Abstract
The purpose of this study was to generate stable cell cultures from head and neck squamous cell carcinomas (HNSCC), and retrospectively analyze the factors associated with successful cell line establishment. Fifty-two HNSCC cell lines were isolated from a series of 199 tumors collected between 1992 and 1997 at the University of Pittsburgh Medical Center. Cell lines were characterized at the molecular and cellular level to determine the features associated with cell line formation. Successful cell line formation was dependent on multiple factors, including gene amplification involving chromosomal band 11q13, local and/or regional involvement of lymph nodes, and alcohol usage. The establishment of HNSCC cell lines enriches the resources available for cancer research. Our findings indicate that generation of stable cell lines from HNSCC is biased towards tumors with a poor prognosis. Our 52 stable lines comprise one of the largest series of HNSCC cell lines in the literature, with complete demographic, histopathologic, clinical, and survival data.
Keywords: head and neck, squamous cell carcinoma, alcohol, tobacco, human papillomavirus, gene amplification, chromosome 11
Introduction
Head and neck squamous cell carcinoma (HNSCC) develops in the squamous epithelial cells of the upper aerodigestive tract, including the oral cavity, oropharynx, hypopharynx and larynx. Based on the incidence rates of HNSCC over the past 20 years in the Surveillance, Epidemiology, and End Results (SEER) database of the National Cancer Institute (NCI), approximately 40,000 new cases and 7,200 deaths are expected in the United States of America each year as a result of this disease.1,2 Worldwide, nearly 500,000 people per year are diagnosed with HNSCC.3,4 The median age at diagnosis across all races is 63 years, with a male to female ratio of 2:1.2,3,5 Despite advances in our understanding of the disease, five-year survival rates have barely exceeded 50% during the past 40 years.3,4 The prognosis of HNSCC is excellent if identified early, but early stage cancer has few symptoms, resulting in delays in diagnosis and treatment.6 Further, recurrence develops in at least one-third of patients with HNSCC, resulting in a poor prognosis.7,8
A number of genetic alterations are known to be important for HNSCC development and progression. A linear sequence of genetic events was first reported by Califano and colleagues.6,9 The concept of field cancerization in HNSCC, first proposed by Slaughter et al. in 1953, suggests that a field of tissue shares the same background carcinogen susceptibility and after exposure, gives rise to multiple “related” primary tumors and/or recurrences.6,10,11 The background susceptibility is thought to be driven by genetic alterations. The most prevalent reported genetic changes in HNSCC include the inactivation of CDKN2A (p16) by methylation, deletion and/or mutation, loss or mutation of TP53, and amplification of 11q13.2,11 The cellular effects of each of these genetic alterations have been extensively reported in the literature (discussed by Califano et al.,6,9 and reviewed by Gollin10).
HNSCC arises through an accumulation of genetic alterations that are thought to result from exogenous exposures.12 Tobacco is considered the most important factor.13,14 This includes tobacco smoking (cigarette, cigar and pipe) and smokeless tobacco use (snuff, chewing tobacco). Smoking has stronger causative links than smokeless tobacco use, related directly to the length of time of use and dose.13,15 Alcohol consumption adds to the risk of head and neck cancer and is thought to act as a promoter, and possibly an initiator of HNSCC.13,14 Alcohol serves as a mucosal irritant, but may also act as a solvent for the carcinogens found in tobacco. It may activate members of the cytochrome p450 family of enzymes, known to activate procarcinogens, thereby exacerbating the effect of alcohol when the individual is also exposed to tobacco.14 In the past few years, the relationship between HPV and head and neck cancer has moved from a strong association to an apparent causal relationship for at least a subset of HNSCC.15-17 Some evidence suggests that HPV has synergistic effects with tobacco products, but we and others have evidence that HPV may be an independent risk factor for HNSCC.17-21 There is also accumulating evidence of genetic predisposition to disease. For example, Fanconi anemia patients have a 500-fold increase in cumulative incidence of HNSCC, and a 14% chance of developing HNSCC by age 40.22 The etiology of HNSCC in patients with no known risk factors remains unclear, and is the topic of substantial investigation.
From our series of 199 head and neck cancers, we have successfully established 52 cell lines from both primary and recurrent tumors. The value of cell lines as models of disease was controversial, but several studies have shown very clearly that HNSCC cell lines are accurate reflections of the original tumor.23-25 We present basic histopathologic and genetic characterization of our series of HNSCC cell lines, the clinical and demographic profiles of the patients, and the histopathologic features of the tumors from which they were derived. We anticipated that the same risk factors that modulate survival and/or treatment failure would be useful in predicting successful culture of HNSCC tissue, and have made an attempt to determine if there are any measurable parameters in our dataset that support this hypothesis. In addition, these cell lines represent an important resource for further investigation into the biology and genetics of head and neck squamous cell carcinoma.
Materials and Methods
From September 1992 to February 1997, tumor, adjacent mucosa and peripheral blood specimens were obtained from consenting patients undergoing surgery for squamous cell carcinoma of the oral cavity at the University of Pittsburgh Medical Center, following Institutional Review Board guidelines. Primary tumors, recurrences and metastases of HNSCC were resected from the upper aerodigestive tract, including sites in the oral cavity, oropharynx, hypopharynx and larynx. In addition, patients provided information via questionnaire, including exposure histories, family histories, and other demographic information. Additional data regarding the tumors were extracted from de-identified pathology reports. Over time, the laboratory of SMG has accumulated data from a variety of molecular and cytogenetic examinations of the HNSCC samples. In brief, 11q13 amplification was assayed by fluorescence in situ hybridization (FISH) with digoxigenin-labeled cosmid probes for INT2, HSTF1 or CCND1 (Oncor, Gaithersburg, MD; D.J. Demetrick, University of Calgary, Canada).24 Exons 5−8 of TP53 were sequenced to screen for mutation by peptide mass signature genotyping; the methods are summarized elsewhere.26-28 HPV status was determined using nested real-time PCR with consensus primers for the L1 sequence of the HPV genome.21
Tissue culture
Resected tissue specimens were collected in α-MEM (Irvine Scientific, Santa Ana, CA) supplemented with 2mM L-glutamine, 10% (v/v) fetal bovine serum (FBS), gentamicin (50μg/ml), chloramphenicol (5μg/ml), clindamycin (10μg/ml), penicillin G (100U/ml), streptomycin (100μg/ml) and amphotericin B (5μg/ml) (supplements from Gibco Invitrogen, Grand Island, NY)29. Specimens were examined by an oral pathologist (primarily, KMR), and then divided for cell culture, histopathology, molecular genetic analysis and direct harvest for molecular cytogenetic analysis. The methods described for culture of head and neck squamous cell carcinomas by Dr. Thomas Carey were used in this study, with minor modifications.30,31 Briefly, each tumor specimen was rinsed in a solution of Hank's balanced salt solution (HBSS; Irvine Scientific) with gentamicin (250μg/ml), chloramphenicol (50μg/ml), clindamycin (100μg/ml), penicillin G (250U/ml) and amphotericin B (50μg/ml), to minimize the risk of microbial contamination. After rinsing, the tissue was minced into 1 mm3 pieces and distributed among three or four T25 flasks (dependent on the tissue mass). To enable the tissue to adhere to the flask surface, the tissue was incubated at 37°C for 5 min prior to adding sufficient medium to cover the bottom of the flask. This medium was comprised of Minimal Essential Medium (Gibco Invitrogen), supplemented with 1% non-essential amino acids, 1% L-glutamine, 50μg/ml gentamicin and 15% (v/v) FBS (FBS from Irvine Scientific, all other supplements from Gibco Invitrogen). Flasks were then returned to a humidified incubator at 37°C in 5% CO2. Long-term cultures are maintained in similar medium, with the substitution of 10% (v/v) FBS (medium designated M-10), and changed twice weekly.
