Abstract
The location of each lipid in a palmitoyloleoylphosphatidylcholine/18:0 sphingomyelin/cholesterol monolayer system is laterally resolved using imaging time-of-flight secondary ion mass spectrometry (TOF-SIMS) without the necessity of adding fluorescent labels. This system of coexisting immiscible liquid phases shows cholesterol domains with sizes and shapes comparable to those in the fluorescence microscopy literature. The results show that SM localizes with cholesterol and that palmitoyloleoylphosphatidylcholine is excluded. Moreover, the segregation is not complete, and there is a small amount of both phospholipids distributed throughout.
Introduction
Upon the basis of the detergent-resistant fraction of cellular membranes, the content of lipid rafts includes sphingomyelin and cholesterol.1 Both cholesterol and sphingomyelin have been hypothesized to be essential for lipid raft formation in the cellular membrane.2 Also, a recent study of the native HIV membrane lipidome provides strong evidence for the existence of lipid rafts in living cells.3 Currently, a substantial body of research is focused on elucidating the interactions between cholesterol and sphingomyelin (ref 4 and references therein). Many methods have been used to investigate sphingomyelin–cholesterol interactions, including nuclear magnetic resonance spectroscopy (NMR)5 and fluorescence microscopy.4,6–13 To obtain images, fluorescence microscopy is the most common technique for visualizing lipid miscibility; however, it requires the addition of fluorescent labels. The degree of influence that the added fluorophores have on domain formation is still in question, but recently Cruz et al. found that trace amounts (<1 mol %) of this probe perturb the morphology of micro- and nanodomains of dipamitoylphosphatidylcholin monolayers.14
One technique that is capable of direct chemical imaging of lipid domains is secondary ion mass spectrometry (SIMS).15–23 In this technique, a focused ion beam is used to desorb secondary ions from the sample, and then the beam is moved across the surface and mass spectra are acquired at each spot on the surface. Using characteristic mass values, the lateral distribution of the individual chemical components are revealed.24 There are two major approaches depending upon the fluence of the primary ion. With static SIMS, a pulsed primary ion beam is used, and the mass spectrum is measured before a significant part of the surface layer has been chemically modified and sputtered away. The low dose of the primary ion allows for high surface sensitivity and molecular information being obtained from the sample without chemical labeling. With dynamic SIMS, a continuous primary ion beam bombards the sample, which leads to sputtering of atoms from the surface. Dynamic SIMS is capable of elemental and isotope analysis and has higher lateral resolution for SIMS imaging. However, isotope labeling is usually required to differentiate molecular species in the sample system. Here we present static time-of-flight (TOF) SIMS studies on monolayers of palmitoyloleoylphosphatidylcholine (POPC)/18:0 sphingomyelin (SM)/cholesterol (CH). Without the use of chemical labels, we find that the ternary mixture of 30:47:23 POPC/18:0 SM/CH produces coexisting liquid phases in which SM is predominantly found within the CH-rich phase and the POPC is predominantly excluded. The location of SM is elucidated without the incorporation of any added labels. Also, TOF-SIMS shows clearly that the lipids do not entirely segregate from one another and illustrates the complexity of lipid interactions.
Materials and Methods
For lipid monolayer characterization by imaging TOF-SIMS, monolayers at the air–water interface were deposited vertically onto solid substrates via the Langmuir–Blodgett (LB) technique. Similar methods have also been utilized for analyses of lipid monolayers by atomic force microscopy,25,26 and supported LB films have been shown to maintain the same lateral distribution of lipids observed at the air–water interface.16,17 Although cellular membranes exist at higher pressures and temperatures, lower pressures are necessary to study lipid miscibility10 in the mixtures. To identify the contents of coexisting phases in our lipid monolayer, we have selected 7 mN/m to ensure the existence of multiple phases in our system because fluorescence microscopy data is already available at similar pressures12 for direct comparison. Also, recent studies suggest that the more relevant parameter to consider when comparing lipid monolayers to cellular membranes is the molecular density.12 The molecular area of our ternary mixture at lower pressure is considerably closer to the molecular area per lipid of an erythrocyte membrane (~40 Å2 for the outer leaflet27 and ~60 Å2 for the inner leaflet28) than that at very high pressure. The materials, substrate preparation, monolayer preparation and deposition, and TOF-SIMS characterization are briefly detailed here. All steps were completed at room temperature (23 ± 2 °C).
Materials
POPC, CH (both from Avanti Polar Lipids, Inc., Alabaster, AL), 18:0 SM (Matreya LLC, Pleasant Gap, PA), 16-mercaptohexadecanoic acid (Sigma-Aldrich, St. Louis, MO), 2-propanol, methanol, and chloroform were used without further purification. A Nanopure Diamond Life Science ultrapure water system (Barnstead International, Dubuque, IA) was used to purify the water used in the production of all monolayers (resistivity of 18.2 MΩ cm).
