Abstract
The treatment of helical objects as a string of single particles has become an established technique to resolve their three-dimensional (3D) structure using electron cryo-microscopy. It can be applied to a wide range of helical particles such as viruses, microtubules and helical filaments. We have made improvements to this approach using Tobacco Mosaic Virus (TMV) as a test specimen and obtained a map from 210,000 asymmetric units at a resolution better than 5 Å. The major improvements include the determination and correction of the contrast transfer function of the microscope. Alignment of helical segments was helped by constraints derived from the helical symmetry of the virus. Furthermore, symmetrization was implemented by multiple inclusions of symmetry-related views in the 3D reconstruction. We used the density map to build an atomic model of TMV. The model was refined using a real-space refinement strategy that accommodates multiple conformers. The atomic model shows significant deviations from the deposited model for the helical form of TMV at the lower-radius region (residues 88 to 109). This region appears more ordered with well-defined secondary structure, compared with the earlier helical structure. The RNA phosphate backbone is sandwiched between two arginine side chains, stabilizing the interaction between RNA and coat protein. A cluster of two or three carboxylates is buried in a hydrophobic environment isolating it from neighboring subunits. These carboxylates may represent the so-called Caspar carboxylates that form a metastable switch for viral disassembly. Overall, the observed differences suggest that the new model represents a different, more stable state of the virus, compared with the earlier published model.
Keywords: high-resolution single-particle electron microscopy, helical reconstruction, refinement, Tobacco Mosaic Virus, viral assembly/disassembly
Introduction
Electron cryo-microscopy (cryo-EM) has become a common technique for three-dimensional (3D) structural analysis that enables the study of a variety of biological specimens ranging from ordered two-dimensional (2D) crystals and helical specimens to single particles with and without symmetry (reviewed in 1). Depending on the order present in these specimens, this method can achieve sufficiently high resolution to build an atomic model. The first such example was bacteriorhodopsin which forms well-ordered 2D crystals 2. The first 3D reconstruction of a helical particle from electron micrographs was obtained from Bacteriophage T4 3. More recently, atomic models of the bacterial flagellum 4 and nicotinic acetylcholine receptor 5 were determined at 4 Å resolution, using electron microscopy of helical specimens. Helical specimens have the advantage that a single view provides sufficient information to reconstruct a 3D object.
Many biological specimens either do not form regular arrays, or the arrays formed contain disorder (deviations from ideal symmetry). For these specimens, high resolution is more difficult to obtain. To deal with disorder and distortions in helical specimens, procedures based on the single-particle approach were developed 6; 7; 8; 9; 10. In the most widely used approach (proposed by Egelman 6), called iterative real-space helical reconstruction (IRSHR) procedure, the helical particle is divided into segments, each of which is treated as an independent single particle. Single-particle electron microscopy (SPEM) does not rely on ordered arrays and has been developed over the last decade to yield structures of symmetrical and non-symmetrical particles that frequently reach sub-nanometer resolution 11; 12; 13. Indeed, IRSHR has led to valuable insights in the structure and function of a variety of helical specimens, up to sub-nanometer resolution 14. However, the segmentation procedure generates additional degrees of freedom in the processing and reconstruction of the particle that may introduce errors and reduce the attainable resolution.
In the present study, we have expanded the original IRSHR procedure by introducing constraints for the alignment parameters of each segment, thus controlling their degree of freedom. Further improvement was achieved by correcting for the contrast transfer function (CTF) of the electron microscope (amplitude and phase correction), and by optimizing the helical symmetrization of the particle.
Tobacco Mosaic Virus
To test the procedure we used Tobacco Mosaic Virus (TMV) as a model system and obtained a 3D reconstruction at a resolution better than 5 Å. TMV is a rod-shaped plant virus with a typical length of 3000 Å, a diameter of 180 Å and a central channel of approximately 40 Å. The virus contains RNA and the coat protein (158 amino acids, ∼17,500 Da) that form a right-handed helix. Three complete turns contain 49 subunits and the helical rise per subunit is 1.408 Å.
TMV exists in a variety of organized structures, such as the disk aggregate and helical rod, depending on the buffer, pH, and ionic strength (reviewed by Klug 15). These structural states are thought to play distinct roles in the genesis of the virus during infection. The so-called protein disk form was crystallized and resolved to atomic detail in 1978 and refined in 1998 16; 17. The structure of the helical form of the virus has been investigated for several decades using electron microscopy 18 and X-ray fiber diffraction 19. The fiber diffraction study led to a structure of 2.9 Å resolution and an atomic model for the helical form. In both structural forms, the coat protein consists predominantly of α-helices oriented perpendicularly to the helical axis of the virus. The genomic RNA is protected from the solvent in its interior and is embedded between subunits making up the upper and lower staircases of the helical rod.
Based on the comparison of the determined 3D structures, a mechanism of the disassembly of the virus was proposed 19: Major conformational differences of the loop region at the inner wall of the virus suggested an involvement of these residues in viral disassembly after host invasion. In early studies, two pH-sensitive carboxylate pairs were postulated to function as the driving force of this structural transition 20. The precise location and identity of these residues, however, is still under debate and, as pointed out recently, the complexity of this interaction might have been underestimated 21. Hence, more direct structural information of these different conformational states is necessary to improve our understanding of the intricacy of these processes.
Results and Discussion
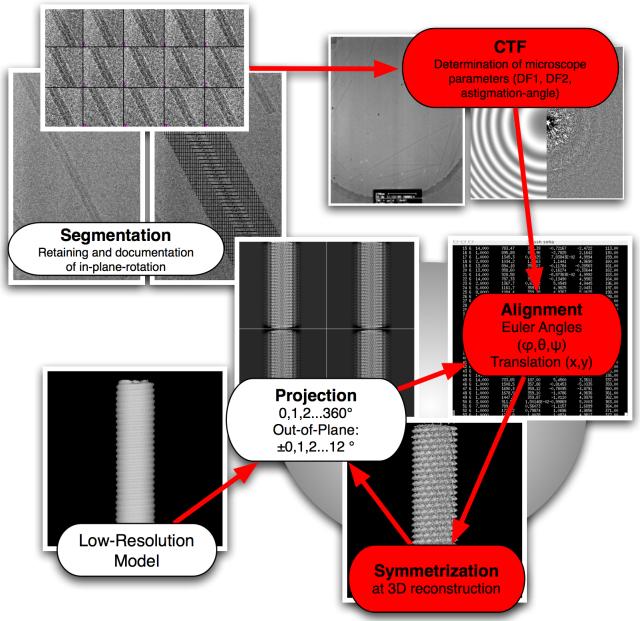
In a conventional single-particle reconstruction method, five alignment parameters have to be determined for each particle to calculate a 3D reconstruction. The alignment of an individual particle is described by the Euler angles (ϕ, ψ, θ) and the in-plane translation (x,y). To obtain a reconstruction at high resolution, a correction for the CTF of the microscope is also required. The refinement of the alignment parameters for each particle is achieved by correlation-based projection matching using a model that is updated after each refinement cycle (Figure 1). These steps are common to most reconstruction and refinement procedures applied to single-particle structures. We have included additional steps specific to helical specimens that exploit the helical symmetry by placing constraints on the alignment parameters. These constraints improve the accuracy of the alignment. The application of helical symmetry in the 3D reconstruction procedure also leads to a gain in signal in the final reconstruction. In our case, helical symmetry is implemented by multiple insertions of segments, which correspond to equivalent views of the particle (see below).
Figure 1.
Segmentation Procedure: (A) A typical TMV rod embedded in vitreous ice. (B) Segmentation of a virus particle into segments with an overlap of 90 % and dimensions of 7.4 × 7.4 nm. Note that the original orientation of the virus is retained and the approximate in-plane rotation angle is recorded in a list. (C) In this example, a virus of 290 nm length yields 28 overlapping segments that are collected on an image stack as a starting point for single-particle processing.
Segmentation Procedure
To make the TMV images compatible with single-particle reconstruction, the virus particles must be segmented. An advantage of the segmentation is that any long-range distortions can be corrected. However, the information about the connectivity between segments is lost because each segment is now treated as a single particle. This information can be partly restored by monitoring the alignment of consecutive segments relative to each other and applying a validation scheme to the five alignment parameters of each segment. (see section on projection matching below).
One important parameter is the size of the segments, which can only be determined empirically. A few considerations should accompany this process. In principle, small segments can correct distortions within the particle more accurately than larger ones. However, there is a practical lower limit in segment size because the alignment accuracy decreases due to the smaller overall signal present in the images of smaller segments compared with images of larger segments. To ascertain a reliable alignment that can be monitored as described below, the minimum segment size for TMV was found to be about 70 nm. We segmented the viruses into image segments of 77.4 nm length to include an additional 10% of the particle needed to apply shifts during 3D reconstruction (see section symmetrization below).
Segmentation procedures described previously apply a rotation to the segment to align the helix axis with the vertical direction of the image to simplify subsequent processing steps 6. If the image of the particle is not scanned in this orientation, interpolation of the originally scanned data becomes necessary, sacrificing some high-frequency information. Our procedure does not require a particular orientation of the particle with respect to the image axes. Instead, segments are extracted along the particle axis while maintaining their original orientation. The images containing the segments are chosen to be square to be compatible with common single-particle processing procedures. During the segmentation, the approximate in-plane angle of the particle axis is determined and used to detect any erroneous alignment of segments within the virus particle in later processing steps (Figure 1).
In the segmentation procedure, consecutive segments overlap by a certain percentage. We chose an overlap of about 90% in accordance with Egelman's originally described procedure 6. This means that a segment was created every 7.0 nm along the particle. This step size is related to the subsequent symmetrization of the reconstruction (see symmetrization section below). Thus, for TMV, a step of 7.0 nm contains about 50 asymmetric units (the helical rise per unit is about 0.14 nm). To take advantage of the symmetry relating these 50 asymmetric units, each segment is included 50 times in the 3D reconstruction using alignment parameters that describe the symmetry-related views. This contrasts with the IRSHR procedure where the structure is symmetrized after an initial reconstruction that includes each segment only once.
The segmentation parameters may be different for other helical specimens. Ideally, the step size will include approximately an integer number of subunits to be compatible with the subsequent symmetrization. For example, an actin filament has a helical rise of 2.75 nm per subunit 22. Therefore, a step size of 5.5 nm or 8.25 nm would be appropriate. The size of the segment, on the other hand, should be larger than that chosen for TMV since the mass per length is significantly smaller compared with TMV.
CTF Determination
High-resolution analysis of cryo specimens requires correction of the CTF of the electron microscope. The first step of this correction is the determination of the defocus and astigmatism present in the image. This was done using the computer programs CTFFIND3 and CTFTILT 23. CTFFIND3 determines average defocus values for an image and does not take into account specimen tilt. Often, a small tilt is introduced unintentionally, however, either due to mechanical tolerances of the microscope goniometer and sample holder, or due to ‘cryo crinkling’ of the carbon film 24. A small specimen tilt leads to variations in the defocus across the image. Using CTFTILT the tilt axis and angle can be determined assuming a planar sample within the field of view. The defocus can then be calculated as a function of location in the image, providing more accurate defocus values for each virus particle. In the present calculation, the location of the virus in the image was taken as the center of gravity of a particle. In addition to the specimen tilt, virus particles often exhibit an out-of-plane tilt with respect to the plane of the image. Therefore, the defocus can also vary along a single particle. The out-of-plane tilt was determined for each virus particle as part of the projection matching procedure described below. This tilt information was used, together with the tilt information of the specimen, to calculate defocus values for each segment of a virus, giving the best possible defocus values for each segment. The defocus difference between the two ends of the virus particles due to out-of-plane tilt showed a Gaussian distribution with a standard deviation of 90 Å.