After culturing, four possible outcomes were observed: 1) microbial contamination, 2) absence of cell growth, 3) fibroblast-like cell growth, or 4) both fibroblast-like cell growth and squamous cell growth. In spite of antibiotic treatment, some cultures became contaminated and those flasks were discarded immediately. If no cell growth was observed after one month, the flasks were also discarded. Cultures that displayed fibroblast-like cell growth in the absence of squamous cells were cryopreserved when they reached confluency for future characterization and possible use as controls. HNSCC cell growth was usually observed 2 to 5 days after primary culture initiation. Most successful primary cultures demonstrated squamous cell growth initially, followed by fibroblast proliferation approximately a week later. In general, two to four weeks after primary culture initiation, cells reached confluency. When growth of fibroblasts obstructed the growth of squamous cells, the flasks were treated with differential trypsinization (DT) to remove the fibroblasts. For DT, the cells were rinsed in HBSS (Irvine Scientific). Then, 1.5 − 2 ml of 0.025% trypsin-EDTA (Gibco Invitrogen) was added to the flask, and the cells were observed under an inverted microscope. The trypsinized cell suspension was removed from the flask when most or all of the fibroblasts detached from the flask surface and the squamous cells remained adherent. Trypsin was inactivated by the addition of M-10. When squamous cells reached at least 70% confluency, the primary culture was subcultured into two T25 flasks. Cell lines were considered established after continuous successful passage of squamous cells and elimination of fibroblast-like cells.
Immunostaining
In preparation for immunohistochemical staining, cultured cells were grown on chamber slides (Nalge Nunc International, Rochester, NY). First, the slides were rinsed in PBS and fixed with 4% paraformaldehyde. To minimize endogenous peroxidase activity, cells were treated with a methanol/hydrogen peroxide solution. The cells were then incubated with one of four different primary antibodies to cytokeratins, which included anti-keratin AE1, anti-keratin AE3, cytokeratin 19 (all from Vector Laboratories), and CAM 5.2 (Beckon Dickinson). For each cell line, one well of the chamber slide was not treated with the primary antibody to control for nonspecific staining with the secondary antibody (Vectastain ABC Elite Kit; Vector Laboratories, Burlingame, CA). The chromagenic substrate was diaminobenzidine (DAB; Vector Laboratories). Slides were lightly counterstained with Mayer's hematoxylin. Finally, the cells were dehydrated in a graded series of ethanols, followed by xylene and mounting with a coverslip.
Statistical Analysis
Fisher's exact test and the Student's t-test were used to compare categorical variables and continuous variables, respectively. Univariate logistic regression was used to screen for potentially significant risk factors. Univariate risk factors with a significance level <0.1 were entered into a multivariate logistic regression analysis to calculate age- and stage-adjusted odds ratios (OR). All tests of significance were two-tailed. Clustering was used in the univariate and multivariate models to account for the influence of subject-level data. A log-rank test for equality of survivor functions was carried out for the observed and disease-free survival models, both of which were sorted by cell line establishment (Y/N). All statistical analyses were performed with the Stata™ 8.0 Statistics/Data Analysis software package (Stata Corporation, College Station, TX).
Results
From September 1992 to February 1997, 199 HNSCC samples were collected from 185 subjects, with patient follow-up available up to as recently as May 2004. All specimens and their information were stripped of any identifiers prior to delivery to the laboratory investigators. Follow-up information has been collected by our ‘honest broker.’ One hundred twenty-four of the 185 subjects were male, which is consistent with the expected 2:1 male:female ratio for HNSCC tumorigenesis. Of the 199 tumor events evaluated in this analysis, 127 of them were initial primary tumors from individuals for which there was no previously recorded history of cancer (HNSCC or otherwise).
Information was collected from patient responses to a questionnaire, including tobacco and alcohol history, family history of cancer, and general demographic information. One hundred forty-one of the subjects (76%) reported use of both alcohol and tobacco during their lifetimes. Of those 141 individuals, 74 had used both substances despite a family history of cancer. A summary of the information for the 48 patients associated with the 52 cell lines that were established is available in Table 1. Among the patients associated with cell line formation, all but one reported use of either alcohol or tobacco products, and only seven reported use of one but not the other. Twenty-two of the 48 patients (46%) also reported a family history of cancer. Additional data were extracted from de-identified pathology reports including the anatomic tumor site, tumor-node-metastasis (TNM) stage, histologic type, histologic grade, resected tumor size, surgery date, status of tissue margins, perineural involvement and extracapsular spread (Table 2). Also available in Table 2 is a compilation of the laboratory data which have been collected over time on the HNSCC samples, including 11q13 amplification status, TP53 mutation status and HPV status. The data have been extracted from previously published and unpublished work on our UPCI:SCC tumors and cell lines.21,24-27 Some of the initial findings on 11q13 amplification were published by Lese et al., and additional results have been accumulated in the laboratory of SMG.24 Of the 52 tumors that eventually formed cell lines, 24 were positive for 11q13 amplification (46%). TP53 mutation analysis was initiated by JCL, and continued by CSI and Dr. Cheryl Telmer.26,27 Somewhat surprisingly, mutation of TP53 was found in only nine of the 52 HNSCC samples used to establish the cell lines (17%). The status was unknown for nine of the samples, but the remaining 35 tumors were wild-type. Tumors and cell lines were screened for HPV by SMG and CCRR.21 Forty-nine of the tumors (25%) and eight of the cell lines (15%) were positive for HPV.
Table 1.
Personal characteristics of the patients associated with the UPCI:SCC cell lines
| Subject ID | UPCI: | Agea | Gender | Ethnicity | Smokingb | Alcoholb | Family Historyb |
|---|---|---|---|---|---|---|---|
| 2 | SCC002 | 29 | F | Caucasian | ○ | • | ○ |
| 3 | SCC003 | 65 | F | Caucasian | • | • | ○ |
| 13 | SCC013 | 70 | F | Caucasian | • | • | ○ |
| 16c | SCC016 | 82 | F | Caucasian | ○ | • | • |
| SCC026 | 82 | Caucasian | |||||
| 29 | SCC029 | 84 | M | Caucasian | • | • | ○ |
| 30 | SCC030 | 38 | M | Caucasian | • | • | • |
| 32 | SCC032 | 60 | M | Caucasian | • | • | • |
| 36c | SCC036 | 56 | M | Caucasian | • | • | ○ |
| SCC104 | 57 | ||||||
| SCC142 | 58 | ||||||
| 40 | SCC040 | 50 | M | Caucasian | ○ | • | • |
| 44 | SCC044 | 57 | F | Caucasian | • | • | ○ |
| 45 | SCC045 | 54 | M | Caucasian | • | • | ○ |
| 51 | SCC051 | 62 | M | Caucasian | • | • | ○ |
| 52 | SCC052 | 58 | F | Caucasian | • | • | • |
| 53 | SCC053 | 63 | M | Caucasian | • | • | ○ |
| 56 | SCC056 | 76 | M | Caucasian | • | • | ○ |
| 60 | SCC099 | 52 | M | Caucasian | • | • | • |
| 62 | SCC062 | 18 | F | Caucasian | • | • | • |
| 65 | SCC065 | 50 | M | Caucasian | • | • | ○ |
| 66 | SCC066 | 75 | F | Caucasian | • | • | • |
| 68 | SCC068 | 60 | M | Caucasian | • | • | • |
| 70 | SCC070 | 34 | F | Caucasian | • | • | • |
| 72 | SCC072 | 61 | F | Caucasian | • | • | • |
| 74 | SCC074 | 51 | F | Indian | ○ | ○ | ○ |
| 75 | SCC075 | 67 | M | Caucasian | • | • | • |
| 77 | SCC077 | 57 | M | Caucasian | • | • | ○ |
| 78 | SCC078 | 60 | M | Caucasian | ○ | • | • |
| 80 | SCC080 | 66 | M | Caucasian | • | • | • |
| 81 | SCC081 | 87 | F | Caucasian | • | • | • |
| 84 | SCC084 | 52 | M | Caucasian | • | • | ○ |
| 89 | SCC089 | 58 | M | Caucasian | • | • | ○ |
| 90c | SCC090 | 46 | M | Caucasian | • | • | • |
| SCC152 | 47 | ||||||
| 103 | SCC103 | 27 | F | Caucasian | • | ○ | ○ |
| 105 | SCC105 | 67 | M | Caucasian | • | • | • |
| 111 | SCC111 | 69 | F | Caucasian | • | • | • |
| 114 | SCC114 | 71 | M | Caucasian | • | • | • |
| 116 | SCC116 | 57 | M | Caucasian | • | • | • |
| 121 | SCC121 | 61 | M | African-American | • | • | ○ |
| 122 | SCC122 | 63 | M | Caucasian | • | • | ○ |
| 125 | SCC125 | 78 | F | Caucasian | • | • | ○ |
| 131 | SCC131 | 73 | M | Caucasian | • | • | ○ |
| 136 | SCC136 | 64 | F | Caucasian | • | ○ | ○ |
| 143 | SCC143 | 76 | F | Caucasian | • | ○ | ○ |
| 153 | SCC153 | 63 | M | Caucasian | • | • | • |
| 154 | SCC154 | 54 | M | Caucasian | • | • | ○ |
| 172 | SCC172 | 68 | M | Caucasian | • | • | • |
| 182 | SCC182 | 71 | M | Caucasian | • | • | ○ |
| 192 | SCC192 | 65 | M | Caucasian | • | • | ○ |
| 200 | SCC200 | 74 | M | Caucasian | • | • | ○ |
Age is indicated at time of HNSCC treatment.
