0.25× TBE. The DNA markers were 11, 18, 80, 102, 174, 257, 267, 298, 434, 458, and 587 basepairs long.
The gels were stained using Sybr Green I for 25 min. The gel was washed with doubly distilled H2O and analyzed with Storm 840 Imager. The gel was excited at 450 nm, and emission was collected at 520 nm with a pixel size of 200 microns. Analysis of IHF binding was based on the band intensity for the free DNA (31), and the binding constants were determined using a 1:1 binding function using Origin 6.0.
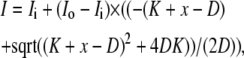 |
where I0 is the initial intensity, Ii is the intensity at saturation, D is the total DNA concentration, K is the KD, and x is the total concentration of IHF.
RESULTS AND DISCUSSION
The 29-nt H1 duplex was placed into an MBEC construct to make MBEC-H1 as depicted in Fig. 1. The binding of IHF to this DNA increases the percentage of the DNA in the closed, low fluorescence state. The temperature dependence of the fluorescence was measured for the MBEC-H1 DNA in the presence and in the absence of IHF. The results in Fig. 1 show that the melting temperature increases by more than 20 K in the presence of IHF. When the reporter duplex contained seven basepairs, the melting temperatures were ∼10 K higher. The binding experiments were carried out at the physiological temperature of 310 K, and at this temperature there is a large difference in the fluorescence between the free and bound states of MBEC-H1. The sample temperature was regulated to within 1 K, and temperature fluctuations of this size will have a small effect on the fluorescence of both free and bound MBEC-H1. The dynamic range of the experiment is determined by the difference in the fluorescence intensity for the free and bound MBEC-H1. The sensitivity of the experiment is high since the fluorescence of the Oregon Green 514 is comparable to that of fluorescein. The MBEC approach was found to offer about three orders of magnitude more sensitivity than the use of the fluorescence of 2-aminopurine. The increase in sensitivity opens up the capability to examine sequence-specific protein-DNA complexes and allows the use of competition experiments that yield accurate ratios of KD values while requiring only a single-labeled DNA. In addition, we have demonstrated that the approach can be used to assay binding under a variety of different solution conditions. With this increase in sensitivity, nanomolar dissociation constants can now be determined by this method.
A 4 nM sample of MBEC-H1 at 310 K was titrated with IHF with the results shown in Fig. 2. The fluorescence decreases as the IHF concentration increases and the plot of the fluorescence intensity versus IHF concentration exhibits the shape of a typical binding curve as shown in Fig. 2. The fit of this fluorescence data gives a KD value for the IHF-MBEC H1 complex of 2.8 nM, as indicated by the results in Fig. 1. The increase in the percentage of the DNA in the closed form may be due to the bending of the DNA in the complex with IHF.
The results in Figs. 1 and 2 were obtained at a DNA concentration of 4 nM. The titration of MBEC-H1 with the IHF experiment was repeated three times with independent samples, the error in the fit of each individual KD value is on the order of 10%, and the reproducibility was also found to be on the order of 10%, as described in the Supplementary Material. These results show that MBEC can be used to monitor the association of IHF with DNA in solution with high sensitivity and reproducibility.