CTF Correction
The correction scheme of the CTF was adapted from the computer program FREALIGN 25; 26. The image FTs were multiplied by the astigmatic CTF determined for each segment. The CTF included a term for amplitude contrast at 7%. Apart from a phase correction, the multiplication leads to an attenuation of amplitudes and noise at resolution values where the CTF is small. For each segment, a non-astigmatic 3D CTF was generated using an average defocus value calculated from the two values describing the astigmatic CTF. The 3D CTFs were squared and summed for all segments in a data set (Figure 2). Projection matching was performed by comparing projections of the reference volume with the CTF-corrected images. After 3D reconstruction the volume from the CTF-corrected images, the amplitudes of the reconstruction were corrected using the summed squared 3D CTFs:
| (1) |
Here, R is the final reconstruction, R̃ represents the reconstruction calculated from N CTF-corrected images, as indicated in the curly brackets, CTFi is the CTF for image Pi, and CTF3D,i is the non-astigmatic CTF calculated in 3D for image Pi. f is related to the Wiener constant, which is a function of noise in R̃. Since we did not determine the noise level here, f was set to a constant to prevent over-amplification of amplitudes at resolution values where the sum in the denominator is small. It was chosen to be 2% of the maximum of the sum. Other values for f, for example 5%, were also used to determine its influence on the final reconstruction. This did not lead to a significant change in the final result, however, demonstrating that the final reconstruction does not depend much on the precise value of f. The main difference between Equation (1) and the implementation in FREALIGN 25; 26 is that the sum of the squared 3D CTFs is calculated from non-astigmatic 3D CTFs. The average astigmatism in the processed images was 300 nm. This introduces an error in the correction of the amplitudes. However, for a sufficiently large data set containing a whole range of defocus values this error is small and can be ignored.
Figure 2.
CTF-correction: (A) Determination of microscope parameters: a stack of micrographs serves as input for the computer programs CTFFIND/CTFTILT 23 to determine the microscope parameters, i.e. image defocus ΔDF1/ΔDF2, astigmation angle α and tilt of the micrograph (left). The graphical output of CTFFIND contains an image that is divided into a simulated power spectrum half of the determined CTF and averaged power spectrum half of the micrograph (right). Micrographs were binned six times before CTF determination in order to enhance the signal. (B) Convolution with determined CTF: the windowed segment Pi on the left (Figure 1) is multiplied with the simulated CTFi in Fourier space (center) resulting in a phase-corrected and amplitude-weighted segment at the right. The corrected image stack is used for alignment and reconstruction. (C) Amplitude correction in 3D: in parallel, for all individual segments, their astigmatic (CTF3D)2 (3D power spectrum in the center) are summed and the final 3D reconstruction of the corrected images (left) is deconvoluted by this sum to yield the corrected 3D reconstruction (right). The CTF correction scheme was adapted from Grigorieff 25; 26.
Projection Matching and Validation
Projection matching 27 was used for the refinement of the TMV structure to high resolution. A cosine-edged rectangular mask was applied to the CTF-corrected images according to the recorded in-plane orientation of the virus particle. The mask was 34 nm wide including 5 nm wide cosine edges on the long sides of the mask. At this point, images were windowed from their intermediate size of 77.4 × 77.4 nm to a final size of 70 × 70 nm for projection matching alignment. A rod was used as an initial reference structure. Projections were generated in 1° increments around the helical axis. The variation of the out-of-plane tilt angle was limited to +/−12° (Figure 3) and was also sampled in 1° steps. The chosen out-of-plane-angle range was determined after analysis of intermediate projection matching results where the tilt of the particles assumed a Gaussian distribution around 0° with a standard deviation of 3.2°.
Figure 3.
Flowchart of data processing. Major adaptations of the IRSHR procedure 6 are highlighted in red: (A) Segments are (B) CTF-corrected and processed (C) in an iterative reconstruction cycle based on projection matching. Projection: A low-resolution model serves as a reference and is projected around its helical axis in 1° increments. As the model becomes more refined out-of-plane tilted projections ±0,1,2…12° are also generated. Alignment: Projections are matched with the overlapping segments extracted from the micrographs. Restraints on alignment are imposed derived from the continuity of the virus particle. 3D reconstruction: The orientational parameters are used to merge the segments into a 3D volume. Each image is inserted multiple times according to its symmetry-equivalent views.
We devised a scheme to validate the alignment results that makes use of geometric relationships between segments known from the helical nature of the specimen. Thus, only those aligned segments showing reasonable in-plane rotational angles and translational parameters were included into the reconstruction. The in-plane angles obtained in the projection matching procedure determined the polarity of each segment. We found that, on average, the in-plane angles for 95% of the segments from a virus indicated the same polarity. The remaining segments were excluded from further processing. An average in-plane angle for the segments was derived from the initial manual identification of the two ends of the virus during segmentation using EMAN's boxer program 28. Segments with an in-plane angle that deviated from the average angle by more than 10° were excluded from the 3D reconstruction.
The projection matching procedure also yielded x,y shifts necessary to align each segment with the reference. In such an alignment, a shift component parallel to the helical axis is not uniquely defined since, due to the helical symmetry, a certain shift along the helical axis is equivalent to a rotation around the axis. In the case of TMV, a shift along the helical axis of 1.408 Å (the helical rise for one subunit) is equivalent to a rotation around the axis of 22.03° (49.02 subunits comprise three 360° helical turns 29). Therefore, we reduced the measured shifts along the helical axis to be within half the helical rise per subunit and calculated the corresponding rotation angle to match a reference projection to a segment. The residual shift along the axis was then applied to the image, together with a shift perpendicular to the axis. The measured shifts perpendicular to the helical axis were monitored to exclude segments with excessive shifts in this direction. The threshold for exclusion of about 50 Å was more generous at the beginning of the refinement. To reduce errors in the perpendicular shifts, a running average of shifts including the current segment and the five preceding and following segments was calculated. These averaged shifts were converted into shifts of integer numbers of pixels to avoid signal loss due to interpolation, and applied to the segments for approximate centering. The centered segments were used as input in following refinement cycles. Therefore, the measured shifts in the alignment became smaller with every refinement cycle, and the threshold for exclusion of segments due to excessive shifts could be lowered to a final value of about 10 Å. However, most measured shifts were much smaller than this limit. At the final stage of refinement, the shifts of the remaining segments assumed a Gaussian distribution with a standard deviation of 2.0 Å, and all of the remaining segments (after the in-plane rotation check) could be included into the final 3D reconstruction (see section on error analysis below).
Symmetrization
For the processing of a helical particle, one of the first steps is usually the determination of the symmetry, i.e. the helical rise per subunit and the distance along the helical axis for a full turn of the helix (pitch). The symmetry is traditionally determined by indexing helical diffraction patterns, a process that requires some experience. By contrast, the IRSHR procedure determines the helical parameters automatically. Although this can be a significant advantage over the traditional method, it is not always guaranteed to yield the most accurate parameters, or it can be biased by the choice of initial parameters. In the case of TMV, the helical parameters were accurately known from previous work using X-ray fiber diffraction 29 and were fixed during image processing.
Most of the published sub-nanometer maps obtained by SPEM take advantage of symmetry present in the particle, for example 60-fold icosahedral symmetry found with many viruses. In TMV, a single segment of 70 nm length contains about 500 subunits that can be used for averaging. For a symmetrical particle with M-fold symmetry, each image is equivalent to M symmetry-related views. This redundancy can be used in the reconstruction process in different ways. In the IRSHR procedure, each image in a data set is assigned one of the possible M symmetry-related views of the particle. Using these assignments, a 3D reconstruction is calculated. As a final step, the symmetry of the particle is imposed on this reconstruction, by averaging over all M symmetry-related reconstructions. Although this procedure results in a reconstruction with the correct symmetry, the procedure is not entirely correct. For example, if weighted back-projection is used for the 3D reconstruction, the weights are usually calculated from the distribution of data in Fourier space. In the reconstruction procedure just described, the distribution of data in Fourier space changes with the imposition of symmetry. The distribution used to calculate the weights in the weighted back projection prior to symmetrization is therefore incorrect 30. If an iterative algebraic reconstruction method (so-called ART) is used, such as the one implemented by Penczek et al. 31, the reconstruction is iteratively modified to match the observed images. Since each image is included in the reconstruction only once, the reconstruction does not necessarily match symmetry-related views. This leads again to errors in the final reconstruction.
We modified the reconstruction process to minimize errors. Many of the reconstruction algorithms implemented in SPIDER do not have the option of observing helical symmetry. Therefore, each image is presented to the reconstruction algorithm M times to include all symmetry-related views. In a back-projection algorithm, this enables correct weighting, while in an iterative algebraic reconstruction algorithm the reconstruction will be constrained by all symmetry-related views.
As described in the segmentation procedure section, each virus was divided into segments with an overlap of 90%. Since each segment included about 500 asymmetric units, the 10% of non-overlapping region of each segment included about 50 asymmetric units. Hence, each asymmetric unit is included in 10 overlapping segments. To take advantage of the full symmetry of the particle, it was therefore necessary to include each segment 50 times in the reconstruction. To do this, 49 copies of each segment were generated, each one of them shifted by a multiple of the helical rise of one asymmetric unit (1.408 Å) and included in the 3D reconstruction with a multiple of the rotation angle between subunits (22.03°). The shifts were applied to the segments of intermediate size of 77.4 × 77.4 nm, which were then windowed to the final size of 70 × 70 nm. Thus, the occurrence of empty areas near the edges of a segment due to the shift operation was avoided.
In principle, a different overlap and step size for segmentation could have been chosen, leading to slightly different symmetrization procedures. For example, if segments were extracted along the helical axis with a step size equivalent to the helical rise of one asymmetric unit, many more segments would be obtained and each segment would have to be included in the reconstruction only once. However, this strategy would produce significantly larger stacks of segments. Furthermore, without interpolation of the image of the virus, the step size is restricted to integer pixels and cannot be easily adjusted to match the helical rise of a single asymmetric unit. In most cases, there is still a mismatch between the step size and an integer number of asymmetric units included in the non-overlapping regions of the segments. In the TMV example, 50 asymmetric units are equivalent to 50 × 1.408 Å = 70.4 Å, while the step size is 70 Å. This small mismatch leads to over-sampling and a certain duplication of data, which is about 0.5% in this case and can be neglected.