Filled circle indicates positivity for given parameter.
Individual contributed multiple UPCI:SCC cell lines. Records for gender, race, smoking and alcohol exposure, and family history are unchanged from first record.
Table 2.
Clinical and histopathologic features of the HNSCC specimens
| UPCI: | Origina | Site | Gradeb | Path Stage | 11q13c | TP53 (codon)d | HPV +e |
|---|---|---|---|---|---|---|---|
| SCC002 | R | TONG | 1 | T4N0 | ○ | WT | ○ |
| SCC003 | NP | TONS | 1 | T1N0 | • | - | ○ |
| SCC013 | NP | MAND | 2 | T4N0 | ○ | WT | ○ |
| SCC016 | NP | TONG | 1 | T1N0 | • | WT | ○ |
| SCC026 | R | ALV RIDGE | 1 | T1NX | • | WT | ○ |
| SCC029 | N | BUCCAL | 3 | T4N2 | • | WT | ○ |
| SCC030f | NP | FOM | 3 | T4N2B | ○ | WT | ○ |
| SCC032 | NP | RMT | 2 | T2N2B | • | WT | ○ |
| SCC036f | NP | TONS | 2 | T3N1 | • | WT | • |
| SCC040 | NP | TONG | 2 | T2N2 | • | WT | ○ |
| SCC044 | NP | BOT | 2 | T2N2C | • | WT | ○ |
| SCC045 | Prior SCC | RMT | 2 | T2N0 | ○ | WT | ○ |
| SCC051f | NP | RMT | - | T3N2B | • | WT | ○ |
| SCC052 | NP | FOM | 2 | T4N2C | ○ | WT | ○ |
| SCC053 | NP | FOM | 2 | T1N0 | ○ | mut (7) | ○ |
| SCC056 | NP | TONG | 2 | T3N2B | • | - | ○ |
| SCC062 | NP | TONG | 1 | T3N0 | • | WT | ○ |
| SCC065 | NP | TONG | 1 | T2N0 | ○ | WT | ○ |
| SCC066 | NP | ALV | 1 | T1N0 | ○ | mut (189) | ○ |
| SCC068 | NP | TONG | 1 | T3N1 | ○ | WT | ○ |
| SCC070 | R | RMT | 2 | T3N1 | • | WT | • |
| SCC072 | NP | TONS | 2 | T3N2B | • | mut (179) | ○ |
| SCC074 | NP | ALV | 3 | T4N1 | • | mut (221) | ○ |
| SCC075 | NP | TONG | 2 | T3N2B | ○ | WT | ○ |
| SCC077 | NP | FOM | 2 | T2N2 | ○ | WT | ○ |
| SCC078 | R | FOM | 2 | T2N0 | • | mut (158) | ○ |
| SCC080f | R | BOT | - | T1N0 | • | WT | ○ |
| SCC081 | NP | ALV RIDGE | 2 | T4N0 | • | WT | ○ |
| SCC084 | R | RMT | 2 | T2N2B | • | WT | ○ |
| SCC089 | NP | TONS | 2 | T4N2B | • | - | ○ |
| SCC090 | R | BOT | 3 | T2N0 | ○ | WT | • |
| SCC099 | R | FOM | 2 | T1N0 | ○ | WT | • |
| SCC103 | NP | TONG | 1 | T1N0 | • | mut (306) | ○ |
| SCC104 | R | FOM | 2 | T4NX | ○ | mut (245) | ○ |
| SCC105 | Prior SCC | FOM | 2 | T2N0 | ○ | mut (155) | ○ |
| SCC111 | NP | FOM | 3 | T1N1 | ○ | WT | ○ |
| SCC114 | Prior SCC (2) | FOM | 2 | T2NX | ○ | mut (248) | ○ |
| SCC116 | NP | ALV RIDGE | 2 | T2N0 | ○ | WT | ○ |
| SCC121 | NP | RMT | 3 | T1N0 | ○ | WT | ○ |
| SCC122 | NP | TONG | 3 | T1N1 | ○ | WT | ○ |
| SCC125 | NP | ALV RIDGE | 2 | T4N2B | ○ | WT | ○ |
| SCC131 | NP | FOM | 1 | T2N2 | • | WT | ○ |
| SCC136 | NP | FOM/RMT | 2 | T3N2 | • | WT | ○ |
| SCC142 | R | FOM/MAND | 2 | T4NX | ○ | WT | ○ |
| SCC143 | R | FOM | 3 | T4N2 | ○ | WT | ○ |
| SCC152 | R | HYPO | 2 | - | ○ | WT | • |
| SCC153 | NP | ALV RIDGE | 2 | T2N1 | • | - | • |
| SCC154 | NP | TONG | 3 | T4N2 | ○ | WT | • |
| SCC172 | R | MAND | 3 | - | ○ | - | ○ |
| SCC182 | NP | RMT | 2 | T2N1 | ○ | - | ○ |
| SCC192 | NP | FOM | 1 | T4N0 | • | - | ○ |
| SCC200 | NP | BOT/TONS | 1 | T2N2B | ○ | - | • |
NP = new primary, R = recurrence, Prior SCC indicates this was not the initial primary (for SCC114 there were 2 previous primaries).
1 = low (well differentiated), 2 = moderate (moderately differentiated), 3 = high (poorly differentiated)
Filled circle indicates 11q13 amplification in the tumor.
If TP53 is mutant, the codon which contains the mutation is specified.
Filled circle indicates HPV-positivity in the tumor.
UPCI:SCC030 and 036 are spindle cell SCCs, UPCI:SCC051 is an adenoid SCC, UPCI:SCC080 is a basaloid SCC.
Tissue culture
Cell culture was carried out on all 199 tumor specimens from the 185 patients. Of the total, 52 (26.1%) were established as cell lines, 101 (51.8%) grew only fibroblasts or were overgrown by fibroblasts, 27 (13.6%) had no cell growth or did not thrive in culture, and 19 (9.5%) became contaminated. Thirty-six of the 52 cell lines (69%) were established from the pool of 127 new primary tumors. Of the remaining 16 cell lines, 13 were derived from recurrences and three came from patients who had a primary tumor prior to entry into the study (Table 3). The successful establishment of a cell line was shown to vary by site of origin of the tumor (Table 4). Alveolar ridge specimens were most successful in producing cell lines (7/11; 64%); soft palate samples were the least successful (0/13; 0%). Oral cavity tumors accounted for 142 of the 199 HNSCC specimens collected, and also 41 of the 52 HNSCC cell lines (78.8%) that were established (Table 4).
Table 3.
Success rate of HNSCC cell line establishment by source of tumor specimens
| Source | No. of specimens | Cell lines established | % Success | % of Total |
|---|---|---|---|---|
| Primary Tumor | 152 | 39 | 25.7 | 75.0 |
| Recurrence | 44 | 13 | 29.6 | 25.0 |
| Metastasis | 3 |
0 |
0 |
0.0 |
| 199 | 52 | 26.1 |
Table 4.