Substrate Preparation
Substrates were 16-mercaptohexadecanoic acid self-assembled monolayers (SAMs) on gold. The gold was deposited onto single-crystal (100) silicon wafers that were first cleaned via piranha etch (3:1 H2SO4/H2O2) to ensure a uniform SiO2 surface. (Extreme caution must be exercised when using piranha etch. An explosion-proof hood should be used.) The silicon substrates were deposited with chromium followed by gold as described by Fisher et al.29 For the formation of the SAMs onto gold, a 1 mM solution of 16-mercaptohexadecanoic acid in 2-propanol was used. Gold deposition, SAM self-assembly, and LB film deposition were confirmed with a single-wavelength (632.8 nm, 1 mm spot size, 70° angle of incidence) Stokes ellipsometer (Gaertner Scientific Corporation, Skokie, IL; model LSE). SAMs on gold substrates were used because these provide higher positive ion SIMS signals of lipid monolayers than do silicon substrates30 and they are also easily reproducible uniformly hydrophilic substrates necessary for our lipid monolayer deposition method.
Monolayer Preparation, Isotherm Analysis, and Sample Deposition
A Kibron μ Trough S-LB (Helsinki, Finland) was used for isotherm acquisition, LB film preparation, and deposition. The subphase was ~65 mL of purified water. All lipid solutions were dissolved in 9:1 chloroform/methanol. At least 15 min was allotted for film equilibration before compression to guarantee complete solvent evaporation. The surface pressure was measured with a Wilhelmy wire interfaced to a personal computer. Uniform compression (7 Å2/molecule/min) of the lipid monolayer was ensured through computer control of the trough barriers, and this also allowed for constant pressure feedback during deposition. The lipid films were each deposited vertically onto SAM substrates at 7 mN/m upon first compression, resulting in a tails-up lipid configuration (Figure 1).
Figure 1.
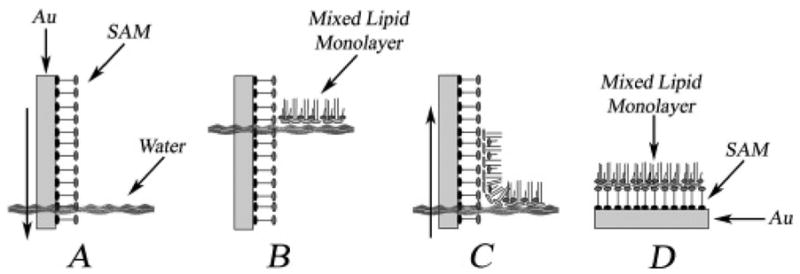
Side view of the vertical deposition of a mixed lipid monolayer onto a substrate. Sample preparation is detailed in the Materials and Methods section. (A) A 16-mercaptohexadecanoic acid self-assembled monolayer (SAM) on a gold (Au) substrate is inserted vertically through the water surface. (B) A mixed lipid monolayer is applied to the water surface and compressed to the desired surface pressure. (C) The substrate is lifted vertically through the water surface, and the mixed lipid monolayer adheres. (D) Schematic representation of the final sample.
To quantify the influence of CH on POPC and SM, the molecular area of an ideal mixture (Aideal) at 7 mN/m is compared to the actual molecular area observed (Aactual). Aideal is the molecular area expected for a noninteracting, immiscible mixture.31 Aideal is calculated by the following equation
| (1) |
where X1 is the mole fraction of component 1, A1 is the molecular area of pure component 1 at 7 mN/m, and A2 is the molecular area of pure component 2 at 7 mN/m. Using Aideal and Aactual, the condensing effect is quantified by determining the percent difference in the two molecular areas by the following equation:
| (2) |
Instrumentation
An imaging TOF-SIMS equipped with a 15 keV Ga+ liquid metal ion gun was used to obtain mass spectrometric data. The mass spectrometer is described in detail by Braun et al.32 Spectra were acquired without the need for charge compensation and with an ion dose no greater than 1012 ions/cm2. The ion beam diameter is ~100 nm; however, for the data presented here, the pixel size (~1 μm2) is dictated by the field of view and the number of pixels. Each scan takes less than 30 s. Total ion images (256 pixels × 256 pixels) were obtained by rastering the ion beam across the surface and taking a mass spectrum at each pixel. For the lipid molecular ion maps, the intensities of the individual lipid-specific ions (i.e., m/z 369 and 385 for CH) were summed at each pixel and then converted to a 64 pixel × 64 pixel image by summing the intensities of surrounding pixels. Summing pixels is reasonable because the chemically resolved lipid domains were larger than 4 pixels × 4 pixels.
Results and Discussion
The lipids under analysis in this study are CH, POPC, and 18:0 SM. A saturated SM was selected because it is biologically more abundant than unsaturated SM.9 Similarly, an unsaturated phosphatidylcholine glycolipid was selected because they are more prevalent than saturated phosphatidylcholine glycolipids in the cellular membrane.9 These specific phospholipids were also chosen because fluorescence microscopy studies of similar monolayer mixtures have recently been presented in the literature.12 This mixture contains a saturated lipid (SM), an unsaturated lipid (POPC), and CH and is known to produce immiscible liquid phases in both monolayers and bilayers.8,12,13 Herein, we illustrate TOF-SIMS as a viable resource for chemically resolving the lateral distribution of immiscible lipid mixtures and thus broadening the resources available for investigating lipid interactions.