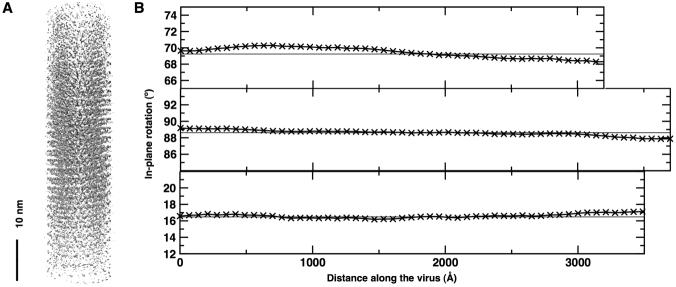
In total, 4251 segments were used from 135 TMV particles on 6 negatives and, given the 50-fold symmetry averaging, the final reconstruction (Figure 4A, 5D) contained 4251 × 50 = 212550 asymmetric units. The structure in Figure 4A was rendered at a higher density threshold to highlight the highest densities in the structure. This representation shows that the density is not uniform along the helical axis but falls off towards both ends of the structure. Therefore, the symmetrization procedure described above does not produce a perfectly symmetrical structure. The deviation from perfect symmetry is a consequence of distortions in the virus particles and inaccuracies in the alignment. For example, if some curvature is present in segments of the virus, it is not possible to bring all parts of these segments into perfect register with the reference projection. Indeed, the curvature of the particles can be visualized by a plot of the in-plane rotation along the virus (Figure 4B). Only part of each segment will be aligned perfectly. The location of this part will, on average, be in the center of a segment. Upon inclusion of segments in the reconstruction, the density half way between the two ends of the reconstruction will receive data from the optimally aligned parts of the segments. It is, therefore, the part with the strongest and best resolved density (Figure 4A). A similar argument can be used for the inaccuracy in the determination of the out-of plane tilt angle. Small deviations from the true angle will result in stronger mismatch the farther the distance from the center of the segment.
Figure 4.
Long-range curvature of a virus. (A) The reconstructed volume displayed at a 3σ threshold. The center of the segment shows stronger density than the upper and lower part due to the long-range curvature of the included viruses. (B) Analysis of the in-plane rotation angle along three viruses from micrographs taken at three different defoci.
Figure 5.
Density map of TMV filtered at about 4.5 Å resolution. (A) Quarter slice of 20×20×2 nm through three adjacent subunits. (B) Density of a single turn comprising 16 subunits and superimposed atomic coordinates viewed parallel to the helical axis. (C) Three turns of 49 subunits. (D) 3D reconstruction of a 70×70×70 nm TMV rod segment from electron micrographs.
In practice, to account for symmetry by multiple inclusions of segments in the 3D reconstruction is computationally more demanding than the imposition of symmetry after reconstruction using each segment only once. In particular, the iterative algebraic reconstruction method requires substantial computing resources. One cycle of the described projection, alignment and 3D reconstruction procedure required about four days on 48 AMD Opteron processors. To make the reconstruction process more efficient, segments matched with the same reference projection were averaged to give one single image, which was then included in the reconstruction. On average, about five segments could be averaged in this way.
For the final reconstruction, additional averaging over neighboring asymmetric units was performed in real space to remove small deviations between asymmetric units created by the reconstruction procedure (see above). Averaged density from the central slice of the reconstructed TMV segment, which has the strongest density, is shown in the presence of the refined atomic coordinates in Figure 5A, B, C. The reconstruction of a complete segment is displayed in Figure 5D.
Error Analysis
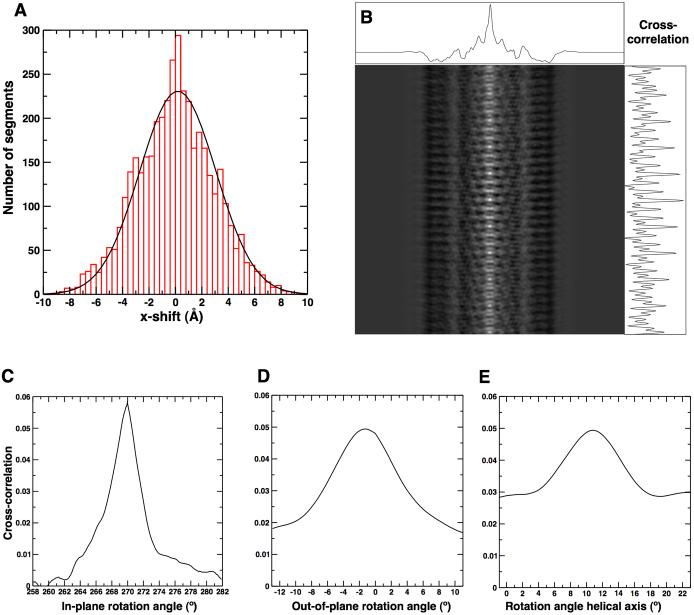
The parameters obtained from the projection matching procedure contain errors that may limit the final resolution of the reconstruction. With the known relationship between neighboring segments, these errors can be estimated. The estimation of errors for the measured shifts perpendicular to the helical axis and the in-plane and out-of-plane rotation angles is straightforward because their values should not vary significantly from segment to segment (see previous section on projection matching and validation). Therefore, the standard deviation for these parameters can be calculated directly from the measured values as the root mean squared (RMS) value of differences between neighboring segments. For TMV the standard deviation of the measured differences between neighboring segments was 2.8 Å (Figure 6A) and, therefore, the standard deviation of the shifts perpendicular to the helical axis of each segment was . The standard deviations of the in-plane and outof-plane rotation angles were 0.1° and 0.5°, respectively. The situation is somewhat more complicated for the shift parallel to the helical axis and the rotation angle around the axis because these two parameters are related by the helical symmetry. In the TMV structure a change in the location z of a segment along the helical axis is equivalent to a rotation Δα around the axis:
| (2) |
Neighboring segments can, therefore, exhibit a range of possible locations along the helical axis, each one corresponding to a different rotation angle around the axis. To estimate errors in the measured values for z and α we assume that deviations from helical symmetry in TMV are very small and can be ignored as a source of deviation from expected values for z and α. This assumption is reasonable since TMV commonly shows layer lines out to 3 to 4 Å 32. Using the measured Δz values between neighboring segments, the corresponding change Δα can be calculated according to Equation (2). These calculated Δα were then compared with those actually measured by the projection matching procedure. The RMS deviation between the calculated and measured values was 44°. This would mean that the angular error for each segment is . This calculation assumes that all contributions to the error originated from the measured rotation angle α and that the measured locations z were error-free. To estimate the actual error in the measured z values we inspected the correlation peak between some of the segments and the corresponding best-matching references from the projection matching procedure (Figure 6B). The measurement error depends on the width and height of the correlation peak. A sharp and high peak is less likely to shift due to a nearby noise peak than a broader and smaller peak where a nearby noise peak could produce a new local maximum more easily. The peak along the helical axis is only slightly narrower (as measured by the full width half maximum, FWHM) than perpendicular to the helical axis and has approximately the same height (measured from the nearest local minimum). We therefore estimate the error in the measured z values to be similar to the error in the measured shifts perpendicular to the helical axis, about 2 Å. According to Equation (2) we expect an RMS deviation between the measured and expected rotation angle Δα of 31°. This is in excellent agreement with the value found in the earlier described measurement. Therefore, most of the observed angular deviation can be ascribed to the measurement error in z. Figures 6C, D, and E plot the correlation coefficient between a segment and the best-matching projection as a function of change in each of the three alignment angles. The peaks are centered on the angles corresponding to the projection. The peak for the in-plane angle is significantly higher and narrower compared with the peak corresponding to the out-of-plane angle, which is in good agreement with the measurement errors given above. The peak height and FWHM for the rotation angle around the helical axis are comparable with those of the out-of-plane angle. Therefore, we estimate the measurement error to be similar in both cases, about 0.5°. Again, this agrees well with the previous finding that most of the deviation between the measured and expected rotation angle Δα is due to the error in the measured location z of each segment along the helical particle.
Figure 6.
Error analysis of alignment parameters. (A) Histogram of x-shift differences from one segment to the next. (B) Correlation map of an image and its matched projection. Correlation peaks along x and y are plotted. Cross-correlation against three Euler angles: (C) in-plane rotation angle (D) out-of-plane rotation angle and (E) angle around the helical axis.
Resolution and scaling of the density map
In order to visualize and interpret a cryo-EM density map appropriately two basic questions have to be addressed: 1. What is the resolution of the map? 2. How is the map scaled correctly to allow a straightforward fitting of an atomic model?
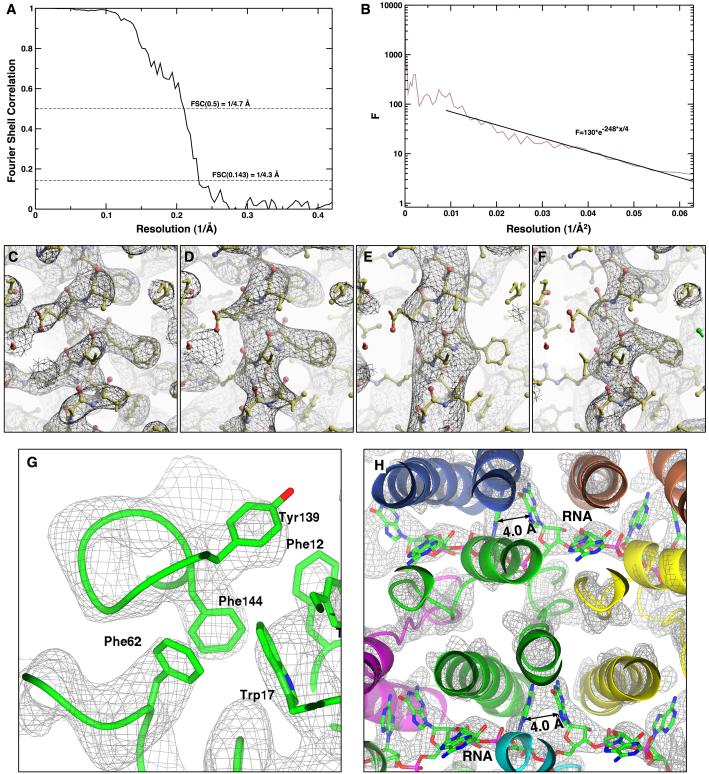
The resolution of the TMV map was estimated in two different ways. The first estimate is based on the commonly used Fourier Shell Correlation (FSC) curve calculated from two reconstructions, each containing one half of the data 33. Only the central segment containing the strongest density of the reconstruction was considered for this calculation. Reconstructions containing this approximately 240 Å long segment were re-calculated using corresponding 240 Å image segments excised from the original stack of virus segments. The FSC curve is shown in Fig. 7A together with commonly used cutoffs for the resolution, i.e. 0.5 11 and 0.143 34. The FSC curve reaches a value of 0.5 at 4.7 Å resolution and drops below 0.143 at 4.3 Å resolution. One problem associated with the resolution estimate based on the FSC curve is a possible bias of the FSC towards higher values 35. A different way to estimate the resolution is based on visible features in the map compared with features visible in maps calculated from atomic models. Figures 7C, D, E, F display the density map rendered at different resolution cutoffs, calculated from an atomic model of TMV that is based on X-ray fiber diffraction (PDB code 2TMV) 19, but was refined to fit into the density map determined by electron microscopy (PDB code 2OM3, see below). In some parts, for example in the α-helical region of the coat protein, the model-based map rendered at 5 Å resolution compares best with the SPEM map when scaled appropriately. In particular, the backbone of α-helices and density corresponding to aromatic side chains is clearly visible in the SPEM reconstruction and the density rendered at 5 Å resolution. Detail at higher resolution is visible in other parts of the map, such as the density corresponding to the RNA (Figure 7H). Therefore, an overall resolution of better than 5 Å appears reasonable.