Success rate of HNSCC cell line establishment by tumor site
| Site | No. of specimens | Cell lines established | % Success | % of Total |
|---|---|---|---|---|
| Base of tongue (BOT) | 12 | 3 | 25 | 5.7 |
| Tongue | 46 | 11 | 23.9 | 21.2 |
| Alveolar ridge (ALV) | 11 | 7 | 63.6 | 13.5 |
| Floor of mouth (FOM) | 33 | 13 | 39.4 | 25 |
| Soft Palate | 13 | 0 | 0 | 0 |
| Buccal mucosa | 17 | 1 | 5.9 | 1.9 |
| Retromolar trigone (RMT) | 26 | 7 | 26.9 | 13.5 |
| Tonsil | 19 | 4 | 21.1 | 7.7 |
| Other | 22 |
6 |
27.3 |
11.5 |
| 199 | 52 | 26.1 | ||
| Oral Cavity | 142 | 41 | 28.9 | 78.8 |
| Oropharynx/Hypopharynx | 48 | 9 | 18.8 | 17.3 |
| Other | 9 |
2 |
22.2 |
3.9 |
| 199 | 52 | 26.1 |
Immunostaining
Thirty-five of the 52 cell lines were completely analyzed by immunostaining, and 16 of these cell lines were positive for all cytokeratins, while five cell lines were negative for all cytokeratins (see Table 5). All of the antibodies tested were made against either a specific cytokeratin or to a panel of cytokeratins. Cytokeratins are intermediate filaments that comprise part of the cytoskeleton, and are characteristic of normal epithelial tissue. There are 20 human cytokeratins that are typed by their molecular weights and isoelectric points. Anti-keratin AE1 recognizes type I keratins, which are members of the acidic subfamily, and AE3 recognizes type II keratins, which are a basic subclass of cytokeratins. Both AE1 and AE3 have been used in previous studies to characterize normal as well as malignant epithelial cells. Since they stain for a wide range of keratins, positive results indicate that the cells are epithelial. In the cell lines tested, 30/35 were positive for both AE1 and AE3. CAM 5.2 tests for cytokeratins 8 and 18, both primary keratins of simple epithelia not normally found in normal stratified epithelium of the oral cavity. Therefore, a positive result indicates that our cultured cells are not normal. Again, 30/35 of our HNSCC cell lines were positive. Cytokeratin 19 is found in the basal layer of some normal squamous cells, but its expression outside the basilar layer is indicative of hyperproliferation (malignant cells). A group of 14 cell lines stained positively for all cytokeratins other than CK19, indicating that although hyperproliferation beyond the basal layer was not detected, the cells were clearly abnormal. Normal fibroblasts did not stain positively for any of the antibodies tested, and thereby served as a negative control.
Table 5.
Immunohistochemical characteristics of the HNSCC cell lines
| Subject ID | UPCI: | CK 19a | keratin IHC AE3a | keratin IHC AE1a | CAM 5.2a |
|---|---|---|---|---|---|
| 70 | SCC070 | • | • | • | • |
| 72 | SCC072 | • | • | • | • |
| 74 | SCC074 | • | • | • | • |
| 78 | SCC078 | • | • | • | • |
| 89 | SCC089 | • | • | • | • |
| 90 | SCC090 | • | • | • | • |
| 103 | SCC103 | • | • | • | • |
| 36b | SCC104 | • | • | • | • |
| 105 | SCC105 | • | • | • | • |
| 111 | SCC111 | • | • | • | • |
| 114 | SCC114 | • | • | • | • |
| 116 | SCC116 | • | • | • | • |
| 121 | SCC121 | • | • | • | • |
| 125 | SCC125 | • | • | • | • |
| 131 | SCC131 | • | • | • | • |
| 136 | SCC136 | • | • | • | • |
| 36 | SCC036 | ○ | • | • | • |
| 40 | SCC040 | ○ | • | • | • |
| 56 | SCC056 | ○ | • | • | • |
| 62 | SCC062 | ○ | • | • | • |
| 65 | SCC065 | ○ | • | • | • |
| 66 | SCC066 | ○ | • | • | • |
| 68 | SCC068 | ○ | • | • | • |
| 75 | SCC075 | ○ | • | • | • |
| 77 | SCC077 | ○ | • | • | • |
| 80 | SCC080 | ○ | • | • | • |
| 81 | SCC081 | ○ | • | • | • |
| 84 | SCC084 | ○ | • | • | • |
| 60 | SCC099 | ○ | • | • | • |
| 122 | SCC122 | ○ | • | • | • |
| 32 | SCC032 | ○ | ○ | ○ | ○ |
| 44 | SCC044 | ○ | ○ | ○ | ○ |
| 45 | SCC045 | ○ | ○ | ○ | ○ |
| 51 | SCC051 | ○ | ○ | ○ | ○ |
| 52 | SCC052 | ○ | ○ | ○ | ○ |
*Filled circle indicates positivity for given antibody.
Subject ID appears multiple times in the table.
Summary of Other Analyses
Amplification of chromosomal band 11q13 in the tumor (p<0.05), involvement of local and/or regional lymph nodes (p<0.01), and features of alcohol use (cumulative (lifetime) exposure and cessation of use, both p<0.05) were the variables related to the tumor that were statistically significantly associated with cell line formation (Table 6). The amplification of 11q13 was positively associated with cell line formation, and 24 of 61 (39.3%) tumors amplified at 11q13 formed cell lines, but only 28 of 126 (22.2%) tumors not amplified at 11q13 were successful in culture (results unknown or missing for 12). Similarly, tumors that had spread to lymph nodes were successful in culture (30/76; 39.5%), but tumors that did not spread to lymph nodes did not succeed in culture (12/73; 16.4%) (data unknown or missing for 50). The patient's cumulative exposure to alcohol was associated with cell line formation (p<0.05; 39 cell lines from long-term or heavy users) as was failure to cease drinking alcohol (p<0.05; 36 cell lines from patients that continued to use alcohol). Similar associations were not found for tobacco use, exposure or cessation patterns.
Table 6.
Variables associated with formation of a cell line
| Variable | Cell Linea | p-valueb | Unadjusted OR (95% CI) | Adjusted OR (95% CI)c | |||
|---|---|---|---|---|---|---|---|
| Y | N | - | |||||
| 199 | 52 | 128 | 19 | ||||
| p53 status | WT | 35 | 74 | 8 | 0.637 | 1.0 | |
| mut | 9 | 15 | 2 | 1.27 (0.50 − 3.19) | |||
| - | 8 | 39 | 9 | ||||
| HPV status | negative | 44 | 88 | 15 | 0.058 | 1.0 | 1.0 |
| positive | 8 | 37 | 4 | 0.43 (0.19 − 1.01) | 0.29 (0.11 − 0.77) | ||
| - | 0 | 3 | 0 | ||||
| AJCC stage (grouped) | I & II | 15 | 46 | 7 | 0.154 | 1.0 | |
| III & IV | 33 | 57 | 7 | 1.78 (0.84 − 3.77) | |||
| - | 4 | 25 | 5 | ||||
| Age group | under 55 | 14 | 34 | 2 | 0.765 | 1.0 | |
| 55−64 | 18 | 38 | 4 | 1.15 (0.47 − 2.82) | |||
| 65−74 | 12 | 39 | 11 | 0.75 (0.30 − 1.88) | |||
| over 75 | 8 | 17 | 2 | 1.14 (0.37 − 3.51) | |||
| 11q13 amp | negative | 28 | 85 | 13 | 0.021 | 1.0 | 1.0 |
| positive | 24 | 32 | 5 | 2.28 (1.15 − 4.51) | 2.14 (1.02 − 4.48) | ||
| - | 0 | 11 | 1 | ||||
| Tissue margin | negative | 44 | 104 | 17 | 1.0 | 1.0 | |
| positive | 6 | 15 | 1 | 0.95 (0.34 − 2.62) | |||
| - | 2 | 9 | 1 | ||||
| Nodal Involvement | negative | 12 | 54 | 7 | 0.005 | 1.0 | 1.0 |
| positive | 30 | 43 | 3 | 3.14 (1.43 − 6.87) | 2.77 (1.06 − 7.22) | ||
| - | 10 | 31 | 9 | ||||
| Extracapsular spread | negative | 11 | 23 | 2 | 0.488 | 1.0 | |
| positive | 20 | 27 | 1 | 1.55 (0.62 − 3.85) | |||
| - | 21 | 78 | 16 | ||||
| Perineural involvement | negative | 16 | 45 | 10 | 0.436 | 1.0 | |
| positive | 21 | 42 | 2 | 1.41 (0.64 − 3.10) | |||
| - | 15 | 41 | 7 | ||||
| Differentiation/Grade | well | 12 | 25 | 6 | 0.678 | 1.0 | |
| mod | 28 | 69 | 8 | 0.85 (0.35 − 2.02) | |||
| poor | 10 | 17 | 0 | 1.23 (0.42 − 3.62) | |||
| - | 2 | 17 | 5 | ||||
| Tobacco use (smoking) | Never | 6 | 18 | 2 | 0.81 | ||
| Ever | 46 | 110 | 17 | ||||
| Alcohol use | Never | 4 | 24 | 5 | 0.072 | 1.0 | 1.0 |
| Ever | 48 | 104 | 14 | 2.77 (0.90 − 8.51) | 2.14 (0.66 − 6.93) | ||
| Cumulative alcohol used | None | 4 | 24 | 5 | 0.027 | 1.0 | 1.0 |
| <30 alc yrs | 7 | 29 | 4 | 1.74 (0.46 − 6.63) | 1.81 (0.46 − 7.12) | ||
| >30 alc yrs | 39 | 65 | 9 | 3.60 (1.15 − 11.27) | 2.43 (0.74 − 7.99) | ||
| - | 2 | 10 | 1 | ||||
| Quit alcohol?e | Never | 4 | 24 | 5 | 0.042 | 1.0 | 1.0 |
| Yes | 8 | 24 | 2 | 2.00 (0.53 − 7.57) | 1.40 (0.33 − 5.92) | ||
| No | 36 | 60 | 9 | 3.66 (1.16 − 11.58) | 2.85 (0.84 − 9.62) | ||
| - | 4 | 21 | 3 | ||||
| Family history (cancer) | No | 28 | 55 | 8 | 0.192 | ||
| Yes | 24 | 73 | 11 | ||||
In the dependent variable (cell line formation), a dashed line indicates cultures that were contaminated and subsequently lost from analysis.