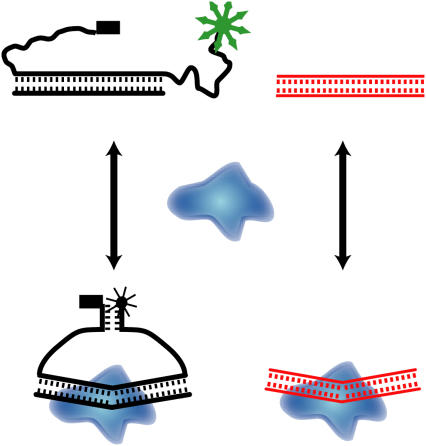
The determination of the KD values of additional DNAs by this approach would require making a MBEC construct for each DNA. In addition, the tail and/or reporter duplex parts of the MBEC constructs may have interactions with IHF that depend on the sequence of the duplex in conjunction with the reporter duplex. Therefore, the KD values for DNA-IHF complexes have been determined by competition experiments. The approach for the competition experiments is depicted in Fig. 3. The binding of IHF to the competing DNA leads to a decrease in the amount of IHF available to bind to the MBEC DNA, and this causes an increase in fluorescence of the MBEC DNA. Two distinct advantages of the competition experiments are that the association of unlabeled DNA with IHF can be monitored with the use of only one MBEC construct DNA, and the KD values for the unlabeled DNAs used in the competition experiments do not depend on the details of the association of IHF with the labeled MBEC-H1, as shown in Fig. 1. In addition, the ratios of the KD values obtained by competition experiments are expected to be highly reliable. In many cases subtle differences in binding behavior are more directly revealed through competition experiments than through direct titration.
FIGURE 3.
The use of MBEC to determine KD values via a competition experiment is depicted. The fluorescence of the MBEC-H1 DNA in the presence of IHF is monitored as a function of the concentration of a competing DNA. The two DNAs compete for IHF binding. The fluorescence as a function of competing DNA concentration can be analyzed to give the KD for the IHF-competing DNA complex using the known KD for the complex of IHF with MBEC-H1 and the known concentrations of IHF, MBEC-H1, and the competing DNA.
In the competition experiments the concentrations of MBEC-H1 and IHF were chosen so that the addition of the competing DNA would induce readily observable changes in fluorescence. The initial conditions, 4 nM MBEC-H1 and 10 nM IHF, of the competition experiments were such that most of the MBEC DNA was bound to IHF. To optimize the experimental conditions and to validate the analyses to determine KD values from the competition data, extensive simulations were carried out.
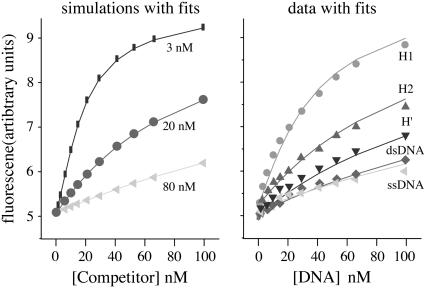
The competition experiments were simulated for a range of KD values using a two-equilibrium model in which the MBEC-H1 and unlabeled DNA compete for IHF binding. Simulations were run using KD values for the competing DNA of 3, 20, and 80 nM, as shown in Fig. 4, using the experimentally determined KD value for the MBEC-H1 complex, 2.8 nM. The simulated data were then fit, using DynaFit, with a two-equilibrium model, and the results are shown in Fig. 4. As described in the Supplementary Material the KD values determined from the fits of the simulated data were found to be in excellent agreement with the input values. These findings indicate that the two-equilibrium model and our analyses of it are appropriate for describing the experimental competition data.
FIGURE 4.
On the left, the simulations of the competition experiment for KD values of the competing DNA of 3, 20, and 80 nM are shown along with the fits of the data. On the right, the experimental results for H1 ( , 8.9 nM KD), H2 (▴, 28 nM KD), H′ (▾, 38 nM KD), a DNA duplex with a scrambled sequence (♦, 71 nM KD), and single-stranded DNA (
, 8.9 nM KD), H2 (▴, 28 nM KD), H′ (▾, 38 nM KD), a DNA duplex with a scrambled sequence (♦, 71 nM KD), and single-stranded DNA ( , 96 nM KD) are shown along with the fits of the data.
, 96 nM KD) are shown along with the fits of the data.
The fluorescence of MBEC-H1 in the presence of IHF as a function of the concentration of the competing H1 duplex DNA is shown in Fig. 4, and these data were used to determine the KD value of the H1-IHF complex. The observed fluorescence data were fit using as inputs the KD value for MBEC-H1 and the known total concentrations of IHF, MBEC-H1, and H1. The KD value for the H1-IHF complex was found to be 8.9 nM. The KD value was determined for three separate sets of data as well as for the average of the three sets of data.