Figure 7.
Assessment of resolution: (A) FSC between two reconstructions calculated from two halves of the data. (B) Fit of an exponential B-factor curve to the average structure factor of the cryo-EM map in resolution zones between 10 and 4 Å resolution. Comparison of density maps calculated from an atomic model at a resolution of (C) 4 (D) 5 (E) 6 Å and (F) the experimental cryo-EM map at a nominal resolution of 4.7 / 4.3 Å (FSC = 0.5 / 0.143). (G) Hydrophobic cluster with strong side-chain density (Phe12, Trp17, Phe62, Tyr139, Phe144) at higher radius of the subunit. (H) RNA located between four-helix bundles of lower and upper subunit neighbors at lower radius. The packed nucleobases exhibit strong density resolving a distance of approximately 4 Å. (α-helical ribbon of the same color originate from a single subunit.)
It is interesting to note that density for many ionizable side chains visible in Figure 7F, such as those of glutamate and arginine, do not produce strong density in the cryo-EM map. This trend was also observed in earlier cryo-EM maps 36; 37. A plausible explanation for these observations is radiation damage of the sample inflicted by the electron beam. This appears to affect carboxyl groups more strongly than aromatic groups. Other ionizable groups can be affected due to the loss of hydrogen-bonding or salt bridges. In the case of arginine side chains, we found strong corresponding density in our cryo-EM map for some and weak density for others (see below). Therefore, the impact of radiation damage must also depend strongly on the local environment. The chemistry of radiation damage has recently been reviewed by Ravelli & Garman 38.
For a comparison of density maps, appropriate resolution-dependent scaling is important. Amplitudes in a reconstruction from electron microscope images usually exhibit a resolution-dependent attenuation due to contrast-degrading effects in the microscope 39 and alignment errors 40. This attenuation of structure factor amplitudes F can be described by an overall temperature factor (B-factor):
| (3) |
Here, g is the resolution in reciprocal space units and F̃ is the unattenuated structure factor amplitude. Note that Equation (3) uses the definition of the B-factor commonly found in X-ray crystallography. To relate this B-factor to the Debye–Waller temperature factor, it has to be divided by 4. The model-based map rendered at 5 Å resolution (Figure 7D) compares best with the SPEM map if the latter is scaled with a B-factor of −280 Å2 and filtered using figure-of-merit weighting 34. This provides an estimate of the overall B-factor describing the attenuation of amplitudes in the electron-microscopy reconstruction. A second estimate of the overall B-factor can be obtained from a Guinier plot of the average amplitudes in a resolution shell against the square of the resolution in reciprocal space units 34. This plot is shown in Figure 7B together with an exponential fit to structure factor amplitudes between 10 and 4 Å resolution. This plot suggests a temperature factor of about 250 Å2, in close agreement with the estimate obtained above.
As shown below, we used the density map obtained for TMV to build a new atomic model. This model could be used as a reference to achieve a more accurate scaling of the density map. This method is similar to a procedure used in 2D crystallography 41. To do this, the atomic model was used to generate a density map with optimal scaling. From this map, average amplitudes were calculated in resolution shells and used to scale the average amplitudes of the experimental map in resolution shells. At low resolution, however, we expect systematic deviations between the density map based on the atomic model and experimental map. In this resolution range (approximately 10 Å and below) solvent effects become important 42. Therefore, we scaled this part of the data using Equation (3) with a B-factor of −280 Å2. The intermediate to high-resolution terms were scaled according to the map calculated from the atomic model followed by figure-of-merit weighting 34. This method produced the best map as judged by the agreement of map features with the atomic model (Figure 7F).
Density and refinement of atomic model
In the literature, the location of structural features within a TMV subunit is described in terms of lower, central, and higher radii corresponding to the radial positions between 20 and 40 Å, 40 and 70 Å, and 70 and 90 Å, respectively 43. We will follow this terminology here. Our 3D reconstruction of TMV (Figure 5) allowed a manual placement of the X-ray atomic model (PDB code 2TMV) 19 into the reconstruction based on the location of the strong α-helical density. The tracing of the peptide backbone through the four-helix bundle motif was straightforward for lower- and central-radius regions. Moreover, characteristic density features of large protruding residue side chains served as reference points to confirm the positions of the α-helices in the density and to align them in proper register. Consequently, the two peripheral helices are packed against a hydrophobic cluster of aromatic side chains (Phe12, Trp17, Phe62, Tyr139, Phe144) visible as strong side-chain density (Figure 7G). The structure also consists of a small three-stranded antiparallel β-sheet at the interface of the four-helix bundle and the two outer helices. Density corresponding to the outer surface of the coat protein at the C- and N-termini is weaker, with four C-terminal residues being disordered and invisible in the density map, which is in agreement with the X-ray structure 19. Furthermore, the RNA of the virus is packed between the four-helix bundles of lower and upper neighboring subunits along the helical staircase. The density corresponding to the nucleobases is well-resolved leading to distinct features separated by 4 Å (Figure 7H). To provide a quantitative measure of the local agreement of the map with the fitted atomic model, we evaluated the local correlation coefficient (CC) per residue along the main chain (Figure 8A) which is also known as the local R-value 44,
| (4) |
Here, Ai are density values from the atomic model generated on a 3D grid representing the structure on a computer, the density average, Bi the experimental density values, and the density average. To obtain CC-values for a specific residue, only density values within a radius of 10 Å from any atoms belonging to the residue are included in the sums.
Figure 8.

Multiconformer refinement of TMV. (A) Local CC-value of the atomic model evaluated for each residue in the protein sequence. The refined structure with a single conformer and an ensemble of five conformers are compared. The presentation of the refined structure as an ensemble of 5 conformers yields higher correlation than a single conformer alone. The ensemble of conformers accounts for the flexibility of the structure and uncertainty in the atomic coordinates. The TMV atomic model determined by X-ray fiber diffraction (PDB code 2TMV) 19 shows significant disagreement with the density between residues 88 and 109 (22 residues). Black bars indicate clearly identified residues corresponding to strong density and high local CC-values due to their bulky side chains. These residues served as guide points in the building of the new atomic model. (B) Protein secondary structure assignment determined by PROCHECK 74. (C) Example of well-defined density of bulky phenylalanine 48. (D) By contrast, side-chain conformations of smaller residues are not clearly defined by the density (Ile21, Glu22, Ile24), here seen along the LS helix. (E, F) Two orthogonal views of a single TMV subunit displayed as an ensemble of five different conformers.
The well-defined α-helical density corresponds to high CC-values (Figure 8A, B). The nomenclature of the four α-helices accounts for the orientation of the helix axis with respect to the radius of the viral helix: right and left slewed (SL and SR) and right and left radial (LR and RR). The positions of bulky visible side-chains, which are highlighted as bars in Figure 8A, highly correlate with the map (Figure 8C), whereas most smaller side chains are not well defined (Figure 8D). Many of the visible side chains contain aromatic groups (see above). Despite the good overall fit of the available X-ray structure we found that 24 residues in the lower radius region of the protein show only poor correspondence with the density map (Figure 8A). We modified this region accordingly, i.e. the LR helix was N-terminally extended by seven residues, now starting at residue 103. Interestingly, the extended LR helix agrees closely with the structure of the disk aggregate form of the virus determined previously 16; 17 (see below) and with density features seen in the structure of a helical form of the virus determined by X-ray fiber diffraction data in 1977 45. This lends confidence to our interpretation of the density in this region of the map.
The best fit for residues 92 to 97 was obtained by assuming a short 310 helical turn at the C-terminus of the RR helix. Residues 90 and 91 assume no defined secondary structure. We will refer to the part of the molecule consisting of the RR helix, residues 90 and 91, and the short 310 helix as the extended RR helix. The extended α-helices were connected by a vertical stretch of polypeptide that was called the V-column (residues 98 to 102), because of its vertical orientation with respect to the virus 43. The new model places all of the deviating 24 residues in the density.
Common X-ray crystallographic refinement minimizes the differences between computed diffraction data from an atomic model and experimental diffraction data in reciprocal space. At the same time an ideal geometry of the polypeptide chain is desired, i.e. bond lengths and dihedral angles of reference amino acids restrain this optimization procedure. In X-ray crystallography reciprocal-space refinement is predominantly used because amplitude data is determined more accurately than phase data. Real-space refinement is a useful technique to fit atomic models into structures resolved at lower resolution (reviewed by DeLaBarre and Brunger 46). Its advantages include a more efficient refinement with faster convergence and a larger convergence radius 47. Although initially developed for X-ray crystallography, its application has recently been extended to structures determined by cryo-EM 48; 49. Hence, we performed an optimization of the target function E(ρ) in real space, to minimize the difference between observed density ρo and calculated density ρc over a molecular volume V:
| (5) |
The real-space molecular dynamics refinement was carried out using X-PLOR 47; 50 with numerous secondary structure restraints (Tables 1 and 2).
Table 1.
Summary of imposed hydrogen bond distance restraints during refinement.
| Secondary structure | Starting and ending residues | Imposed pattern | |
|---|---|---|---|
| Helix N (7−13) | 7 | n (CO) and n+3 (NH) | |
| 8 − 9 | n (CO) and n+4 (NH) | ||
|
Helix LS (20−31) |
20 − 27 |
n (CO) and n+4 (NH) |
|
| β-sheet at intermediate radius | 139 (CO) and 68 (NH) | ||
| 139 (NH) and 68 (CO) | |||
| 69 (CO) and 18 (NH) | |||
| 69 (NH) and 18 (CO) | |||
|
Helix RS (37−50) |
37 − 46 |
n (CO) and n+4 (NH) |
|
| Helix RR (75−89) |
75 − 85 |
n (CO) and n+4 (NH) |
|
| Extended RR 310 turn (92−97) |
92 − 94 |
n (CO) and n+3 (NH) |
|
| Helix LR (107−134) |
107 − 130 | n (CO) and n+4 (NH) | |
|
Helix C (141−146) |
141 |
n (CO) and n+4 (NH) |
|
| 143 | n (CO) and n+3 (NH) |
Table 2.