P-values are from Fisher's exact test.
Odds ratios are adjusted for age (age group) and stage (grouped stage).
Cumulative use determined by multiplying the drink equivalent per day (1 shot = I beer = 1 glass wine) by years of alcohol use. Missing if quantity or duration was omitted on questionnaire.
Dependent upon patient reporting cessation of alcohol use prior to treatment for HNSCC and also providing a date of cessation.
The presence of HPV was associated with a “protective effect,” since 44/147 (29.9%) HPV-negative tumors formed cell lines, whereas only 8/49 (16.3%) HPV-positive tumors formed cell lines (status unknown for three). Tumor stage was not significantly associated with cell line formation by the Fisher's exact test, but we anticipate confounding with regard to this variable since recurrences were not routinely staged by the Pathologist, resulting in non-random missing data. A test of proportions was used to confirm the expected relationship between tumor stage and cell line formation, wherein late stage (III and IV) tumors were more aggressive and therefore, more likely to thrive in culture (borderline significance, p=0.12). The mean age at diagnosis for patients whose HNSCC grew into a cell line (60.6 yrs) was not significantly different from the age at diagnosis for those patients whose tumors did not yield a cell line (61.5 yrs).
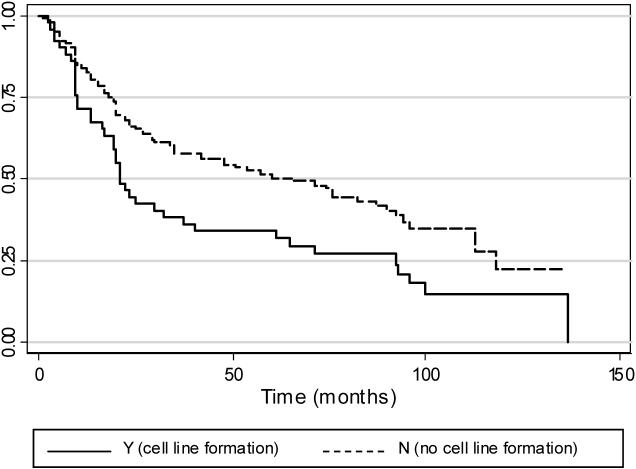
Among all 185 patients, median observed survival was 48 months; five-year observed survival was 47.1% and ten-year observed survival was 20.6%. The median observed survival time, defined as the length of time from a patient's initial presentation in the clinic with an HNSCC until they either died or otherwise exited the study (censored, lost to follow-up), was 22 months if the tumor yielded a cell line and 60 months if it did not (log-rank test for equality of survivor functions; p<0.05). This survival function, dependent on successful establishment of a cell line from the HNSCC specimen, is shown in Figure 1.
Figure 1.
Kaplan-Meier estimates of observed survival in 185 patients presenting with HNSCC, based on cell line formation. Failure is defined as death, and for patients with multiple HNSCC, the date of the first tumor is used to estimate survival time. The median observed survival time, defined as the length of time from the patient's presentation in the clinic with an HNSCC until they either die or otherwise exit the study (censored, lost to follow-up), is 22 months if the tumor yields a cell line and 60 months if it does not (log-rank test for equality of survivor functions; p<0.05).
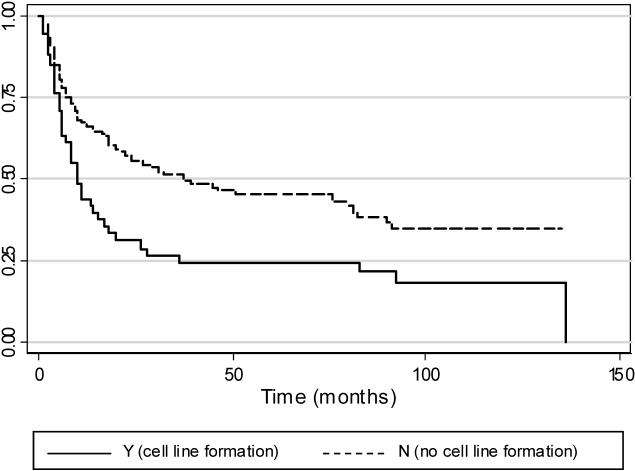
Median disease-free survival was 24 months, five-year disease-free survival was 40.0%, and ten-year disease-free survival was 32.3%. Failure is defined as the occurrence of a new primary, recurrence, metastasis or death. The median disease-free survival time for an individual presenting with an HNSCC that yielded a cell line was 10 months, and the mean time for an HNSCC that did not yield a cell line was 37 months (log-rank test for equality of survivor functions; p<0.05). This survival function, dependent on successful establishment of a cell line from the HNSCC specimen, is shown in Figure 2.
Figure 2.
Kaplan-Meier estimates of disease-free survival for 185 patients presenting with HNSCC, based on cell line formation. Failure is defined as the occurrence of a new primary, recurrence, metastasis or death, and includes any subsequent tumors (HNSCC or otherwise) or death. For patients with multiple HNSCC, the date of the first tumor is used to estimate survival time. The median disease-free survival time for an individual presenting with an HNSCC that yielded a cell line is 10 months, and the mean time for an HNSCC that did not yield a cell line is 37 months (log-rank test for equality of survivor functions; p<0.05).
Discussion
The 52 HNSCC cell lines that we established, with an overall success rate of 26.1% are attributable to 48 patients diagnosed with head and neck squamous cell carcinoma between September 1992 and February 1997. Three patients had multiple HNSCC, including one with three HNSCC during the observed time period. Our overall success rate for cell line establishment is comparable to others who have attempted to culture SCC cell lines. Heo et al. observed an overall success rate of 24%.31 Many of the tumors and cell lines have been examined for human papillomavirus (HPV) infection, TP53 mutation status, and 11q13 gene amplification, and the majority of cell lines have been karyotyped.21,24-27
We tested a range of variables that have been shown previously or were suspected to have prognostic significance, and found a statistically significant association between 11q13 amplification, nodal status, and features of alcohol consumption and the establishment of cell lines (Table 6). Our findings suggest that more aggressive tumors are more likely to adhere and divide under idealized and controlled culture conditions. The variable most strongly associated with tumors that yield a cell line is the presence of positive lymph nodes (adjusted-OR 2.82, 1.01 − 7.86). The involvement of local and regional lymph nodes is known to be one of the most significant clinical indicators of poor prognosis. Amplification of chromosomal band 11q13 (adjusted-OR 2.21, 1.07 − 4.57) is also a significant predictor of cell line formation, and 11q13 amplification has long served as a cytogenetic and molecular marker of poor prognosis.2,11 Considerable research has been devoted to 11q13 in our lab and in others, and it is increasingly clear that CCND1, the gene for cyclin D1 found within this amplicon, is not the only critical gene driving 11q13 amplification. However, CCND1 overexpression has been shown to reduce dependence on normal physiological growth stimuli and promote hyperproliferation even in growth factor-deprived conditions, providing cells with a selective advantage through dysregulation of the cell cycle.32-34 It is not clear whether these oncogenic effects are mediated through enhanced proliferative capacity or if CCND1 also contributes to cellular differentiation in concert with other cell cycle-related genes and pathways.35-38 Further, TP53 mutation or loss and overexpression of CCND1 are independently sufficient to abrogate normal cell cycle control, perhaps explaining why we did not observe an association between p53 status and cell line formation in our study.39
A test of trend showed observed survival (time from study entry to failure or study conclusion) is associated with cell line formation (p<0.05; Figure 1). A similar test of trend in disease-free survival (time from study entry to death or study conclusion) was also significant (p<0.05; Figure 2). While it seems that knowing which variables tend to assort with cell line formation has limited predictive value in the clinic, we find that those same variables are hallmarks of aggressive, late-stage tumors and poor prognosis for the patient.