Cholesterol Condenses a 2:3 POPC/18:0 Sphingomyelin Mixture
Pressure–area isotherms for each of the pure lipids, the binary mixtures, and the ternary mixture are presented in Figure 2. The actual and ideal molecular areas and the percent difference between the two are presented in Table 1. Isothermal data for the pure lipids are similar to literature values (i.e., for CH,23 SM,33 and POPC34). It is clear that CH has a condensing effect on both POPC (Figure 2A) and 18:0 SM (Figure 2B). The 2:1 SM/CH mixture has a molecular area that is nearly equal to that of pure CH at 7 mN/m and is −20% different from ideal. However, POPC has a much higher molecular area than 18:0 SM at 7 mN/m and a smaller CH condensing effect (percent difference of −8%). These results are similar to literature values (i.e., for POPC/CH34 and SM/CH35). The binary mixture of 2:3 POPC/SM has an actual molecular area that is larger than its ideal and shows no condensation. The differences between the CH-induced condensation of 18:0 SM and POPC arise from the unsaturated acyl chain of POPC that reduces the efficiency of the molecular packing and leads to a higher molecular area for both its pure and binary mixtures. Interestingly, the ternary mixture behaves more like the binary phospholipid/CH mixtures with the addition of the second phospholipid (percent difference of −5% for 4:3 POPC/CH + 47% SM and 4% for 2:1 SM/CH + 30% POPC), rather than the binary phospholipid mixture plus CH (percent difference of −20%).
Figure 2.
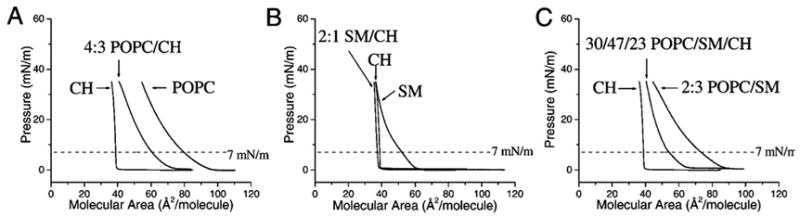
Pressure–area isotherms of mixtures containing POPC, CH, and 18:0 SM. (A) CH, POPC, and 4:3 POPC/CH. (B) CH, SM, and 2:1 SM/CH. (C) CH, 2:3 POPC/SM, and 30:47:23 POPC/18: 0SM/CH. The dotted line in each demarks a pressure of 7 mN/m.
Table 1.
Actual Average Molecular Area Observed at 7 mN/m (Using an Average of at Least Three Isotherms) and the Calculated Ideal Average Molecular Area of Each of the Mixtures as Well as the Percent Difference between the Two Areas for Each of the Mixtures
| Sample | actual area (Å2/molecule) | ideal area (Å2/molecule) | percent difference (%) |
|---|---|---|---|
| POPC | 82 ±2.7 | ||
| 18:0 sphingomyelin (SM) | 53 ± 1.8 | ||
| cholesterol (CH) | 39 ± 1.3 | ||
| 2:3 POPC/SM | 70 ± 3.5 | 65 | 9 |
| 4:3 POPC/CH | 57 ± 6.3 | 63 | −8 |
| 2:1 SM/CH | 37 ± 0.2 | 48 | −20 |
| 2:3 POPC/SM + 23% CH | 63 | −20 | |
| 4:3 POPC/CH + 47% SM | 53 ± 3.3 | 55 | −5 |
| 2:1 SM/CH + 30% POPC | 51 | 4 |
The ternary mixture contains a saturated lipid (SM), an unsaturated lipid (POPC), and CH. Saturated lipids are more readily condensed by CH and will therefore be more prevalent in the CH-rich condensed phase,12 whereas unsaturated lipids have a more limited condensation due to rigid double bonds. TOF-SIMS allows for the visualization of this within the ternary system and is evidenced in the following sections.
TOF-SIMS Identifies Each Lipid in a Mixture
Using TOF-SIMS, all lipids are identified within each of the monolayer systems using mass fragments unique to each lipid. The mass spectra for the pure lipids are presented in Figure 3. The molecular structures of each of the lipids, along with their corresponding unique positive ion SIMS fragments, are presented in Figure 4. The gold substrate is detected in each of the spectra (Au+ at m/z 197). The CH fragments are [M − H]+ at m/z 385 and [M − OH]+ at m/z 369. Although phosphocholine ([C5H15NPO4]+ at m/z 184) is a prevalent fragment, it arises from both POPC and 18:0 SM and thus cannot be used to differentiate the location of one from the other. However, the protonated molecular ions, [M + H]+, are detected and used for both phospholipids: m/z 731 for 18:0 SM and m/z 760 for POPC. Also, POPC headgroup fragment [C8H19NPO4]+ at m/z 224 is unique to POPC because it includes a portion of the glycerol backbone. The sphingosine backbone fragment [C17H30ON]+ at m/z 264 is a second fragment unique to 18:0 SM. Although not shown, all lipids were detected in each of the binary mixtures, and the ion images were found to be homogeneous with each lipid present throughout. The 16-mercaptohexadecanoic acid SAM is also observed in the mass spectra with most of its respective peaks below m/z 200. More importantly, none of the SAM peaks overlap with the lipid peaks of interest.29
Figure 3.