Summary of the imposed ϕ/ψ restraints during refinement. First attempts of coordinate refinement resulted in numerous violations in the Ramachandran plots for these residues. They are mostly found in loops that do not possess a clear secondary structure. Hence, we used the deposited 2TMV atomic coordinates to obtain additional restraints for those parts of the structure.
| Residue | ϕ (°) | ψ (°) |
|---|---|---|
| 14 | −76.77 | −176.94 |
| 16 | −120.45 | −3.72 |
| 31 | −54.58 | −10.05 |
| 32 | −109.01 | 81.54 |
| 33 | −165.01 | −161.91 |
| 34 | −145.83 | 116.92 |
| 35 | −85.90 | 21.17 |
| 36 | −125.27 | 10.46 |
| 53 | −125.72 | 137.62 |
| 55 | −120.20 | 72.41 |
| 56 | −79.27 | 87.42 |
| 57 | −102.83 | −172.29 |
| 58 | −58.84 | −64.47 |
| 59 | −55.11 | −15.02 |
| 60 | −116.21 | −178.39 |
| 61 | −158.89 | 159.40 |
| 62 | −65.37 | 120.35 |
| 63 | −51.17 | 146.99 |
| 64 | −72.62 | −67.85 |
| 65 | −77.55 | 20.79 |
| 71 | −40.46 | −62.51 |
| 72 | −82.50 | 74.44 |
| 73 | −109.90 | 113.65 |
| 74 | −38.95 | −22.40 |
| 89 | −153.23 | 121.746 |
| 95 | −50.00 | −27.00 |
| 96 | −50.00 | −27.00 |
| 105 | −58.00 | −47.00 |
| 136 | −48.37 | 135.49 |
| 139 | −80.95 | −166.63 |
| 140 | −157.82 | 155.97 |
| 152 | −105.07 | 140.15 |
At about 5 Å resolution, however, the conformation of most of the side chains remains uncertain. To address this uncertainty, we used a multiconformer refinement approach where the atomic coordinates are represented by an ensemble of structures. This strategy is common practice in NMR structure determination 51 and has also been applied in the past to X-ray crystallography to model motional disorder in proteins 52; 53. In our case, it provides an intuitive way of describing the coordinate uncertainty arising from two factors: 1. limitation of experimental resolution and 2. intrinsic disorder in the protein. We generated five copies of the atomic coordinates, assigned a fractional occupancy to each conformer and excluded any possible inter-conformer interactions. The increased degrees of freedom introduced by the additional atomic models led to an increase in the local CC-values (Figure 8A). The refinement produced an ensemble of structures, which exhibits an RMS deviation of 0.7 Å for the main chain atoms, and 1.1 Å for main and side chain atoms. For display purposes a single model was generated from the five conformers by averaging their atomic coordinate, followed by an energy minimization step that restored bond lengths and angles to allowed values.
It should be noted that regions of weaker density are traditionally accommodated in a refinement by assigning high B-factors to atoms in these regions. Using multiple conformers the degrees of freedom added to the refinement can be more explicitly controlled compared with the traditional B-factor refinement. Furthermore, the RMS deviation between atomic coordinates from different conformers can be directly interpreted as an uncertainty in the coordinates. In the case of a B-factor refinement the interpretation in terms of a coordinate uncertainty is not straightforward.
Taken together, the tertiary structure of the coat protein of TMV consists of an antiparallel four-helix-bundle at central radius and two peripheral helices at higher radius connected via a short antiparallel β-sheet (Figure 9A, B, F). Interactions at central radius are known to stabilize the packing between adjacent subunits (Figure 5A). The virus undergoes a single turn of 360° with 16 1/3 subunits (Figure 5B), i.e. three turns consist of 49 subunits (Figure 5C). A linear repetition of the three-turn building block gives rise to the rod-shaped virus of around 300 nm in length (Figure 5D).
Figure 9.

(A, B) Two orthogonal views of the average structure and adjacent subunits including RNA. (C) RMS deviation per residue between earlier atomic models of TMV and the refined average coordinates derived from the cryo-EM map. Residues 88 to 109 show major deviations. (D) Superimposition of TMV structures. Green: refined average structure derived from the cryo-EM map (PDB code 2OM3), blue: PDB code 2TMV – the helical virus derived from X-ray fiber diffraction 19, red: PDB code 1EI7 – four-layer-disk-aggregate 17. (E) Comparison of the radial density distribution filtered to about 10 Å. Green: PDB code 2OM3, blue: PDB code 2TMV, black: experimental distribution determined by Franklin and Holmes 1958 54. The calculated density distributions for the atomic models include a correction for solvent scattering 75. (F) Simplified topology of tertiary structure based on the cryo-EM map. The names of the helices are assigned according to the 2TMV structure. New elements include an extended RR helix with an additional 310 helical turn and an extended LR helix. The V-column forms a high-density stretch at the inner wall of the virus 43.
Comparison of the Cryo-EM structure with other high-resolution structures of TMV
The new atomic model (PDB code 2OM3) built using the cryo-EM density map contains most of the main structural features that were also seen in previous structures of TMV (see above). In early low-resolution X-ray fiber diffraction studies on TMV, the radial density distribution was determined 54, and it shows closer agreement with the radial density calculated for the 2OM3 model than with the calculated radial density of the 2TMV model (Figure 9E). The main difference between the cryo-EM structure and earlier structures occurs in the loop region at lower radius, which includes residues 88 to 109 (Figure 9C). Residues in this part of the structure, together with the RNA that binds in this region of the molecule, are important for the conversion between the helical and disk aggregate form of the virus. The following discussion of a possible assembly/disassembly mechanism will, therefore, focus on this part of the structure.
Four principal structural elements can be identified in the cryo-EM structure at lower radius that play important parts in the assembly/disassembly mechanism: 1) a carboxylate cluster formed by Glu95 in one subunit, and Asp109 and Asp116 from the adjacent subunit, 2) a carboxylate cluster consisting of Glu97, Glu106 and Asp109, which are part of the same subunit, 3) a hydrophobic patch consisting of Ile93/95 and Val96, neutral residues (Asn98, Gln99, Ala100, Asn101) of the V-column making up the inner wall, and residues Ala105, Leu108, and 4) an RNA-arginine sandwich formed by Arg90 and Arg92 in the extended RR helix, and RNA. The configurations of these structural elements differ significantly from those found in earlier structures determined for TMV. A comparison of the cryo-EM structure with earlier structures is, therefore, useful for elucidating the role of the different structural elements in the assembly/disassembly mechanism. For this comparison, two earlier structures have to be considered. The first is the structure of a helical form of the virus, determined by X-ray fiber diffraction to 2.9 Å resolution 19 and interpreted by an atomic model (PDB code 2TMV). The second is the structure of the disk aggregate, identical in protein composition but without RNA, that was determined and refined by X-ray crystallography methods to 2.4 Å resolution 16; 17 (PDB code 1EI7). The disk-aggregate structure differs significantly at lower radius compared with the helical 2TMV structure. These differences were interpreted as different conformational states of the virus. The present cryo-EM structure shows significant deviations from both of the previously determined structures in the region at lower radius with an RMS deviation per residue of up to 7 Å (Figure 9C, D). We will consider it, therefore, a third distinct structure of the virus that may represent a third state in its assembly/disassembly mechanism. A summary of the differences of the three structures is listed in Table 3.
Table 3.
All of the determined structures of TMV proteins show a high degree of similarity at intermediate and higher radius of the virus. Summary of major differences in the region at lower radius are given.
| Method/Resolution | Structural features at lower radius (residues 85 to 107) |
|---|---|
| X-ray fiber diffraction at 2.9 Å 16 (PDB code 2TMV) | 1. Presence of RNA |
| 2. Ordered Cα-trace without distinct secondary structure | |
| 3. Glu95 and Glu106 are putative Ca ligands | |
|
X-ray crystal diffraction at 2.8 Å 13 and refined at 2.4 Å 14 (PDB code 1E17) |
1. Absence of RNA |
| 2. Disordered Cα-trace at lower radius with an extended LR helix (residues 104 to 134) | |
| 3. Arg90 and 92 point in opposite directions | |
|
Cryo-EM at 4.4 Å (PDB code 2OM3) |
1. Presence of RNA |
| 2. Ordered Cα-trace at lower radius: RR helix interrupted by RNA at residues 90 and 91 and connected to extended RR 310-turn (residues 92 to 97), V-column (residues 98 to 102) and extended LR helix (residues 104 to 134) | |
| 3. Arg90 and Arg92 sandwich RNA | |
| 4. Hydrophobic carboxyl cage shielding Caspar carboxylates | |
| 5. Putative candidates for Caspar carboxylates: Glu97, Glu106 and Asp109 |
(1) Intersubunit carboxylate cluster
Multiple studies have addressed the question of how the disassembly of the helical virus occurs - a process indispensable for viral infection. The dissociation of the TMV rod can be replicated in dilute alkaline solution 55 and point mutations in the lower-radius region have identified residues Glu95 and Asp116 as essential for the destabilization of the virus 56. In the present atomic model based on the cryo-EM structure, these potentially negatively charged residues from adjacent subunits are only separated by about 7 Å. They are found in close proximity to the ribose moiety and thought to form a destabilizing element of the helical assembly. Asp109, which is separated from Asp116 by two α-helical turns, may participate in this interaction depending on its side-chain orientation, which is not precisely known due to the limited resolution of the cryo-EM map. However, so long as other stabilizing factors are in place, such as the RNA-arginine sandwich and the hydrophobic patch (see below), a repulsive charge interaction between these carboxylates does not lead to disassembly. In the helical 2TMV structure, Glu95 and Glu106 from adjacent subunits are separated by about 7 Å and repel each other when charged. The carboxyl groups of these residues were identified as putative calcium ligands 19, thus conveying calcium sensitivity to the helical assembly. Hence, although the participating residues are not the same, both the cryo-EM and earlier helical structures contain carboxylates that potentially destabilize the helical assembly. In the disk aggregate form carboxylates from adjacent subunits are disordered resulting in a highly mobile loop containing residues 92 to 108 with B-factors greater than 100 17.
(2) Intrasubunit carboxylate cluster (Caspar carboxylates)
A group of two carboxylates per subunit, the so-called Caspar carboxylates, were postulated to act as a metastable switch to initiate disassembly upon a change in the environmental milieu 20. In the present cryo-EM structure, the extended LR and RR α-helices and their connection by the V-column brings Glu97 and Glu106 into close proximity, about 6 Å, within the same subunit. This is sufficient for a possible electrostatic interaction. However, unlike in the case of the intersubunit carboxylate cluster, this pair does not have a destabilizing effect on the helical assembly because pairs in neighboring subunits are spaced too far apart for significant electrostatic interaction. Electrostatic interaction is further reduced by the hydrophobic patch (see below).
Asp109 in the same subunit is also located close to Glu106 and could participate in this interaction, increasing the concentration of negative charge. The cluster consisting of Glu97, Glu106 and Asp109 does not exist in the other two TMV structures, 2TMV and 1EI7, where the participating carboxylates are further removed from each other. Instead, in the case of the helical 2TMV structure, Glu106 is positioned close to Glu95 in the neighboring subunit (see above).