Tumors positive for HPV were less likely to form a cell line (adjusted-OR 0.31, 0.11 − 0.82). We and others have shown that HPV infection associates with less aggressive tumors that have a better prognosis.2,16,21,40,41 Rose Ragin et al. showed the frequency of 11q13 amplification stratified by tumor site and HPV status.21 Overall, they detected a lower prevalence of 11q13 amplification in the HPV-positive tumors than in the HPV-negative group. A higher proportion of HPV-positive tumors were seen in the oropharynx compared to the oral cavity by Rose Ragin et al., suggesting that the higher frequency of cell line formation in the oral cavity compared to the oropharynx may be due to the site preference of HPV infection. In our study, tumors from the oral cavity accounted for 41 of the 52 HNSCC cell lines (78.8%) that were established (Table 4). In contrast, oropharynx tumors contributed only nine cell lines (17.3%). It should also be noted that 41 of the 142 oral cavity specimens were successful in culture (28.9% success), whereas only nine of the 48 oropharynx specimens were successful in culture (18.8% success). The success rate of cell line formation according to tumor site was included primarily to illustrate the range of sites within the oral cavity from which our cell lines were derived, but it also provides the opportunity to question why certain sites were more successful than others. It is most likely a combination of factors, including, but not limited to, HPV infection, tissue type, tumor stage, p53 status, other genetic alterations in the tumor, and exogenous exposures. Additional research is necessary to evaluate the effects of these combinations of variables. It is also conceivable that this observation resulted from difficulties in isolating cell lines from the various soft tissues in these sites (dissociation, manipulation, contaminating normal tissue, etc.), although human error is probably not sufficient to explain the observed relationship between tumor site and success in culture.
Tobacco use was not found to be significantly associated in our analysis, possibly because of the intricacies in accounting for exposure levels based on type of tobacco used (cigarettes, cigars, pipes, snuff and chewing tobacco). In addition to the type of exposure, the “strength” of a given exposure may be brand or product-related. Another consideration is that the majority of patients that yielded a cell line were smokers (only five non-smokers yielding six cell lines, and only 23 of the 185 patients with no exposure history), making it difficult to test for significance in this variable. A similar problem was encountered for alcohol exposure, wherein only four cell lines came from patients with no exposure history, and only 32 of the 185 patients had no exposure history. In spite of this, alcohol use was of borderline significance (p<0.1) in our analyses, and alcohol use (adjusted-OR 2.14, 0.66 − 6.93), lifetime exposure (adjusted-OR 2.43, 0.74 − 7.99) and failure to stop using alcohol (adjusted-OR 2.85, 0.84 − 9.62) appeared to be associated with cell line formation. The significance of either exposure variable is likely obscured by the preponderance of patients having reported use of one or both of these agents prior to HNSCC diagnosis. It should also be noted that 46% of the patients whose tumors led to cell lines had a family history of cancer. It is difficult to distinguish the genetic contribution of a cancer-predisposing syndrome or even an aberrant allele, given that the appropriate gene is affected, from a behavior such as smoking, and to some extent alcohol use, as these behaviors tend to have ‘inheritance’ patterns of their own. The interaction between a pre-existing genetic alteration, such as a defect in the DNA damage response, and tobacco and/or alcohol exposure would be expected to compound the impact on cell growth pathways, resulting in more aggressive tumors. These effects are difficult to quantify and/or predict at this time.
Less than half (77/185; 41.6%) of the enrolled patients were diagnosed with a single tumor (HNSCC or otherwise) during the observed time period (before failure or conclusion of the study), and this same group yielded only 12 of the 52 HNSCC cell lines (15.6% success rate). This observation is subject to bias due to the open enrollment study design, but may have importance with regard to etiology of disease. The relatively small difference in percentages between five- and 10-year disease-free survival (40.0% and 32.3%, respectively) in all 185 patients, indicate that most individuals that were disease-free at five years remained disease-free after another five years (overall disease-free survival; data not shown). This suggests an underlying difference in disease features between individuals who develop multiple tumors (HNSCC or otherwise) and those with only a single tumor. There has been some discussion in the literature regarding second neoplasms in patients presenting with HNSCC, which has led to the conclusion that preventative treatment would lower the rate of second tumors.42-45 This has also driven speculation that there may be a mechanism to identify patients that are at an elevated risk to develop multiple malignancies. Although our study was not designed appropriately to answer this question, the results are certainly supportive of differential HNSCC pathogenesis and development.
The establishment of 52 HNSCC cell lines has value for both basic and translational research. Our UPCI:SCC collection represents one of the larger such resources, and complements available series of HNSCC cell lines, including the UM-SCC, SCC-, PCI-, BICR-, and UT-SCC series, among others.31,46-51 As evidence of their utility, many papers have already been published utilizing a subset of our available UPCI:SCC cell lines.28,52-68 The establishment of cell lines for research purposes is a worthwhile process, but is biased towards more aggressive tumors, which are likely the result of interactions between multiple relevant exposures and consequent genetic alterations. Hence, it is critical that as much information as possible is documented about the tumor and the patient from which a cell line was derived, especially if it is to be used for studies that have translational significance.
Acknowledgements
This work was supported by generous grants from the National Institutes of Health (NIH) (R01DE10513, R01DE12008, R01DE14729 to SMG) and the American Cancer Society (EDT-44 to SMG). JSW and CCRR are supported by the UPCI Cancer Education and Career Development grant (CECD; R25CA089507) to William S. Bigbee. Molecular cytogenetic analyses were carried out in the UPCI Cytogenetics Facility, supported in part by NIH grant P30CA47904 to Ronald B. Herberman. Some of the studies were carried out in the context of the Oral Cancer Center at the University of Pittsburgh (P60DE13059 to ENM). The authors are grateful to Dr. Jennifer Grandis for scientific leadership, Dr. William S. Saunders for stimulating discussions and Dr. Roslyn Stone for sharing her expertise in statistical analysis. The authors are grateful to Ms. Jaya Reddy and Seia Comsa for growing and caring for many of these cell cultures, Ms. Robin Wagner and Ms. Jennifer Ridge-Hetrick for clinical, demographic and follow-up information, and to Dr. Billy Appel for assistance with the histopathology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest None of the authors have any financial or personal relationships with other people or organizations that could inappropriately influence or bias their contribution to this paper.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56(2):106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Forastiere A, Koch W, Trotti A, Sidransky D. Head and neck cancer. N Engl J Med. 2001;345(26):1890–900. doi: 10.1056/NEJMra001375. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, et al. Cancer statistics, 2005. CA Cancer J Clin. 2005;55(1):10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 4.Kim MM, Califano JA. Molecular pathology of head-and-neck cancer. Int J Cancer. 2004;112(4):545–53. doi: 10.1002/ijc.20379. [DOI] [PubMed] [Google Scholar]
- 5.Ries LAG, Eisner MP, Kosary CL, Hankey BF, Miller BA, Clegg L, et al. SEER Cancer Statistics Review, 1975−2002. 2005 http://seer.cancer.gov/csr/1975_2002 SEER website.