Since the observed KD value for H1 from the competition experiments is more than twice that of MBEC-H1, it appears that IHF has modest favorable interactions with the tails and reporter duplex of the MBEC construct. Bending of the MBEC construct relative to the duplex DNA as well as the formation of the reporter duplex could also enhance the affinity of the MBEC-HI-IHF interaction. Additional validation came from competition experiments that gave the same KD value for the H1 DNA when carried out at different IHF concentrations. We note that the details of the interaction of IHF with the MBEC-H1 DNA will not affect the KD values determined for the competing DNAs.
Extensive simulations and fittings of the experimental data, shown in the Supplementary Material, indicate that the KD values can be determined to within ∼10% from MBEC competition data. As described in the Supplementary Material the reproducibility of the competition experiments was found to be within 10%. In addition, the KD values obtained using the average of three experiments are the same, within experimental error, as that obtained from averaging the KD values determined from the separate experiments.
This competition procedure has also been used to determine the KD values for the H′ and H2 DNAs as well as a duplex DNA with a scrambled sequence and a single-stranded DNA, with the results shown in Fig. 4. The KD values determined here for H1, H2, and H′ are 8.9, 28, and 38 nM, respectively. The H′ sequence exhibits a lower affinity relative to H1 and H2 than previously observed by other methods probably because of the absence of the A-tract sequence (30). The KD values for a random sequence duplex DNA, 71 nM, and a ssDNA, 96 nM, were also determined. This range of KD values is consistent with prior reports on the specificity of IHF (30).
These results show that MBEC can allow DNA-protein complex formation to be monitored under equilibrium conditions with high sensitivity and reproducibility using currently available fluorimeters and plate readers. Since the KD values are determined by competition experiments, only a single MBEC DNA is needed, which enhances the cost effectiveness of the method as well as reducing the time and effort needed. It is noted that the MBEC method reports on how much of the DNA-protein complex is present and not on the details of the structure of the complex. This makes the method useful for determination of KD values as a function of sequence, or other variable, which can be used to infer structural information. The temperature range of MBEC can be tuned by varying the number of basepairs in the reporter duplex.
In this study protein binding induces further bending of the DNA in the MBEC construct. It is noted that bending is not required for this technique to determine KD values. For example, if protein binding caused the opening of the reporter duplex, due to steric or other effects, then the fluorescence would increase upon protein binding whereas it decreases upon IHF binding.
Comparison of MBEC results with those obtained by gel mobility shift experiments
Since gel mobility shift experiments are widely used, we have compared the MBEC results with those from gel shifts. Although there are literature reports on the binding of H1, H2, and H′ DNAs to IHF (15–17,19,21,27,30,32,33), these prior results were obtained on a variety of constructs that are not the same as those used here for the MBEC experiments; so a direct comparison of KD values is not appropriate. Therefore we have carried out gel mobility shift experiments using the same DNAs as used in the MBEC experiments. These experiments were designed to compare the two techniques more than to gain new information about the sequence specificity of IHF.
The gel shift experiments were carried out with each of the same DNAs and over the same range of IHF concentrations as used in the MBEC experiments so that the results of the two approaches can be directly compared. A typical gel is shown in Fig. 5. The KD values for the DNA-IHF complexes obtained by gel mobility shift assays and by MBEC are listed in Table 1. The results from the two methods are in good general agreement with each other especially with respect to the ratio of any pair of KD values. The KD values obtained by gel mobility shift experiments are larger than those obtained by MBEC. Because of dilution effects and the nonequilibrium nature of the experiment, gel mobility shift experiments tend to underestimate the strength of complex formation (31). Also, the errors in the fits of the KD values from gel mobility shift experiments are significantly larger than those determined from MBEC experiments.