(3) Hydrophobic patch and carboxyl cage
In the cryo-EM structure, a destabilizing effect of the intrasubunit carboxylate cluster is reduced by the hydrophobic patch (Ile93, Ile95, Val96, Asn98, Gln99, Ala100, Asn101, Ala105 and Leu108) that isolates clusters in neighboring subunits. As a consequence of the hydrophobic patch, the major contribution to the tight packing of subunits at lower radius originates from hydrophobic interactions. This agrees with predictions based on thermodynamic studies of the polymerization of TMV 57. The charge architecture at lower radius, consisting of the hydrophobic patch and six carboxylates (Glu95, Glu97, Glu106, Asp109, Asp115 and Asp116) was termed “carboxyl cage” 45 (Figure 10B, E and H). A similar situation was hypothesized by Caspar to explain the observed anomalous pKa values of 7.5 of the Caspar carboxylates due to their low dielectric environment 20. In the cryo-EM structure, only Glu97 and Glu106 are clearly buried by the hydrophobic patch. This structure suggests, therefore, that only these two residues, and possibly Asp109 that is close to Glu106, are part of the metastable switch formed by the carboxyl cage. This carboxylate cluster, when protonated and uncharged, is in the “off” position of the metastable switch, thus avoiding electrostatic repulsion between the carboxylates. The switch is operated by an increase in pH that initiates proton release and a buildup of negative charge (see below). The close proximity of Glu97, Glu106 and Asp109 in the present cryo-EM structure suggests that it represents the stable “off” position of the carboxylates. This characteristic charge architecture was neither found in the helical 2TMV structure nor in the disk structure.
Figure 10.

(A, B) Electrostatic potential of the atomic model based on the cryo-EM structure in two orthogonal views. The phosphates of the RNA are neutralized by two arginines at position 90 and 92. The extended RR-helix, V-column and extended RL-helix form the boundaries of the “carboxyl cage” which harbors negatively charged residues Glu97 and Glu106 pointing towards the interior of the subunit. Glu95 and Asp116/Asp109 from the adjacent subunit form another carboxylate cluster. Comparison of (C) disk-aggregate (PDB code 1EI7), (D) helical TMV structure from X-ray fiber diffraction (PDB code 2TMV), and (E) helical TMV structure from cryo-EM (PDB code 2OM3) at lower radius. Positively and negatively charged residues are highlighted in blue and red, respectively. (E) The experimental cryo-EM density is superimposed on the refined atomic coordinates in chicken-wire style. Upon RNA-binding the Arg92 switches in conformation to sandwich the negatively charged phosphate groups with Arg90. Magnified inner-radius region of the solvent-accessible surface colored with the electrostatic potential calculated from the atomic coordinates of the disk aggregate (F), helical TMV from X-ray fiber diffraction (G), and helical TMV from the cryo-EM density (H).
(4) RNA-arginine sandwich
The cryo-EM density map shows clear density for the side chains of Arg90, Arg92, Arg112 and Arg113 (Figure 10E). Arg90 and Arg92 sandwich the phosphate backbone of the RNA. Their positions suggest that they contribute positive charge to stabilize the negative charge of the RNA phosphates, together with Arg112 and Arg113 (Figure 10A and E). The intimate interaction between the arginines and RNA is thought to have a stabilizing effect on the helical form of the virus over a wide range of buffer conditions 55. The RNA-arginine sandwich is not present in the helical 2TMV structure, reducing bonding interactions between RNA and the protein. The different bonding pattern in 2TMV is accompanied by a change in secondary structure in this region. While in the cryo-EM structure the RR helix is extended by one turn of a 310 helix through residues 97 (see above), positioning Arg90 and Arg92 into a sandwich, the RR helix extends only to residue 89 in the 2TMV structure. In the disk aggregate form that does not contain RNA, the Arg92 side chain points approximately in the opposite direction compared with its orientation in the cryo-EM and 2TMV structures. Again, the trace of the backbone differs markedly in this region (Figure 10A and C).
Towards an assembly/disassembly mechanism of TMV
Based on the structural elements of TMV as they appear in different structures of the virus, a possible mechanism for the assembly and disassembly of the virus can be discussed. It must be noted, however, that differences in the experimental conditions used here and in previous experiments may be partly responsible for the structural differences described here (see below). The following discussion is, therefore, somewhat speculative.
We assume that the assembly is a reversible process that proceeds in the opposite direction of the disassembly process. This assumption agrees with previous results that dissociated viruses can re-aggregate in the test tube under appropriate buffer conditions and re-polymerize into one third of the length of a helical rod 58. Starting with the helical form of the virus, two main events can be identified that lead to disassembly. The first is a conformational change in the region at lower radius of the virus caused by the Caspar carboxylates, and the second is the dissociation of RNA and, subsequently, protein subunits to form the disk aggregate form of the virus (Figure 11).
Figure11.
Summary of the putative disassembly/assembly mechanism of TMV. The structural conversion from the helical into the disk-aggregate form can be induced by a change in pH. The atomic models of two adjacent subunits of the coat protein from the lower and upper layer of the three determined structures are displayed. Negatively and positively (red and blue) charged residues are highlighted because of their importance in the assembly/disassembly process. The grey color of the disk-structure main chain at the inner wall presents the disordered backbone with a B-factor greater than 100. (A) The cryo-EM structure (PDB code 2OM3) might represent the stable state of the helical TMV because of its high secondary structure order: the intrasubunit carboxylate cluster (Glu97, Glu106 and possibly Asp109) acts as a metastable switch - here in the spring-loaded 'off' position. Upon a change in the environmental milieu (rise in pH) protons are lost from this cluster of residues and the switch is turned 'on', initiating the opening of the extended RR helical turn at lower radius. (B) The 2TMV structure can be interpreted as a transitional state between the helical form of TMV and the disk form: the loss of secondary structure in the lower-radius region affects the binding between adjacent subunits because of electrostatic repulsion between residues Glu95, Asp109 and Asp116 that are located in adjacent subunits. The loss of secondary structure also loosens the RNA binding to the coat protein that is mediated by the positively charged Arg90, Arg92, Arg112 and Arg113. The conformational change in the lower-radius region ultimately leads to RNA dissociation. (C) 1EI7 structure of the disk aggregate: the lower-radius regions of the protein subunits in the disk structure are further removed from each other compared with their arrangement in the helical forms of the virus. A decrease in pH is thought to promote RNA binding and reversal of the disassembly process.
(1) Conformational change in the lower radius region
The absence of RNA in the disk protein reflects the main difference between the disk aggregate and helical forms of the virus. Addition of RNA to the former was shown to initiate structural conversion to the later 59. Comparison of the disk aggregate protein structure with the cryo-EM structure reveals a striking difference in the positioning of the Arg92 side chain (Figure 10C and E). This suggests that it participates in the metastable switch that changes the protein conformation from one that binds RNA tightly (the RNA-arginine sandwich) to another that is more likely to release the RNA. This conformational change is initiated by proton release from the intrasubunit carboxylate cluster (the Caspar carboxylates, see above) that has a pKa of 7.5. The release of two protons per subunit was observed at a pH of 7.5 upon detergent-induced denaturation of the virus 60. This leads to electrostatic repulsion of carboxyl groups within the cluster. These forces may lead to a partial breakup of secondary structure in the extended RR and LR helices. This loss of secondary structure would constitute a change of the metastable switch from the “off” position described earlier to the “on” position where RNA release is possible. A loss of ordered structure as a necessary step for RNA release was already proposed earlier, based on a comparison of the helical 2TMV virus structure with the disk aggregate protein structure (1EI7): inspection of the density in the lower-radius region (residues 88 to 109) suggests it is better defined, and therefore more ordered in the former, compared with the latter 61. In the cryo-EM structure, there is clear density for the entire lower-radius region and the extent of recognizable secondary structure in this part of the protein is significantly larger than in the helical 2TMV structure. This indicates that the degree of order in the lower-radius region is still higher in the cryo-EM structure.
(2) Dissociation
The transition between order and disorder represents a metastable switch that is driven by the Glu97/Glu106 carboxylates. The 310 helical turn (residues 92 to 97) that forms an extension to the RR helix is a central part of the structure forming the switch, presumably because it is less stable than an α-helical turn. When the 310 helical turn opens due to repulsion between intramolecular negative charges, the intricate stabilizing charge network between the RNA, Arg90 and Arg92 is loosened. A similar element consisting of one aspartic acid within a 310 helical turn and positioned in a hydrophobic environment was identified in EF-Tu/EF-T and Mss4 in the X-ray crystal structures of these Rab GTPases 62; 63. The 310 helical turn is essential for the function of this switch.
Dissociation of the RNA and protein subunits is required for the conversion of the helical virus into the disk aggregate form. The disruption of the RNA-arginine sandwich accompanying the conformational change in the lower radius region must ultimately lead to the dissociation of RNA and, with it, the loss of a major stabilizing factor of the helical virus structure. The loss of secondary structure also affects the hydrophobic patch at the interface between subunits. A disruption of this interface may further facilitate disassembly. Electrostatic repulsion between carboxylates in the intersubunit carboxylate cluster (Glu95, Glu116 and possibly Asp109) drives subunits apart once the helical structure is sufficiently destabilized. Repulsion between subunits might be aided by other carboxylates, for example those from the broken carboxyl cage (Glu97 and Glu106), as well as a carboxylate pair consisting of Glu50 and Asp77 in the helical region of the virus 64. A repulsive interaction between Glu95 and Glu106 was already proposed by Namba and co-workers 19. Mutations of Glu95 and Asp116 into residues with neutral side chains corroborates their crucial role for the dissociation of the helical virus 56.
Disassembly sequence
Using the observed degree of order in the lower-radius region and the aggregation state, the three structures could be viewed as representing different stages of viral disassembly. Thus, the cryo-EM structure would represent the stable helical form of the virus with the highest degree of order. The helical 2TMV structure may represent an intermediate towards disassembly where some of the structural ordering is lost. The disk-aggregate form represents the final stage of disassembly. It is interesting to note that some of the secondary structure that is lost in the LR helix in the 2TMV structure, compared with the cryo-EM structure, is restored in the disk-aggregate structure. This suggests that some reordering might take place once the disassembly process is completed.
A note on structural differences and experimental conditions
The discussion of a possible disassembly mechanism hinges to a large extent on the degree of order seen in the loop region (residues 88 to 109) of the TMV coat protein. Structural changes in this region of the protein are dependent on pH, as discussed above, but also on calcium concentration 65. These factors were put forward to explain differences between the helical 2TMV structure and the structurally related helical U2 strain, which has 72 % sequence homology. On the other hand, differences in temperature during sample preparation or data acquisition were shown to be responsible for the degree of order in the loop region of the disk protein. Data for the first disk structure of TMV was recorded at room temperature, and density of the loop region was not observed 16. Data collected under cryo conditions, however, strengthened the density at lower radius and facilitated the tracing of the main chain 17. Our sample was plunge-frozen in the cold room at 4° C, and also exhibits strong density at lower radius. By contrast, samples used in X-ray fiber diffraction experiments to determine the helical 2TMV structure were prepared at room temperature 66 and show weaker density at lower radius. Density in that region may also have been weakened by the fact that it was represented mainly by a single Bessel function 43. Taken together, factors such as pH, ionic strength, and temperature are known to influence the morphology of TMV and led to the formulation of a “phase diagram” of the virus in analogy to changes between thermodynamically distinct phases of chemical compounds 15.