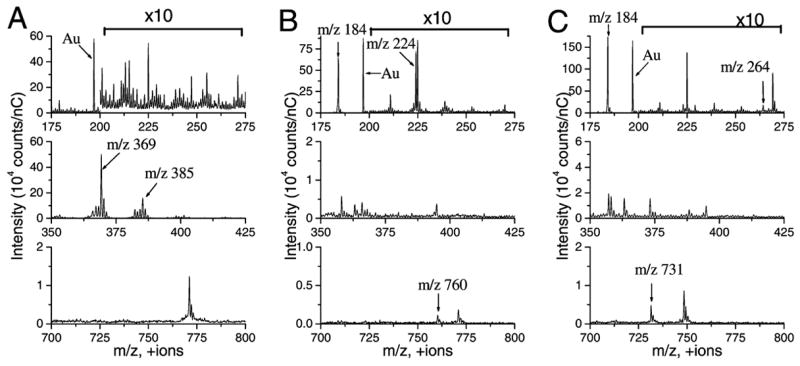
Mass spectra and significant + SIMS fragments of the pure lipids: (A) CH, (B) POPC, and (C) 18:0 SM. The characteristic ions are the POPC headgroup fragment [C8H19NPO4]+ at m/z 224, SM backbone fragment [C17H30ON]+ at m/z 264, CH fragments [M − OH]+ at m/z 369 and [M − H]+ at m/z 385, 18:0 SM protonated molecular ion [M + H]+ at m/z 731, and POPC protonated molecular ion [M + H]+ at m/z 760. Phosphocholine ([C5H15NPO4]+ at m/z 184) results from both POPC and SM. The intensity within m/z 200–275 has been multiplied by a factor of 10. The total ion dose was less than 1012 ion/cm2 for each.
Figure 4.
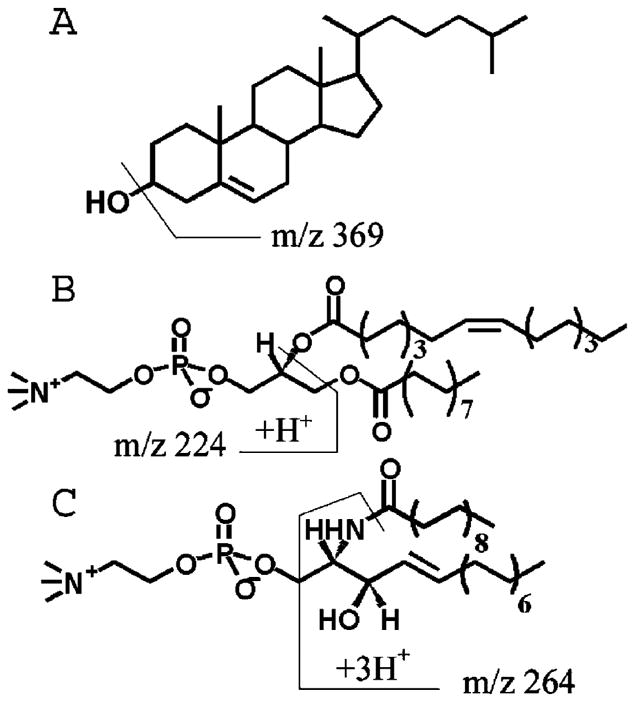
Molecular structures of (A) cholesterol (CH), (B) POPC, and (C) 18:0 SM with +SIMS fragment ions labeled. CH has two main fragments: [M − H]+ at m/z 385 and [M − OH]+ at m/z 369. POPC has two main fragments: [M + H]+ at m/z 760 and [C8H19NPO4]+ at m/z 224. SM has two main fragments: [M + H]+ at m/z 731 and [C17H30ON]+ at m/z 264.
The mass spectrum of the ternary 30:47:23 POPC/SM/CH mixture is presented in Figure 5A. Each of the unique lipid fragments mentioned above is detected in this mixture. Although their intensities are low, the 18:0 SM peaks ([M + H]+ at m/z 731 and [C17H30NO]+ at m/z 264) are detected. The lower intensity in the upper part of the images is likely due to a slight tilt of the sample in the instrument. The tilt is caused by mounting the sample onto a copper stage for SIMS analysis after it is made. Thus, it does not affect the domain segregation that occurs at the air–water interface before the sample is transferred onto the substrate and subsequently mounted onto the copper stage. The quantitative examination presented later included samples with and without tilt artifacts, and no statistical difference was seen between them. The SIMS image of samples without a tilt artifact is presented in Figure 1 of Supporting Information. Note that the total ion image (Figure 5B) and phosphocholine image (Figure 5C) both hint at nearly circular domain formation in the slight variation of intensity across the surface. Most importantly, the addition of CH induces domain formation, and this will be more clearly evidenced in the next section.
Figure 5.