- 6.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56(11):2488–92. [PubMed] [Google Scholar]
- 7.Schwartz GJ, Mehta RH, Wenig BL, Shaligram C, Portugal LG. Salvage treatment for recurrent squamous cell carcinoma of the oral cavity. Head Neck. 2000;22(1):34–41. doi: 10.1002/(sici)1097-0347(200001)22:1<34::aid-hed6>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 8.Xi S, Grandis JR. Gene therapy for the treatment of oral squamous cell carcinoma. J Dent Res. 2003;82(1):11–6. doi: 10.1177/154405910308200104. [DOI] [PubMed] [Google Scholar]
- 9.Califano J, Westra WH, Meininger G, Corio R, Koch WM, Sidransky D. Genetic progression and clonal relationship of recurrent premalignant head and neck lesions. Clin Cancer Res. 2000;6(2):347–52. [PubMed] [Google Scholar]
- 10.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6(5):963–8. doi: 10.1002/1097-0142(195309)6:5<963::aid-cncr2820060515>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 11.Gollin SM. Chromosomal alterations in squamous cell carcinomas of the head and neck: window to the biology of disease. Head Neck. 2001;23(3):238–53. doi: 10.1002/1097-0347(200103)23:3<238::aid-hed1025>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 12.Reshmi SC, Gollin SM. Chromosomal instability in oral cancer cells. J Dent Res. 2005;84(2):107–17. doi: 10.1177/154405910508400203. [DOI] [PubMed] [Google Scholar]
- 13.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3(10):733–44. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 14.Hunter KD, Parkinson EK, Harrison PR. Profiling early head and neck cancer. Nat Rev Cancer. 2005;5(2):127–35. doi: 10.1038/nrc1549. [DOI] [PubMed] [Google Scholar]
- 15.Gillison ML, Koch WM, Shah KV. Human papillomavirus in head and neck squamous cell carcinoma: are some head and neck cancers a sexually transmitted disease? Curr Opin Oncol. 1999;11(3):191–9. doi: 10.1097/00001622-199905000-00010. [DOI] [PubMed] [Google Scholar]
- 16.Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–20. doi: 10.1093/jnci/92.9.709. [DOI] [PubMed] [Google Scholar]
- 17.Herrero R, Castellsague X, Pawlita M, Lissowska J, Kee F, Balaram P, et al. Human papillomavirus and oral cancer: the International Agency for Research on Cancer multicenter study. J Natl Cancer Inst. 2003;95(23):1772–83. doi: 10.1093/jnci/djg107. [DOI] [PubMed] [Google Scholar]
- 18.Koch WM, Lango M, Sewell D, Zahurak M, Sidransky D. Head and neck cancer in nonsmokers: a distinct clinical and molecular entity. Laryngoscope. 1999;109(10):1544–51. doi: 10.1097/00005537-199910000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Mork J, Lie AK, Glattre E, Hallmans G, Jellum E, Koskela P, et al. Human papillomavirus infection as a risk factor for squamous-cell carcinoma of the head and neck. N Engl J Med. 2001;344(15):1125–31. doi: 10.1056/NEJM200104123441503. [DOI] [PubMed] [Google Scholar]
- 20.Braakhuis BJ, Snijders PJ, Keune WJ, Meijer CJ, Ruijter-Schippers HJ, Leemans CR, et al. Genetic patterns in head and neck cancers that contain or lack transcriptionally active human papillomavirus. J Natl Cancer Inst. 2004;96(13):998–1006. doi: 10.1093/jnci/djh183. [DOI] [PubMed] [Google Scholar]
- 21.Ragin CCR, Taioli E, Weissfeld JL, White JS, Rossie KM, Modugno F, Gollin SM. 11q13 amplification status and HPV in relation to p16 expression defines two distinct etiologies of head and neck tumors. Brit J Cancer. 2006 doi: 10.1038/sj.bjc.6603394. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, et al. High incidence of head and neck squamous cell carcinoma in patients with Fanconi anemia. Arch Otolaryngol Head Neck Surg. 2003;129(1):106–12. doi: 10.1001/archotol.129.1.106. [DOI] [PubMed] [Google Scholar]
- 23.Worsham MJ, Wolman SR, Carey TE, Zarbo RJ, Benninger MS, Van Dyke DL. Chromosomal aberrations identified in culture of squamous carcinomas are confirmed by fluorescence in situ hybridisation. Mol Pathol. 1999;52(1):42–6. doi: 10.1136/mp.52.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lese CM, Rossie KM, Appel BN, Reddy JK, Johnson JT, Myers EN, et al. Visualization of INT2 and HST1 amplification in oral squamous cell carcinomas. Genes Chromosomes Cancer. 1995;12(4):288–95. doi: 10.1002/gcc.2870120409. [DOI] [PubMed] [Google Scholar]
- 25.Lese CM, Reshmi SC, Ried T, Gottberg W, Wilson JW, Reddy JK, et al. Genomic imbalances and patient outcome in oral squamous cell carcinoma. manuscript in preparation.
- 26.Law JC, Whiteside TL, Gollin SM, Weissfeld J, El Ashmawy L, Srivastava S, et al. Variation of p53 mutational spectra between carcinoma of the upper and lower respiratory tract. Clin Cancer Res. 1995;1(7):763–8. [PubMed] [Google Scholar]
- 27.Ishwad CS, Shuster M, Bockmuhl U, Thakker N, Shah P, Toomes C, et al. Frequent allelic loss and homozygous deletion in chromosome band 8p23 in oral cancer. Int J Cancer. 1999;80(1):25–31. doi: 10.1002/(sici)1097-0215(19990105)80:1<25::aid-ijc6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 28.Telmer CA, An J, Malehorn DE, Zeng X, Gollin SM, Ishwad CS, et al. Detection and assignment of TP53 mutations in tumor DNA using peptide mass signature genotyping. Hum Mutat. 2003;22(2):158–65. doi: 10.1002/humu.10248. [DOI] [PubMed] [Google Scholar]
- 29.Fogh J. Human tumor lines for cancer research. Cancer Invest. 1986;4(2):157–84. doi: 10.3109/07357908609038260. [DOI] [PubMed] [Google Scholar]
- 30.Carey TE. Head and Neck Tumor Cell Lines. 1994:79–120. [Google Scholar]
- 31.Heo DS, Snyderman C, Gollin SM, Pan S, Walker E, Deka R, et al. Biology, cytogenetics, and sensitivity to immunological effector cells of new head and neck squamous cell carcinoma lines. Cancer Res. 1989;49(18):5167–75. [PubMed] [Google Scholar]
- 32.Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci U S A. 1994;91(2):709–13. doi: 10.1073/pnas.91.2.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musgrove EA, Lee CS, Buckley MF, Sutherland RL. Cyclin D1 induction in breast cancer cells shortens G1 and is sufficient for cells arrested in G1 to complete the cell cycle. Proc Natl Acad Sci U S A. 1994;91(17):8022–6. doi: 10.1073/pnas.91.17.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwijsen RM, Klompmaker R, Wientjens EB, Kristel PM, van der Burg B, Michalides RJ. Cyclin D1 triggers autonomous growth of breast cancer cells by governing cell cycle exit. Mol Cell Biol. 1996;16(6):2554–60. doi: 10.1128/mcb.16.6.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang W, Kahn SM, Zhou P, Zhang YJ, Cacace AM, Infante AS, et al. Overexpression of cyclin D1 in rat fibroblasts causes abnormalities in growth control, cell cycle progression and gene expression. Oncogene. 1993;8(12):3447–57. [PubMed] [Google Scholar]
- 36.Bartkova J, Lukas J, Muller H, Strauss M, Gusterson B, Bartek J. Abnormal patterns of D-type cyclin expression and G1 regulation in human head and neck cancer. Cancer Res. 1995;55(4):949–56. [PubMed] [Google Scholar]
- 37.Robles AI, Larcher F, Whalin RB, Murillas R, Richie E, Gimenez-Conti IB, et al. Expression of cyclin D1 in epithelial tissues of transgenic mice results in epidermal hyperproliferation and severe thymic hyperplasia. Proc Natl Acad Sci U S A. 1996;93(15):7634–8. doi: 10.1073/pnas.93.15.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Jong JS, van Diest PJ, Michalides RJ, Baak JP. Concerted overexpression of the genes encoding p21 and cyclin D1 is associated with growth inhibition and differentiation in various carcinomas. Mol Pathol. 1999;52(2):78–83. doi: 10.1136/mp.52.2.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu L, Davidson BJ, Murty VV, Li RG, Sacks PG, Garin-Chesa P, et al. TP53 gene mutations and CCND1 gene amplification in head and neck squamous cell carcinoma cell lines. Int J Cancer. 1994;59(3):383–7. doi: 10.1002/ijc.2910590316. [DOI] [PubMed] [Google Scholar]