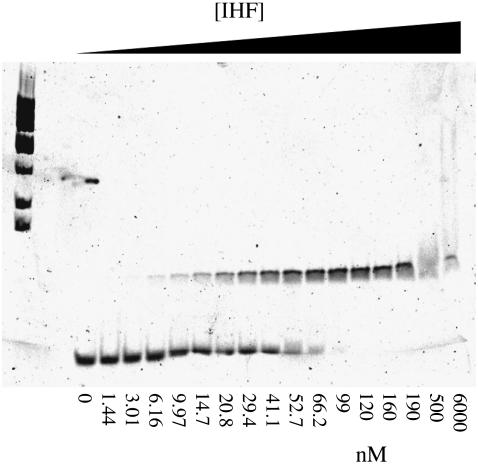
FIGURE 5.
Gel mobility shift assay results for the 29-nt duplex H1 DNA at a concentration of 10 nM with IHF are shown. The KD value determined from this gel is 27 ± 7 nM. At high IHF concentrations smearing of the bands is observed that may be due to formation of complexes containing more than one IHF.
TABLE 1.
The KD values and errors, in nM, for the formation of the IHF complexes with DNA, as determined by MBEC and GMSA are listed
| MBEC | error | GMSA | error | |
|---|---|---|---|---|
| MBEC-H1 | 2.8 | 0.3 | ||
| H1 | 8.9 | 0.5 | 31 | 14 |
| H2 | 28 | 1.3 | 98 | 12 |
| H′ | 38 | 1.3 | 110 | 16 |
| scrambled duplex | 71 | 2.3 | 155 | 41 |
| scrambled ssDNA | 96 | 6.4 | 166 | 75 |
The gel shift results indicate that higher order complexes are observed at protein concentrations near 1 × 10−6 M, as shown in the Supplementary Material. The MBEC experiments were carried out at concentrations up to 10−7 M. Fits of the MBEC data did not improve significantly when higher order complexes were included, indicating that these are not present at high percentage at concentrations of up to 10−7 M. Additional complexes due to nonspecific binding are observed at protein concentrations higher than 10−6 M.
Determination of the KD values as a function of salt concentration
The formation of DNA-protein complexes has been studied as a function of salt concentration to determine the net number of salt bridges formed in the complex. This information is of interest in gaining understanding about the structures and interactions present in DNA-protein interactions as well as the sequence dependence of the interactions. Record and co-workers have used calorimetry to investigate the salt dependence of the KD value of DNA-IHF complexes as a function of potassium concentration (2,16). They have found that the net number of salt bridges, about seven, is much smaller than the number predicted by the crystal structure of the DNA-IHF complex (2,16). The specificity of the formation of DNA-IHF complexes is also dependent on the concentration of potassium (2,16). These and other results have led to a model of DNA-IHF complex formation in which IHF loses many salt bridges to accommodate DNA binding (2,16).
The MBEC experiment is particularly well suited for the determination of KD values as a function of potassium concentration and can do so at concentrations orders of magnitude lower than those used in calorimetry experiments. The fluorescence of MBEC-H1 was determined as a function of IHF concentration over a range of potassium concentration, with the results shown in Fig. 6 and Table 2. As expected, increases in the potassium concentration lead to increases in the KD values for both specific and nonspecific binding. The results also indicate that the MBEC approach is suitable over a wide range of solvent conditions.
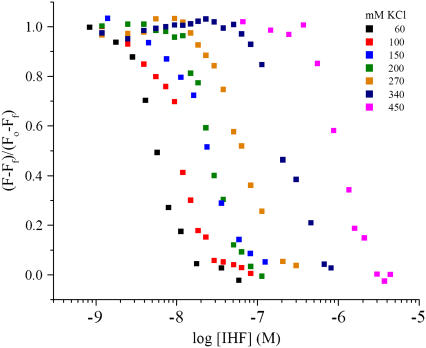
FIGURE 6.