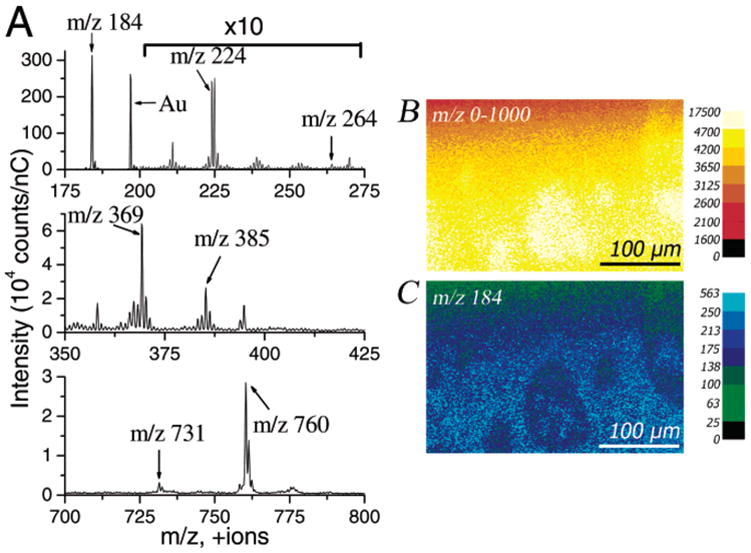
(A) Mass spectra with significant + SIMS fragments labeled for the ternary mixture 30:47:23 POPC/18:0 SM/CH. The characteristic ions are POPC headgroup fragment [C8H19NPO4]+ at m/z 224, SM backbone fragment [C17H30ON]+ at m/z 264, cholesterol fragments [M − OH]+ at m/z 369 and [M − H]+ at m/z 385, 18:0 SM protonated molecular ion [M + H]+ at m/z 731, and POPC protonated molecular ion [M + H]+ at m/z 760. Phosphocholine ([C5H15NPO4]+ at m/z 184) results from both POPC and SM. The intensity within m/z 200–275 has been multiplied by a factor of 10. The total ion dose was less than 1012 ion/cm2. (B) Total ion image and (C) phosphocholine ion image ([C5H15NPO4]+ at m/z 184). Images are 256 pixels × 256 pixels with the scale bar representing 100 μm, ion intensity scales in counts/nC, and a total ion dose of less than 1012 ion/cm2.
Sphingomyelin Localizes with Cholesterol, and POPC is Excluded
The individual lipid localizations for the ternary mixture (shown in Figure 5A) are presented in Figure 6. The data clearly show multiple liquid phases and the location of each lipid. The size and shape of the CH domains are similar to those shown in previous work,9 with the central domain being approximately 90 μm in diameter. The ion signal for POPC is lower in the CH-rich phase, indicating that it is excluded, whereas the ion signal for SM is most intense in the regions containing CH. The localization of SM is less clear as a result of the overall lower ion signal of the SM fragments. An intensity line scan through the ion images (Figure 6B) shows the varying intensity of each lipid signal. The CH ion yield begins at zero counts and rises to show a single ~40 μm CH domain. The ion signal for POPC begins high but falls sharply at the CH domain, indicating that it is excluded from this densely packed region. In contrast, the ion yield for SM varies in a spatial pattern similar to the ion signal of CH. From Figure 6B, the relative yield of each phospholipid in the CH-rich phase versus that in the CH-poor phase is 2:1 for SM and 1:2 for POPC. Thus, SM is localized within the CH-rich phase. Four samples of the ternary mixture were made and analyzed, and similar results were obtained for each.
Figure 6.
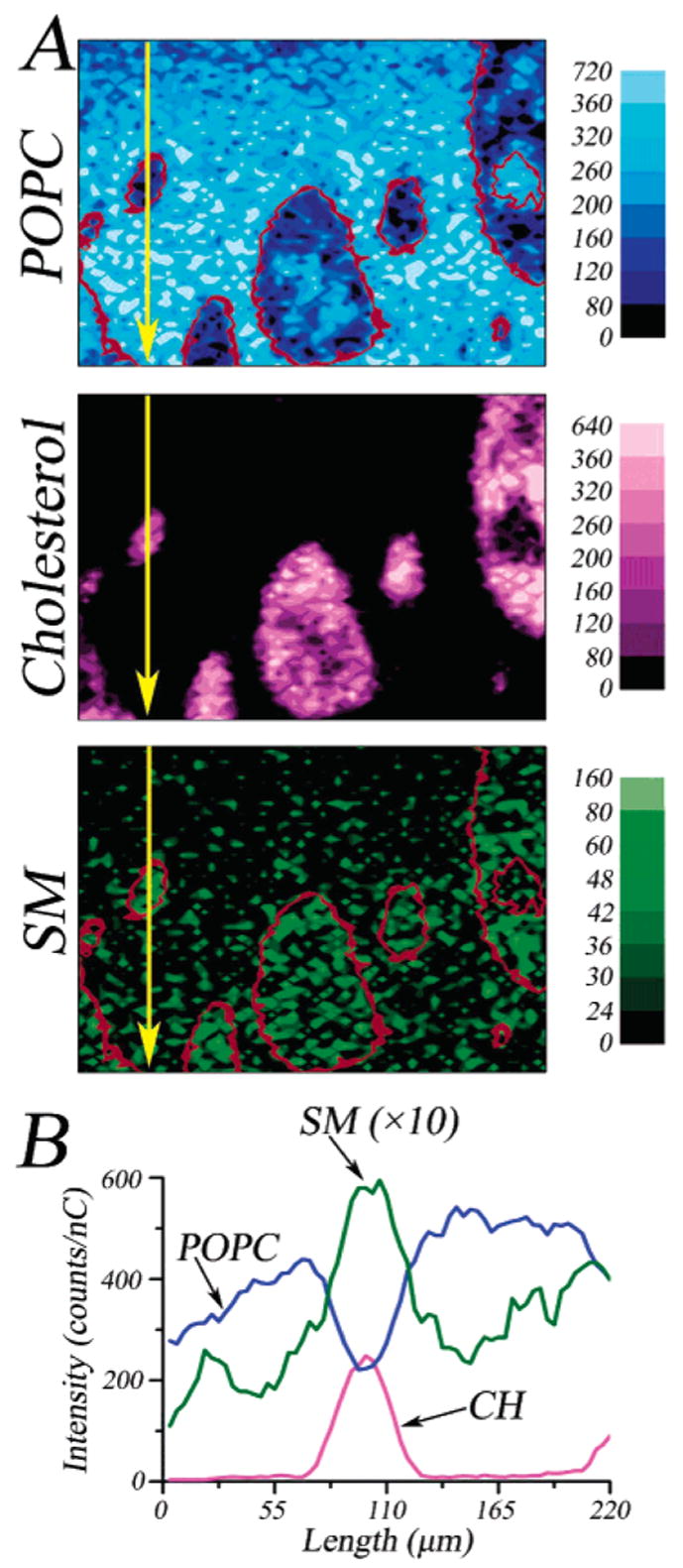
(A) Lipid molecular ion intensity mappings for 30:47:23 POPC/18:0 SM/CH. The +SIMS total ion image is presented in Figure 5B. The POPC signal ([M + H]+ at m/z 760 and [C8H19NPO4]+ at m/z 224) is represented in blue, the CH signal ([M − H]+ at m/z 385 and [M − OH]+ at m/z 369) is represented in pink, and the SM signal ([M + H]+ at m/z 731 and [C17H30ON]+ at m/z 264) is represented in green. The outlines of the cholesterol domains (red) in the POPC and SM images are shown for emphasis. The ion intensity scales are in counts/nC, and the field of view is 320 μm × 220 μm. (B) Ion intensity line scan for each of the lipid ion maps. The region of the line scan is indicated by the yellow arrow in A.
Lipid Segregation Not Complete