The influence of different experimental factors on the observed structure of the virus somewhat complicates the interpretation of the various structures. Interestingly, a 4 Å resolution map of the helical virus was already published in 1977 45 but without interpretation by an atomic model which, at the time, was technically difficult. These earlier data are in good agreement with the cryo-EM density presented here, in particular at lower radius. This map, therefore, supports our data and justifies its interpretation with a new atomic model. A precise location of important carboxylates, however, has to be confirmed by a structure that resolves the exact details of these side-chain conformations at different assembly/disassembly states of the virus.
Conclusion
We describe a new method for the processing of helical particles using the single-particle approach. This method is based on the IRSHR procedure described by Egelman 6 but includes an improved correction for the CTF of the electron microscope, a new symmetrization procedure and the use of alignment parameter constraints derived from the helical symmetry of the particles. Using the new method, a structure of TMV at a resolution better than 5 Å was obtained from 135 viruses collected from 6 micrographs. The method is generally applicable to helical specimens, and particularly useful for those filaments with high degree of intrinsic disorder, such as amyloid fibrils that are difficult or impossible to reconstruct by more traditional helical procedures 67.
The TMV reconstruction obtained with our new method is comparable in resolution with the reconstruction obtained by Yonekura et al. of the bacterial flagellum 4. These authors used stringently selected particles based on the strength of their layer lines, a process that could also improve the resolution of the present structure. Based on the work on the bacterial flagellum, another improvement of the resolution can be expected from data collected on a liquid-helium microscope with top-entry stage. More recently, Yonekura and co-workers demonstrated that the use of electron energy filtering enhances the signal-to-noise ratio of cryo-vitrified bacterial flagella68, which represents a promising strategy to improve alignment accuracy of the segmented helical specimens in single-particle based approaches.
We tested several methods of scaling density maps and assessing their resolution. With new automated data acquisition methods underway that make the collection of larger data sets a routine task 69 and a foreseeable increase in computational capacity, sub-nanometer to atomic resolution of many helical specimens may become a routine task in 3D cryo-EM.
The modification of the existing atomic model of the helical virus (PDB code 2TMV) 19 was necessary to accommodate all residues in the density. We performed refinement of the atomic coordinates (PDB code 2OM3) and identified novel structural elements that may be important in the assembly/disassembly mechanism of the virus. These elements include a carboxylate cluster with carboxylates from the adjacent subunit, a metastable pH-sensitive switch consisting of a carboxylate cluster buried in a hydrophobic cage, and an RNA-arginine sandwich that stabilizes RNA-protein interaction. The new structural elements correlate with other properties of TMV observed earlier 20; 45; 56; 57. Comparison of the cryo-EM structure with structures determined earlier, of the helical form of the virus 19 and the RNA-free disk aggregate form (PDB code 2EI7) 17, suggests that the cryo-EM structure represents a third distinct state of the virus. Based on these three structures, we suggest a possible disassembly pathway.
Material and methods
Electron microscopy
TMV was isolated according to the standard procedure 66. The TMV stock solution was diluted five-fold. The buffer of the diluted sample contained 5 mM EDTA and was applied to a copper grid at pH 7.4. Cryo-EM samples were prepared as described elsewhere 70. The vitrified specimens were imaged on a Technai F30 microscope at 200 kV, with a magnification of 59,000 and a dose of 15 e-/A2. Images were recorded at underfocus of 1.5, 3.0 and 4.0 μm on Kodak ISO153 films.
Image processing
6 micrographs were scanned with a raster size of 7 μm resulting in a pixel size of 1.163 Å. 135 virus particles were boxed yielding 4355 segments of 77.4 × 77.4 nm with a step soze of 7 nm along the helical axis by EMAN's boxer program 28. The original in-plane-orientation of each virus was retained in the segment and recorded in a list. Further image processing was carried out with the SPIDER image processing package 71. The approximate magnification of the images was determined as follows. Virus particles were cropped into rectangular boxes (EMAN's helixboxer), apodized and Fourier-transformed. A calibrated magnification of 60190 was determined by measuring the 23 Å layer line from 49 viruses and corrected by the determined out-of-plane tilt angle.
Model building and refinement
The TMV molecule with PDB code 2TMV 19 was manually placed into the density. Residues 85 – 110 had to be adjusted to fit into the EM density using the computer program Coot 72. The atomic coordinates were refined using a real-space molecular dynamics module of X-PLOR 48; 50 by imposing strong constraints on the geometry of secondary structural elements. Recently updated dihedral angle restraints 73 that are based on high-resolution X-ray structures were included in the refinement. Carbonyl oxygen/amide proton distances were enforced to match the characteristic hydrogen-bonding pattern of α-helices. An initial refinement was carried out using three adjacent polypeptide chains and a single continuous chain of 3×3=9 RNA residues to represent the covalent connection between asymmetric RNA units. Therefore, non-crystallographic symmetry (NCS) was imposed between these three asymmetric units. Additional strict NCS was enforced for all neighboring subunits of the three central subunits to account for their van-der-Waals environment. First attempts of coordinate refinement resulted in numerous violations in the Ramachandran plots for residues without a clear secondary structure. Therefore, these ϕ/ψ angles were constrained according to the X-ray reference structure 2TMV (list of constraints in Table 2). Furthermore, the ϕ/ψ angels of the 310 helical turn (extended RR-helix residues 92−97) had to be fixed to −50° and −27°. According to PROCHECK 79.0, 18.7, 1.7, 0.6% of the ϕ/ψ angles occupied the most favored, additionally allowed, generously allowed, and disallowed region, respectively, of the Ramachandran plot 74. Once the structure showed good agreement with the density, each chain was duplicated four times and assigned an occupancy of one fifth. Again, the ensemble of five structures was refined in real space with the respective secondary structure restraints. The structures were validated with PROCHECK and showed very similar ϕ/ψ statistics as indicated for the single conformer above.
Comparison of PDB coordinates
The superpositions and RMS deviations of the PDB coordinates (2TMV, 2EI7, 2OM3) were calculated using the X-PLOR package 50. The radial density distributions of 2TMV and 2OM3 were calculated by including a correction for solvent scattering according to an algorithm proposed by Langridge et al. 75, and low-pass filtered at about 10 Å resolution.
Figure preparation
Chimera was used to display density maps and atomic models (Figure 5) 76. Coot in conjunction with Raster3D generated close-up views of atomic models in the presence of density to evaluate the fit (Figure 7 and 8) 72; 77. CCP4MG assisted in the visualization of atomic models and rendered the surface of the atomic models superimposed with the electrostatic potential (Figure 9 and 10) 78. Topdraw helped to sketch the topology of the protein (Figure 9 and 10) 79.
Protein Data Bank accession number
Coordinates and experimental density map of the helical TMV structure have been deposited in the RCSB Protein Data Bank (code 2OM3).
Acknowledgements
N.G. gratefully acknowledges financial support from the National Institutes of Health, grant 1 P01 GM-62580. M.F. was supported by a grant from the Bundesministerium für Bildung und Forschung (BMBF, BioFuture). We thank David DeRosier for valuable discussions on the manuscript. Special thanks go to Ken Holmes for the critical reading of the manuscript, a correction of a magnification error in our data, and help with the calculation of radial density distribution from atomic models.
Abbreviations used:
- 2D
two-dimensional
- 3D
three-dimensional
- B-factor
temperature factor
- CC
cross correlation
- cryo-EM
electron cryo-microscopy
- CTF
contrast transfer function
- FSC
Fourier shell correlation
- FT
Fourier transform
- FWHM
full width half maximum
- IRSHR
iterative real-space helical reconstruction
- NCS
non-crystallographic symmetry
- RMS
root mean squared
- SPEM
single-particle electron microscopy
- TMV
Tobacco Mosaic Virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Henderson R. Realizing the potential of electron cryo-microscopy. Quarterly Reviews of Biophysics. 2004;37:3–13. doi: 10.1017/s0033583504003920. [DOI] [PubMed] [Google Scholar]
- 2.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 3.De Rosier DJ, Klug A. Reconstruction of Three Dimensional Structures from Electron M icrographs. Nature. 1968;217:130–134. doi: 10.1038/217130a0. [DOI] [PubMed] [Google Scholar]
- 4.Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–50. doi: 10.1038/nature01830. [DOI] [PubMed] [Google Scholar]
- 5.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–89. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–34. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 7.Jimenez JL, Guijarro JI, Orlova E, Zurdo J, Dobson CM, Sunde M, Saibil HR. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. Embo J. 1999;18:815–21. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beroukhim R, Unwin N. Distortion correction of tubular crystals: improvements in the acetylcholine receptor structure. Ultramicroscopy. 1997;70:57–81. doi: 10.1016/s0304-3991(97)00070-3. [DOI] [PubMed] [Google Scholar]
- 9.Holmes K, Angert I, Kull FJ, Jahn W, Schröder RR. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 10.Morgan DG, DeRosier D. Processing images of helical structures: a new twist. Ultramicroscopy. 1992;46:263–85. doi: 10.1016/0304-3991(92)90019-g. [DOI] [PubMed] [Google Scholar]
- 11.Böttcher B, Wynne SA, Crowther RA. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature. 1997;386:88–91. doi: 10.1038/386088a0. [DOI] [PubMed] [Google Scholar]
- 12.Ludtke SJ, Chen DH, Song JL, Chuang DT, Chiu W. Seeing GroEL at 6 A resolution by single particle electron cryomicroscopy. Structure (Camb) 2004;12:1129–36. doi: 10.1016/j.str.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Halic M, Becker T, Frank J, Spahn CMT, Beckmann R. Localization and dynamic behavior of ribosomal protein L30e. Nature Structural & Molecular Biology. 2005;12:467–468. doi: 10.1038/nsmb933. [DOI] [PubMed] [Google Scholar]
- 14.Wang YA, Yu X, Overman S, Tsuboi M, Thomas GJ, Jr, Egelman EH. The Structure of a Filamentous Bacteriophage. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.06.027. [DOI] [PubMed] [Google Scholar]
- 15.Klug A. The tobacco mosaic virus particle: structure and assembly. Philosophical Transactions: Biological Sciences. 1999;354:531–535. doi: 10.1098/rstb.1999.0404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloomer AC, Champness JN, Bricogne G, Staden R, Klug A. Protein disk of tobacco mosaic virus at 2.8 Å resolution showing the interactions within and between subunits. Nature. 1978;276:362–368. doi: 10.1038/276362a0. [DOI] [PubMed] [Google Scholar]