Fluorescence of the MBEC-H1 DNA as a function of IHF concentration was obtained at 60, 100, 150, 200, 270, 340, and 450 mM KCl. The KD values of complex formation were determined at each KCl concentration and are given in Table 2. The plots are of the observed fluorescence minus the final fluorescence divided by the initial fluorescence minus the final fluorescence.
TABLE 2.
The KD values as a function of the concentration of KCl for the formation of the complexes of IHF with duplex MBEC-H1 and single-stranded MBEC-H1 are listed
| [KCl], mM | Duplex MBEC-H1 | ssMBEC-H1 |
|---|---|---|
| 60 | 3.7 | 42 |
| 100 | 9.3 | 160 |
| 150 | 21 | 250 |
| 200 | 27 | 900 |
| 270 | 64 | |
| 340 | 230 | |
| 450 | 1080 |
Visual inspection of the results in Fig. 6 suggests that the binding is partially cooperative. However, the quality of the fit does not improve substantially upon inclusion of an additional parameter for cooperativity. The increase in the size of the occluded site as a function of salt concentration (2,34) will also mimic the effect of cooperativity. The apparent cooperativity may arise in part because of the smaller dynamic range of the data at high salt concentrations. However, even at the highest salt concentrations at 37°C, the difference in fluorescence intensity between free and bound forms is sufficient for determining KD values.
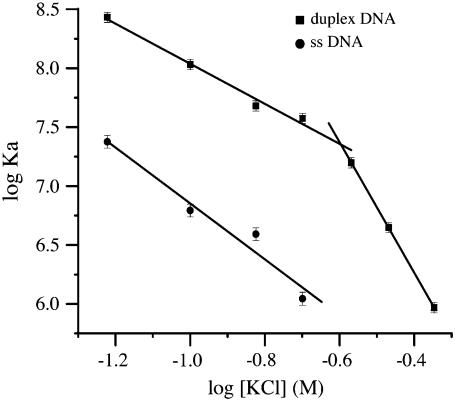
The determination of the net number of salt bridges comes from plots of the log of Ka versus the log of the potassium concentration, as shown in Fig. 7. The results for specific binding are similar to those obtained by calorimetry (2,16) with the KD value having a small slope at salt concentrations below 100 mM, −1.7, with the slope increasing at higher salt concentrations to −5.5. The value of −5.5 is in good agreement with the slope of −7 derived from calorimetry data (2,16). Results were also obtained for nonspecific binding, which gave a slope of −2.4, and these are included in Fig. 7.
FIGURE 7.
The logs of the Ka values for duplex and single-stranded MBEC-H1 DNAs are plotted as a function of the log of the KCl concentration along with the error bar for each data point. The plot of log Ka versus log KCl indicates the number of cations released during complex formation. The slopes for the duplex DNA case are −1.7 in low salt and −5.5 in high salt. The slope for nonspecific, single-stranded DNA is −2.4.
It is seen that the specificity of IHF-DNA complex formation increases with increasing salt concentration, up to ∼250 mM KCl, since the Ka values of the nonspecific complex formation have a larger slope than for specific complex formation. The calorimetry experiments were carried out at much higher concentrations than the MBEC experiments, and more nonspecific binding was observed by calorimetry than by MBEC, presumably due to the large difference in sample concentration. At KCl concentrations above ∼250 mM, the specificity begins to decrease as the slope of the specific binding becomes greater than that of the nonspecific binding at these high KCl concentrations.
Future directions
The MBEC methodology is being adapted to high throughput screening of sequence specificity, both of the protein and the DNA, as well as inhibition of DNA-protein complex formation as a function of sequence. It is noted that bending of the DNA is not the only way that protein binding can alter the equilibrium. The MBEC approach is now being applied to MutS, a large protein.
SUPPLEMENTARY MATERIAL
To view all of the supplemental files associated with this article, visit www.biophysj.org.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation Grant MCB 0316625 (I.M.), an investigator grant from the Patrick and Catherine Weldon Donaghue Medical Research Foundation (I.M.), and National Institutes of Health Grant GM 076550 (P.H.B.). The National Institutes of Health Molecular Biophysics training grant GM 008272 and the Howard Hughes Medical Institute supported J.V. and L.A.