- 40.Ritchie JM, Smith EM, Summersgill KF, Hoffman HT, Wang D, Klussmann JP, et al. Human papillomavirus infection as a prognostic factor in carcinomas of the oral cavity and oropharynx. Int J Cancer. 2003;104(3):336–44. doi: 10.1002/ijc.10960. [DOI] [PubMed] [Google Scholar]
- 41.Schlecht NF. Prognostic value of human papillomavirus in the survival of head and neck cancer patients: an overview of the evidence. Oncol Rep. 2005;14(5):1239–47. [PubMed] [Google Scholar]
- 42.Cooper JS, Pajak TF, Rubin P, Tupchong L, Brady LW, Leibel SA, et al. Second malignancies in patients who have head and neck cancer: incidence, effect on survival and implications based on the RTOG experience. Int J Radiat Oncol Biol Phys. 1989;17(3):449–56. doi: 10.1016/0360-3016(89)90094-1. [DOI] [PubMed] [Google Scholar]
- 43.Sturgis EM, Miller RH. Second primary malignancies in the head and neck cancer patient. Ann Otol Rhinol Laryngol. 1995;104(12):946–54. doi: 10.1177/000348949510401206. [DOI] [PubMed] [Google Scholar]
- 44.Bhattacharyya N, Nayak VK. Survival outcomes for second primary head and neck cancer: a matched analysis. Otolaryngol Head Neck Surg. 2005;132(1):63–8. doi: 10.1016/j.otohns.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 45.Leon X, Quer M, Diez S, Orus C, Lopez-Pousa A, Burgues J. Second neoplasm in patients with head and neck cancer. Head Neck. 1999;21(3):204–10. doi: 10.1002/(sici)1097-0347(199905)21:3<204::aid-hed4>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 46.Carey TE, Kimmel KA, Schwartz DR, Richter DE, Baker SR, Krause CJ. Antibodies to human squamous cell carcinoma. Otolaryngol Head Neck Surg. 1983;91(5):482–91. doi: 10.1177/019459988309100503. [DOI] [PubMed] [Google Scholar]
- 47.Buchhagen DL, Worsham MJ, Dyke DL, Carey TE. Two regions of homozygosity on chromosome 3p in squamous cell carcinoma of the head and neck: comparison with cytogenetic analysis. Head Neck. 1996;18(6):529–37. doi: 10.1002/(SICI)1097-0347(199611/12)18:6<529::AID-HED7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 48.Weichselbaum RR, Dahlberg W, Beckett M, Karrison T, Miller D, Clark J, et al. Radiation-resistant and repair-proficient human tumor cells may be associated with radiotherapy failure in head- and neck-cancer patients. Proc Natl Acad Sci U S A. 1986;83(8):2684–8. doi: 10.1073/pnas.83.8.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prime SS, Nixon SV, Crane IJ, Stone A, Matthews JB, Maitland NJ, et al. The behaviour of human oral squamous cell carcinoma in cell culture. J Pathol. 1990;160(3):259–69. doi: 10.1002/path.1711600313. [DOI] [PubMed] [Google Scholar]
- 50.Grenman R, Carey TE, McClatchey KD, Wagner JG, Pekkola-Heino K, Schwartz DR, et al. In vitro radiation resistance among cell lines established from patients with squamous cell carcinoma of the head and neck. Cancer. 1991;67(11):2741–7. doi: 10.1002/1097-0142(19910601)67:11<2741::aid-cncr2820671105>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 51.Masters JRW, Palsson B. Cancer Cell Lines Part 2. 1999;II [Google Scholar]
- 52.Virgilio L, Shuster M, Gollin SM, Veronese ML, Ohta M, Huebner K, et al. FHIT gene alterations in head and neck squamous cell carcinomas. Proc Natl Acad Sci U S A. 1996;93(18):9770–5. doi: 10.1073/pnas.93.18.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shuster MI, Han L, Le Beau MM, Davis E, Sawicki M, Lese CM, et al. A consistent pattern of RIN1 rearrangements in oral squamous cell carcinoma cell lines supports a breakage-fusion-bridge cycle model for 11q13 amplification. Genes Chromosomes Cancer. 2000;28(2):153–63. [PubMed] [Google Scholar]
- 54.Saunders WS, Shuster M, Huang X, Gharaibeh B, Enyenihi AH, Petersen I, et al. Chromosomal instability and cytoskeletal defects in oral cancer cells. Proc Natl Acad Sci U S A. 2000;97(1):303–8. doi: 10.1073/pnas.97.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun PC, Uppaluri R, Schmidt AP, Pashia ME, Quant EC, Sunwoo JB, et al. Transcript map of the 8p23 putative tumor suppressor region. Genomics. 2001;75(1−3):17–25. doi: 10.1006/geno.2001.6587. [DOI] [PubMed] [Google Scholar]
- 56.Huang X, Gollin SM, Raja S, Godfrey TE. High-resolution mapping of the 11q13 amplicon and identification of a gene, TAOS1, that is amplified and overexpressed in oral cancer cells. Proc Natl Acad Sci U S A. 2002;99(17):11369–74. doi: 10.1073/pnas.172285799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lango MN, Dyer KF, Lui VW, Gooding WE, Gubish C, Siegfried JM, et al. Gastrin-releasing peptide receptor-mediated autocrine growth in squamous cell carcinoma of the head and neck. J Natl Cancer Inst. 2002;94(5):375–83. doi: 10.1093/jnci/94.5.375. [DOI] [PubMed] [Google Scholar]
- 58.Xi S, Zhang Q, Dyer KF, Lerner EC, Smithgall TE, Gooding WE, et al. Src kinases mediate STAT growth pathways in squamous cell carcinoma of the head and neck. J Biol Chem. 2003;278(34):31574–83. doi: 10.1074/jbc.M303499200. [DOI] [PubMed] [Google Scholar]
- 59.Toomes C, Jackson A, Maguire K, Wood J, Gollin S, Ishwad C, et al. The presence of multiple regions of homozygous deletion at the CSMD1 locus in oral squamous cell carcinoma question the role of CSMD1 in head and neck carcinogenesis. Genes Chromosomes Cancer. 2003;37(2):132–40. doi: 10.1002/gcc.10191. [DOI] [PubMed] [Google Scholar]
- 60.Ragin CC, Reshmi SC, Gollin SM. Mapping and analysis of HPV16 integration sites in a head and neck cancer cell line. Int J Cancer. 2004;110(5):701–9. doi: 10.1002/ijc.20193. [DOI] [PubMed] [Google Scholar]
- 61.Reshmi SC, Saunders WS, Kudla DM, Ragin CR, Gollin SM. Chromosomal instability and marker chromosome evolution in oral squamous cell carcinoma. Genes Chromosomes Cancer. 2004;41(1):38–46. doi: 10.1002/gcc.20064. [DOI] [PubMed] [Google Scholar]
- 62.Hewitt C, Wilson P, McGlinn E, MacFarlane G, Papageorgiou A, Woodwards RT, et al. DLC1 is unlikely to be a primary target for deletions on chromosome arm 8p22 in head and neck squamous cell carcinoma. Cancer Lett. 2004;209(2):207–13. doi: 10.1016/j.canlet.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 63.Reing JE, Gollin SM, Saunders WS. The occurrence of chromosome segregational defects is an intrinsic and heritable property of oral squamous cell carcinoma cell lines. Cancer Genet Cytogenet. 2004;150(1):57–61. doi: 10.1016/j.cancergencyto.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM, Saunders WS. Resolution of anaphase bridges in cancer cells. Chromosoma. 2004;112(8):389–97. doi: 10.1007/s00412-004-0284-6. [DOI] [PubMed] [Google Scholar]
- 65.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307(5706):127–9. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 66.Ferris RL, Martinez I, Sirianni N, Wang J, Lopez-Albaitero A, Gollin SM, et al. Human papillomavirus-16 associated squamous cell carcinoma of the head and neck (SCCHN): a natural disease model provides insights into viral carcinogenesis. Eur J Cancer. 2005;41(5):807–15. doi: 10.1016/j.ejca.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 67.Sirianni N, Wang J, Ferris RL. Antiviral activity of Cidofovir on a naturally human papillomavirus-16 infected squamous cell carcinoma of the head and neck (SCCHN) cell line improves radiation sensitivity. Oral Oncol. 2005;41(4):423–8. doi: 10.1016/j.oraloncology.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 68.Lin CJ, Grandis JR, Carey TE, Gollin SM, Whiteside TL, Koch WM, et al. Head and neck squamous cell carcinoma cell lines: established models and rationale for selection. Head Neck. 2006 doi: 10.1002/hed.20478. In Press. [DOI] [PubMed] [Google Scholar]