The degree of lipid segregation between phases is evident in Figure 6. The line scan in Figure 6B shows that CH has nearly complete segregation into circular micrometer-size domains; however, neither the POPC nor SM signals fall to zero in the phase where they are least concentrated, thus neither SM nor POPC is completely segregated from these phases. This is a significant observation and demonstrates a unique capability of TOF-SIMS imaging. The lack of complete lipid segregation further illustrates the complexity of the lipid interactions. Strong physicochemical interactions between SM and CH should draw these lipids closer together and increase their segregation from POPC. The phospholipids in this system are not drawn completely into one phase or the other but rather are more prevalent in the phases where interactions are more favorable. Hence the structure of SM does not lead to interactions with CH that are dominant enough to entice complete segregation.
Quantification of Lipid Content
It would be valuable to extract quantitative compositional information directly from the images shown in Figure 6. For mass spectrometry experiments in general36–39 and for SIMS experiments in particular, 30,40,41 ion signal intensity is not typically proportional to concentration because of what are generally referred to as matrix effects. For the system studied here, for example, it has been shown that when CH is co-localized with phosphocholine–phospholipids, proton transfer can increase the intensity of phosphocholine ([C5H15NPO4]+ at m/z 184).30 However, there are strategies that can be employed to take into account these matrix effects and provide at least an estimate of the composition of lipids inside and outside the CH domains shown in Figure 6. Our approach is to calculate a relative sensitivity factor (RSF) for each of the three lipid components in the LB films. RSF is used most commonly in the elemental analysis of doped materials.42–44 When the matrix elemental concentration is constant, then RSF is defined by
| (3) |
where RSFx is the relative sensitivity factor for component x; Cx is the concentration of component x; IM is the secondary ion intensity of the matrix reference ion; and Ix is the secondary ion intensity for the relevant ion of component x. A number of different reference ions, including Au+ and several major peaks from 16-mercaptohexadecanoic acid SAM, were examined. We finally chose Au+ at m/z 197 because its intensity is most constant from sample to sample. The values for the single-component lipid films are given in Table 2. Note that the RSF value for CH is more than 50 times larger than for SM and 6.6 times larger than for POPC.
Table 2.
Relative Sensitivity Factors (RSF) for Each of the Lipid Components in the One-, Two-, and Three-Component Lipid LB Filmsa.
| CH
|
SM
|
POPC
|
|
|---|---|---|---|
| sample composition | [M − OH]+ (m/z 369) | [C17H30ON]+ (m/z 264) | [C8H19NPO4]+ (m/z 224) |
| Relative Sensitivity Factor (RSF) | |||
| cholesterol (CH) | 0.72 ± 0.1 | ||
| 18:0 sphingomyelin (SM) | 0.013 ± 0.001 | ||
| POPC | 0.11 ± 0.005 | ||
| 2:3 POPC/SM | 0.026 ± 0.001 | 0.17 ± 0.01 | |
| 4:3 POPC/CH | 0.34 ± 0.002 | 0.13 ± 0.01 | |
| 2:1 SM/CH | 0.38 ± 0.005 | 0.23 ± 0.001 | |
| 30:47:23 POPC/SM/CH | 0.31 ± 0.003 | 0.021 ± 0.002 | 0.21 ± 0.02 |
All the calculations included at least three measurements of different areas in one or two samples.