Editor: Jonathan B. Chaires.
References
- 1.Kozlov, A. G., and T. M. Lohman. 2000. Large contributions of coupled protonation equilibria to the observed enthalpy and heat capacity changes for ssDNA binding to Escherichia coli SSB protein. Proteins. 4(Suppl.):8–22. [DOI] [PubMed] [Google Scholar]
- 2.Saecker, R. M., and M. T. Record Jr. 2002. Protein surface salt bridges and paths for DNA wrapping. Curr. Opin. Struct. Biol. 12:311–319. [DOI] [PubMed] [Google Scholar]
- 3.Overman, L. B., and T. M. Lohman. 1994. Linkage of pH, anion and cation effects in protein-nucleic acid equilibria. Escherichia coli SSB protein-single-stranded nucleic acid interactions. J. Mol. Biol. 236:165–178. [DOI] [PubMed] [Google Scholar]
- 4.Petri, V., M. Hsieh, E. Jamison, and M. Brenowitz. 1998. DNA sequence-specific recognition by the Saccharomyces cerevisiae “TATA” binding protein: promoter-dependent differences in the thermodynamics and kinetics of binding. Biochemistry. 37:15842–15849. [DOI] [PubMed] [Google Scholar]
- 5.Petri, V., M. Hsieh, and M. Brenowitz. 1995. Thermodynamic and kinetic characterization of the binding of the TATA binding protein to the adenovirus E4 promoter. Biochemistry. 34:9977–9984. [DOI] [PubMed] [Google Scholar]
- 6.Jen-Jacobson, L., L. E. Engler, and L. A. Jacobson. 2000. Structural and thermodynamic strategies for site-specific DNA binding proteins. Struct. Fold. Des. 8:1015–1023. [DOI] [PubMed] [Google Scholar]
- 7.Arthanari, H., K. Wojtuszewski, I. Mukerji, and P. H. Bolton. 2004. Effects of HU binding on the equilibrium cyclization of mismatched, curved, and normal DNA. Biophys. J. 86:1625–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnet, G., O. Krichevsky, and A. Libchaber. 1998. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc. Natl. Acad. Sci. USA. 95:8602–8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnet, G., S. Tyagi, A. Libchaber, and F. R. Kramer. 1999. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc. Natl. Acad. Sci. USA. 96:6171–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang, X., Y. Mi, J. J. Li, T. Beck, S. Schuster, and W. Tan. 2002. Molecular beacons: fluorogenic probes for living cell study. Cell Biochem. Biophys. 37:71–81. [DOI] [PubMed] [Google Scholar]
- 11.Tan, W., X. Fang, J. Li, and X. Liu. 2000. Molecular beacons: a novel DNA probe for nucleic acid and protein studies. Chemistry (Easton). 6:1107–1111. [DOI] [PubMed] [Google Scholar]
- 12.Swinger, K. K., and P. A. Rice. 2004. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 14:28–35. [DOI] [PubMed] [Google Scholar]
- 13.Azam, T. A., S. Hiraga, and A. Ishihama. 2000. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 5:613–626. [DOI] [PubMed] [Google Scholar]
- 14.Bonnefoy, E., and J. Rouviere-Yaniv. 1991. HU and IHF, two homologous histone-like proteins of Escherichia coli, form different protein-DNA complexes with short DNA fragments. EMBO J. 10:687–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zulianello, L., P. van Ulsen, P. van de Putte, and N. Goosen. 1995. Participation of the flank regions of the integration host factor protein in the specificity and stability of DNA binding. J. Biol. Chem. 270:17902–17907. [DOI] [PubMed] [Google Scholar]
- 16.Holbrook, J. A., O. V. Tsodikov, R. M. Saecker, and M. T. Record Jr. 2001. Specific and non-specific interactions of integration host factor with DNA: thermodynamic evidence for disruption of multiple IHF surface salt-bridges coupled to DNA binding. J. Mol. Biol. 310:379–401. [DOI] [PubMed] [Google Scholar]