- 17.Bhyravbhatla B, Watowich SJ, Caspar DLD. Refined Atomic Model of the Four-Layer Aggregate of the Tobacco Mosaic Virus Coat Protein at 2.4-A Resolution. Biophysical Journal. 1998;74:604–615. doi: 10.1016/S0006-3495(98)77819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jeng TW, Crowther RA, Stubbs G, Chiu W. Visualization of alpha-helices in tobacco mosaic virus by cryo-electron microscopy. J Mol Biol. 1989;205:251–7. doi: 10.1016/0022-2836(89)90379-3. [DOI] [PubMed] [Google Scholar]
- 19.Namba K, Pattanayek R, Stubbs G. Visualization of protein-nucleic acid interactions in a virus. Refined structure of intact tobacco mosaic virus at 2.9 A resolution by X-ray fiber diffraction. J Mol Biol. 1989;208:307–25. doi: 10.1016/0022-2836(89)90391-4. [DOI] [PubMed] [Google Scholar]
- 20.Caspar DL. Assembly and Stability of the Tobacco Mosaic Virus Particle. Adv Protein Chem. 1963;18:37–121. doi: 10.1016/s0065-3233(08)60268-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Planchart A, Stubbs G. Caspar Carboxylates: The Structural Basis of Tobamovirus Disassembly. Biophysical Journal. 1998;74:633–638. doi: 10.1016/S0006-3495(98)77822-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 23.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–47. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 24.Vonck J. Parameters affecting specimen flatness of two-dimensional crystals for electron crystallography. Ultramicroscopy. 2000;85:123–129. doi: 10.1016/s0304-3991(00)00052-8. [DOI] [PubMed] [Google Scholar]
- 25.Grigorieff N. FREALIGN: High-resolution refinement of single particle structures. J Struct Biol. 2006 doi: 10.1016/j.jsb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. J Mol Biol. 1998;277:1033–46. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- 27.Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–70. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 28.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 29.Stubbs G, Makowski L. Coordinated use of isomorphous replacement and layer-line splitting in the phasing of fiber diffraction data. Acta Cryst A. 1982;38:417–425. [Google Scholar]
- 30.Paul D, Patwardhan A, Squire JM, Morris EP. Single particle analysis of filamentous and highly elongated macromolecular assemblies. J Struct Biol. 2004;148:236–50. doi: 10.1016/j.jsb.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 31.Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- 32.Jeng TW, Chiu W. High resolution cryo system designed for JEM 100CX electron microscope. Ultramicroscopy. 1987;23:61–6. doi: 10.1016/0304-3991(87)90227-0. [DOI] [PubMed] [Google Scholar]
- 33.Harauz G, van Heel M. Exact filters for general geometry three dimensional reconstruction. OPTIK. 1986;73:146–156. [Google Scholar]
- 34.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Grigorieff N. Resolution measurement in structures derived from single particles. Acta Crystallographica Section D Biological Crystallography. 2000;56:1270–1277. doi: 10.1107/s0907444900009549. [DOI] [PubMed] [Google Scholar]
- 36.Grigorieff N, Ceska TA, Downing KH, Baldwin JM, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. Journal of Molecular Biology. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 37.Mitsuoka K, Hirai T, Murata K, Miyazawa A, Kidera A, Kimura Y, Fujiyoshi Y. The Structure of Bacteriorhodopsin at 3.0 A Resolution Based on Electron Crystallography: Implication of the Charge Distribution. Journal of Molecular Biology. 1999;286:861–882. doi: 10.1006/jmbi.1998.2529. [DOI] [PubMed] [Google Scholar]
- 38.Ravelli RB, Garman EF. Radiation damage in macromolecular cryocrystallography. Curr Opin Struct Biol. 2006 doi: 10.1016/j.sbi.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Henderson R. Image contrast in high-resolution electron microscopy of bio logical macromolecules: TMV in ice. Ultramicroscopy. 1992;46:1–18. doi: 10.1016/0304-3991(92)90003-3. [DOI] [PubMed] [Google Scholar]
- 40.Jensen GJ. Alignment Error Envelopes for Single Particle Analysis. Journal of Structural Biology. 2001;133:143–155. doi: 10.1006/jsbi.2001.4334. [DOI] [PubMed] [Google Scholar]
- 41.Havelka WA, Henderson R, Oesterhelt D. Three-dimensional structure of halorhodopsin at 7 A resolution. J Mol Biol. 1995;247:726–38. doi: 10.1006/jmbi.1995.0176. [DOI] [PubMed] [Google Scholar]
- 42.Jiang JS, Brunger AT. Protein hydration observed by X-ray diffraction. Solvation properties of penicillopepsin and neuraminidase crystal structures. J Mol Biol. 1994;243:100–15. doi: 10.1006/jmbi.1994.1633. [DOI] [PubMed] [Google Scholar]
- 43.Holmes KC. Protein-RNA interactions during TMV assembly. J Supramol Struct. 1979;12:305–20. doi: 10.1002/jss.400120304. [DOI] [PubMed] [Google Scholar]
- 44.Chapman MS. Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron-density function. Acta Crystallographica Section D Biological Crystallography. 1995;A51:69–80. [Google Scholar]
- 45.Stubbs G, Warren S, Holmes K. Structure of RNA and RNA binding site in tobacco mosaic virus from 4-Å map calculated from X-ray fibre diagrams. Nature. 1977;267:216–221. doi: 10.1038/267216a0. [DOI] [PubMed] [Google Scholar]
- 46.DeLaBarre B, Brunger AT. Considerations for the refinement of low-resolution crystal structures. Acta Crystallogr D Biol Crystallogr. 2006;62:923–32. doi: 10.1107/S0907444906012650. [DOI] [PubMed] [Google Scholar]
- 47.Chen Z, Blanc E, Chapman MS. Real-space molecular-dynamics structure refinement. Acta Crystallographica Section D Biological Crystallography. 1999;55:464–468. doi: 10.1107/s090744499801097x. [DOI] [PubMed] [Google Scholar]
- 48.Chen JZ, Furst J, Chapman MS, Grigorieff N. Low-resolution structure refinement in electron microscopy. J Struct Biol. 2003;144:144–51. doi: 10.1016/j.jsb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A. Study of the Structural Dynamics of the E. coli 70S Ribosome Using Real-Space Refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 50.Brünger AT. X-PLOR 3.1, A System for X-ray Crystallography and NMR. Yale University Press; New Haven, CT: 1992. [Google Scholar]
- 51.Wuthrich K. Protein structure determination in solution by NMR spectroscopy. 1990. [PubMed]
- 52.Gros P, van Gunsteren WF, Hol WG. Inclusion of thermal motion in crystallographic structures by restrained molecular dynamics. Science. 1990;249:1149. doi: 10.1126/science.2396108. [DOI] [PubMed] [Google Scholar]
- 53.Chen Z, Chapman MS. Conformational Disorder of Proteins Assessed by Real-Space Molecular Dynamics Refinement. Biophysical Journal. 2001;80:1466–1472. doi: 10.1016/S0006-3495(01)76118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Franklin RE, Holmes KC. Tobacco mosaic virus: application of the method of isomorphous replacement to the determination of the helical parameters and radial density distribution. Acta Crystallographica. 1958;11:213–220. [Google Scholar]
- 55.Schramm G, Schumacher G, Zillig W. An Infectious Nucleoprotein from Tobacco Mosaic Virus. Nature. 1955;175:549–550. doi: 10.1038/175549a0. [DOI] [PubMed] [Google Scholar]
- 56.Lu B, Stubbs G, Culver JN. Carboxylate interactions involved in the disassembly of tobacco mosaic tobamovirus. Virology. 1996;225:11–20. doi: 10.1006/viro.1996.0570. [DOI] [PubMed] [Google Scholar]
- 57.Lauffer MA. Polymerization-Depolymerization of Tobacco Mosaic Virus Protein. VII. A Model*. Biochemistry. 1966;5:2440–2446. doi: 10.1021/bi00871a038. [DOI] [PubMed] [Google Scholar]
- 58.Harrington WF, Schachman HK. Studies on the alkaline degradation of tobacco mosaic virus. I. Ultracentrifugal analysis. Arch Biochem Biophys. 1956;65:278–95. doi: 10.1016/0003-9861(56)90194-1. [DOI] [PubMed] [Google Scholar]
- 59.Fraenkel-Conrat H, Singer B. Reconstitution of tobacco mosaic virus. III. Improved methods and the use of mixed nucleic acids. Biochim Biophys Acta. 1959;33:359–70. doi: 10.1016/0006-3002(59)90126-x. [DOI] [PubMed] [Google Scholar]
- 60.Koshland DE, Jr, Simmons NS, Watson JD. Absence of Phosphotriester Linkages in Tobacco Mosaic Virus. Journal of the American Chemical Society. 1958;80:105–107. [Google Scholar]
- 61.Namba K, Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science. 1986;231:1401–6. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- 62.Zhu Z, Dumas JJ, Lietzke SE, Lambright DG. A helical turn motif in M ss4 is a critical determinant of Rab binding and nucleotide release. Biochemistry. 2001;40:3027–3036. doi: 10.1021/bi002680o. [DOI] [PubMed] [Google Scholar]
- 63.Kawashima T, Berthet-Colominas C, Wulff M, Cusack S, Leberman R. The structure of the Escherichia coli EF-Tu EF-Ts complex at 2.5 Å resolution. Nature. 1996;379:511–518. doi: 10.1038/379511a0. [DOI] [PubMed] [Google Scholar]
- 64.Culver JN, Dawson WO, Plonk K, Stubbs G. Site-directed mutagenesis confirms the involvement of carboxylate groups in the disassembly of tobacco mosaic virus. Virology. 1995;206:724–30. doi: 10.1016/s0042-6822(95)80096-4. [DOI] [PubMed] [Google Scholar]
- 65.Pattanayek R, Stubbs G. Structure of the U2 strain of tobacco mosaic virus refined at 3.5 A resolution using X-ray fiber diffraction. J Mol Biol. 1992;228:516–28. doi: 10.1016/0022-2836(92)90839-c. [DOI] [PubMed] [Google Scholar]
- 66.Gregory J, Holmes KC. Methods of preparing oriented tobacco mosaic virus sols for X-ray diffraction. J. Mol. Biol. 1965;13:796. [Google Scholar]
- 67.Sachse C, Xu C, Wieligmann K, Diekmann S, Grigorieff N, Fandrich M. Quaternary structure of a mature amyloid fibril from Alzheimer's abeta (1−40) Peptide. J Mol Biol. 2006;362:347–54. doi: 10.1016/j.jmb.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 68.Yonekura K, Braunfeld M, Maki-Yonekura S, Agard D. Electron energy filtering significantly improves amplitude contrast of frozen-hydrated protein at 300kV. J Struct Biol. 2006;156:524–36. doi: 10.1016/j.jsb.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 69.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: The new Leginon system. J Struct Biol. 2005 doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 70.Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 71.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–9. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 72.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D Biological Crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 73.Priestle JP. Improved dihedral-angle restraints for protein structure refinement. Journal of Applied Crystallography. 2003;36:34–42. [Google Scholar]
- 74.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 75.Langridge R, Marvin DA, Seeds WE, Wilson HR, Hooper CW, Wilkins MHF, Hamilton LD. The molecular configuration of deoxyribonucleic acid. II. Molecular models and their Fourier transforms. J. Mol. Biol. 1960:38–64. [Google Scholar]
- 76.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 77.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular images. Methods Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 78.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP 4 molecular-graphics project. Acta Crystallographica Section D Biological Crystallography. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 79.Bond CS. TopDraw: a sketchpad for protein structure topology cartoons. Bioinformatics. 2003;19:311–312. doi: 10.1093/bioinformatics/19.2.311. [DOI] [PubMed] [Google Scholar]