The magnitude of matrix effects can be discerned by calculating the RSF values for all combinations of binary components, that is, 2:3 POPC/SM, 4:3 POPC/CH, and 2:1 SM/CH for conditions where there is no observable domain formation. The molar ratios of the lipid components in these binary systems are the same as the ratios utilized in the ternary system. For the three-component system, the RSF values may be obtained directly from a region of the monolayer that represents the macroscopic stoichiometry. These calculations assume that the concentrations reported for the two- and three-component systems are identical to the concentrations of the lipid mixtures applied to the LB trough. These values are also reported in Table 2. Note that the RSF values for CH are lower by about a factor of 2 and the RSF values for SM are higher by about a factor of 2. These changes are consistent with the proton-transfer mechanism noted above.
The two-component RSF values have been used to estimate the average concentration of each component in a three-component film. The RSF values of the two-component mixtures were averaged and applied to the signal intensities measured for the three-component mass spectrum. These results are shown in Table 3. The agreement between the expected values and the calculated values provides a sense of the reliability of the numbers. Similarly, using the RSF values calculated from the two- and three-component films, it is possible to estimate the molar concentrations of each species inside and outside the CH domains shown in Figure 6. Two methods were employed. In the first method (a), the two-component RSF values were applied to the measured ion intensity inside and outside a CH domain shown in Figure 6. In the second method (b), the RSF values of the three-component film were applied directly to these secondary ion intensities. The results are shown in Table 3. The magnitude of the numbers certainly suggests that SM is more concentrated in the CH domains and that POPC is presently largely outside the CH domain, as is evident from an inspection of Figure 6.
Table 3.
Concentration of Each of the Lipid Components within and outside the Cholesterol Domains of Figure 5a
| 30/47/23 POPC/SM/CH | CH
|
18:0 (SM)
|
POPC
|
|||
|---|---|---|---|---|---|---|
| [M − OH]+ (m/z 369)
|
[C17H30ON]+ (m/z 264)
|
[C8H19NPO4]+ (m/z 224)
|
||||
| a | B | a | b | a | b | |
| Concentration (%) | ||||||
| entire sample | 19 ± 2 | 43 ± 2 | 38 ± 2 | |||
| within CH domains | 20 ± 2 | 21 ± 2 | 61 ± 2 | 65 ± 6 | 19 ± 1 | 14 ± 1 |
| outside CH domains | 0 | 0 | 20 ± 1 | 31 ± 3 | 80 ± 4 | 69 ± 7 |
The concentration was determined in (a) using the RSF values for the two-component systems and in (b) using the RSF values for the three-component system. Note that the error in concentration arises from the RSF value used in the calculation.
Implications
The location and relative amounts of each of the lipid components in a mixture of immiscible liquid phases have been determined using TOF-SIMS imaging without the need for labels or markers. The ternary mixture of 30:47:23 POPC/18:0 SM/CH produces coexisting liquid phases in which the SM localizes with CH while POPC is antilocalized with CH. However, the degree of lipid segregation is not complete, and further investigation of this phenomenon could help explain how lipid rafts on the order of a few molecules combine to form larger rafts in the cellular membrane. These results show the promise that TOF-SIMS imaging holds for resolving co-localized lipids and lipids present at varying concentrations.
Acknowledgments
Financial support was obtained from the National Institutes of Health and the National Science Foundation under Grant #CHE-0555314. We acknowledge Dr. David L. Allara and his research group for gold deposition and the use of their ellipsometer.
Footnotes
Supporting Information Available: Lipid molecular ion intensity mappings for another sample of 30:47:23 POPC/18:0 SM/CH without the sample tilt artifact. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Brown DA, Rose JK. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 3.Brugger B, Blass B, Haberkant P, Leibrecht I, Wieland FT, Krausslich H. Proc Natl Acad Sci USA. 2006;103:2641–2646. doi: 10.1073/pnas.0511136103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holopainen JM, Metso AJ, Mattila JP, Jutila A, Kinnunen PKJ. Biophys J. 2004;86:1510–1520. doi: 10.1016/S0006-3495(04)74219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epand RM, Epand RF. Chem Phys Lipids. 2004;132:37–46. doi: 10.1016/j.chemphyslip.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Slotte JP. Biochim Biophys Acta. 1995;1235:419–427. doi: 10.1016/0005-2736(95)80031-a. [DOI] [PubMed] [Google Scholar]
- 7.Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E. Biophys J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veatch SL, Keller SL. Phys Rev Lett. 2002;89:268101–268104. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]