- 17.Yang, C. C., and H. A. Nash. 1989. The interaction of E. coli IHF protein with its specific binding sites. Cell. 57:869–880. [DOI] [PubMed] [Google Scholar]
- 18.Lee, E. C., L. M. Hales, R. I. Gumport, and J. F. Gardner. 1992. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 11:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lynch, T. W., E. K. Read, A. N. Mattis, J. F. Gardner, and P. A. Rice. 2003. Integration host factor: putting a twist on protein-DNA recognition. J. Mol. Biol. 330:493–502. [DOI] [PubMed] [Google Scholar]
- 20.Rice, P. A., S. Yang, K. Mizuuchi, and H. A. Nash. 1996. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 87:1295–1306. [DOI] [PubMed] [Google Scholar]
- 21.Travers, A. 1997. DNA-protein interactions: IHF—the master bender. Curr. Biol. 7:R252–R254. [DOI] [PubMed] [Google Scholar]
- 22.Ellenberger, T., and A. Landy. 1997. A good turn for DNA: the structure of integration host factor bound to DNA. Structure. 5:153–157. [DOI] [PubMed] [Google Scholar]
- 23.Rice, P. A. 1997. Making DNA do a U-turn: IHF and related proteins. Curr. Opin. Struct. Biol. 7:86–93. [DOI] [PubMed] [Google Scholar]
- 24.Drlica, K., and J. Rouviere-Yaniv. 1987. Histonelike proteins of bacteria. Microbiol. Rev. 51:301–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freundlich, M., N. Ramani, E. Mathew, A. Sirko, and P. Tsui. 1992. The role of integration host factor in gene expression in Escherichia coli. Mol. Microbiol. 6:2557–2563. [DOI] [PubMed] [Google Scholar]
- 26.Dame, R. T., J. van Mameren, M. S. Luijsterburg, M. E. Mysiak, A. Janicijevic, G. Pazdzior, P. C. van der Vliet, C. Wyman, and G. J. Wuite. 2005. Analysis of scanning force microscopy images of protein-induced DNA bending using simulations. Nucleic Acids Res. 33:e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hales, L. M., R. I. Gumport, and J. F. Gardner. 1996. Examining the contribution of a dA+dT element to the conformation of Escherichia coli integration host factor-DNA complexes. Nucleic Acids Res. 24:1780–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nash, H. A., C. A. Robertson, E. Flamm, R. A. Weisberg, and H. I. Miller. 1987. Overproduction of Escherichia coli integration host factor, a protein with nonidentical subunits. J. Bacteriol. 169:4124–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Filutowicz, M., H. Grimek, and K. Appelt. 1994. Purification of the Escherichia coli integration host factor (IHF) in one chromatographic step. Gene. 147:149–150. [DOI] [PubMed] [Google Scholar]
- 30.Wang, S., R. Cosstick, J. F. Gardner, and R. I. Gumport. 1995. The specific binding of Escherichia coli integration host factor involves both major and minor grooves of DNA. Biochemistry. 34:13082–13090. [DOI] [PubMed] [Google Scholar]
- 31.Carey, J. 1991. Gel retardation. Methods Enzymol. 208:103–117. [DOI] [PubMed] [Google Scholar]
- 32.Hales, L. M., R. I. Gumport, and J. F. Gardner. 1994. Determining the DNA sequence elements required for binding integration host factor to two different target sites. J. Bacteriol. 176:2999–3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azam, T. A., and A. Ishihama. 1999. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J. Biol. Chem. 274:33105–33113. [DOI] [PubMed] [Google Scholar]
- 34.Record, M. T. Jr., W. Zhang, and C. F. Anderson. 1998. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 51:281–353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.