- 9.de Almeida RFM, Fedorov A, Prieto M. Biophys J. 2003;85:2406–2416. doi: 10.1016/s0006-3495(03)74664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McConnell HM, Radhakrishnan A. Biochim Biophys Acta. 2003;1610:159–173. doi: 10.1016/s0005-2736(03)00015-4. [DOI] [PubMed] [Google Scholar]
- 11.Crane JM, Tamm LK. Biophys J. 2004;86:2965–2979. doi: 10.1016/S0006-3495(04)74347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stottrup BL, Stevens DS, Keller SL. Biophys J. 2005;88:269–276. doi: 10.1529/biophysj.104.048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veatch SL, Keller SL. Phys Rev Lett. 2005;94:148101–148104. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 14.Cruz A, Vazquez L, Velez M, Perez-Gil J. Langmuir. 2005;21:5349–5355. doi: 10.1021/la046759w. [DOI] [PubMed] [Google Scholar]
- 15.Biesinger MC, Paepegaey PY, McIntyre NS, Harbottle RR, Petersen NO. Anal Chem. 2002;74:5711–5716. doi: 10.1021/ac020311n. [DOI] [PubMed] [Google Scholar]
- 16.Bourdos N, Kollmer F, Benninghoven A, Ross M, Sieber M, Galla HJ. Biophys J. 2000;79:357–369. doi: 10.1016/S0006-3495(00)76297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harbottle RR, Nag K, McIntyre NS, Possmayer F, Petersen NO. Langmuir. 2003;19:3698–3704. [Google Scholar]
- 18.Kraft ML, Weber PK, Longo ML, Hutcheon ID, Boxer SG. Science. 2006;313:1948–1951. doi: 10.1126/science.1130279. [DOI] [PubMed] [Google Scholar]
- 19.Marxer CG, Kraft ML, Weber PK, Hutcheon ID, Boxer SG. Biophys J. 2005;88:2965–2975. doi: 10.1529/biophysj.104.057257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McQuaw CM, Sostarecz AG, Zheng L, Ewing AG, Winograd N. Langmuir. 2005;21:807–813. doi: 10.1021/la0479455. [DOI] [PubMed] [Google Scholar]
- 21.McQuaw CM, Zheng L, Ewing AG, Winograd N. Appl Surf Sci. 2006;252:6716–6718. doi: 10.1016/j.apsusc.2006.02.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ross M, Steinem C, Galla HJ, Janshoff A. Langmuir. 2001;17:2437–2445. [Google Scholar]
- 23.Sostarecz AG, McQuaw CM, Ewing AG, Winograd N. J Am Chem Soc. 2004;126:13882–13883. doi: 10.1021/ja0472127. [DOI] [PubMed] [Google Scholar]
- 24.Henry CM. Chem Eng News. 2004;82:33–35. [Google Scholar]
- 25.Yuan C, Johnston LJ. Biophys J. 2002;82:2526–2535. doi: 10.1016/S0006-3495(02)75596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence JC, Saslowsky DE, Edwardson JM, Henderson RM. Biophys J. 2003;84:1827–1832. doi: 10.1016/s0006-3495(03)74990-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Deenen LLM, De Gier J. In: The Red Blood Cell. Surgenor DM, editor. Academic Press; New York: 1974. p. 147. [Google Scholar]
- 28.Marsh D. CRC Handbook of Lipid Bilayers. CRC Press; Boca Raton, FL: 1990. [Google Scholar]
- 29.Fisher GL, Hooper AE, Opila RL, Allara DL, Winograd N. J Phys Chem B. 2000;104:3267–3273. [Google Scholar]
- 30.Sostarecz AG, Cannon DM, Jr, McQuaw CM, Sun S, Ewing AG, Winograd N. Langmuir. 2004;20:4926–4932. doi: 10.1021/la0496892. [DOI] [PubMed] [Google Scholar]
- 31.Cadenhead DA, Müeller-Landau F. J Colloid Interface Sci. 1980;78:269–270. [Google Scholar]
- 32.Braun RM, Blenkinsopp P, Mullock SJ, Corlett C, Willey KF, Vickerman JC, Winograd N. Rapid Commun Mass Spectrom. 1998;12:1246–1252. doi: 10.1002/(SICI)1097-0231(19980930)12:18<1246::AID-RCM316>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 33.Vaknin D, Kelley MS, Ocko BM. J Chem Phys. 2001;115:7697–7704. [Google Scholar]
- 34.Smaby JM, Brockman HL, Brown RE. Biochemistry. 1994;33:9135–9142. doi: 10.1021/bi00197a016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smaby JM, Momsen M, Kulkarni VS, Brown RE. Biochemistry. 1996;35:5696–5704. doi: 10.1021/bi953057k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schiller J, Arnhold J, Benard S, Müller M, Reichl S, Arnold K. Anal Biochem. 1999;267:46–56. doi: 10.1006/abio.1998.3001. [DOI] [PubMed] [Google Scholar]
- 37.Benard S, Arnhold J, Lehnert M, Schiller J, Arnold K. Chem Phys Lipids. 1999;100:115–125. [Google Scholar]
- 38.Petkoviæ M, Schiller J, Müller M, Benard S, Reichl S, Arnold K, Arnhold J. Anal Biochem. 2001;289:202–216. doi: 10.1006/abio.2000.4926. [DOI] [PubMed] [Google Scholar]
- 39.Schiller J, Zschörnig O, Petkoviæ M, Müller M, Arnhold J, Arnolk K. J Lipid Res. 2001;42:1501–1508. [PubMed] [Google Scholar]
- 40.Cannon DM, Jr, Winograd N, Ewing AG. Annu Rev Biophys Biomol Struct. 2000;29:39–263. doi: 10.1146/annurev.biophys.29.1.239. [DOI] [PubMed] [Google Scholar]
- 41.Roddy TP, Cannon DM, Jr, Ostrowski SG, Winograd N, Ewing AG. Anal Chem. 2003;75:4087–4094. doi: 10.1021/ac026353t. [DOI] [PubMed] [Google Scholar]
- 42.Wilson RG. J Appl Phys. 1998;63:5122–5125. [Google Scholar]
- 43.Novak SW, Wilson RG. J Appl Phys. 1991;69:463–465. [Google Scholar]
- 44.Wilson RG, Novak SW. J Appl Phys. 1991;69:466–474. [Google Scholar]


