Abstract
A class of antibacterials has been discovered that inhibits the growth of Gram-positive pathogenic bacteria. RWJ-49815, a representative of a family of hydrophobic tyramines, in addition to being a potent bactericidal Gram-positive antibacterial, inhibits the autophosphorylation of kinase A of the KinA∷Spo0F two-component signal transduction system in vitro. Analogs of RWJ-49815 vary greatly in their ability to inhibit growth of bacteria and this ability correlates directly with their activity as kinase A inhibitors. Compared with the potent quinolone, ciprofloxacin, RWJ-49815 exhibits reduced resistance emergence in a laboratory passage experiment. Inhibition of the histidine protein kinase∷response regulator two-component signal transduction pathways may present an opportunity to depress chromosomal resistance emergence by targeting multiple proteins with a single inhibitor in a single bacterium. Such inhibitors may represent a class of antibacterials that potentially may represent a breakthrough in antibacterial therapy.
Infectious disease is the number one cause of mortality in the world despite the armamentarium of antibacterial agents developed over the last half century. Bacterial strains have evolved during this time that are recalcitrant to our best efforts to destroy them, and they have begun to compromise the treatment of infectious disease particularly in the hospital setting. Resistance to antibacterials takes many forms and is a probable consequence of widespread use and misuse of antibiotics. Thus, the search for new antibacterials directed toward new targets is not only a continuous process but also, at this time, an urgent necessity.
Discovery of new antibacterial agents has met with limited success in recent years (1) with some noteworthy exceptions still in the early stage of development such as the oxazolidinones (2–5), glycylcyclines (6–8), and the lipid A deacetylase inhibitors (9). Most of these compounds inhibit processes that are known to be essential for bacterial cell viability, e.g., translation or outer membrane assembly. Few attempts have been made to specifically target the mechanisms by which pathogenic bacteria establish an infection within the host. In particular, the inhibition of expression of bacterial virulence factors offers an opportunity for specific intervention at the level of host invasion through biochemical processes, which are clearly unique to the bacterial cell (10).
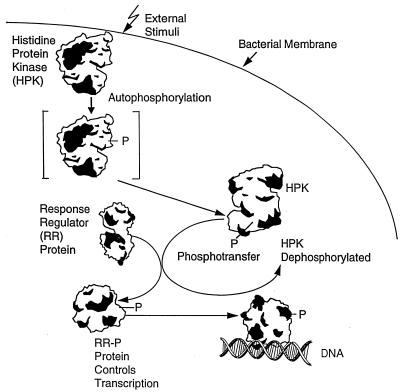
It is now clear that virulence is an adaptive genetic response requiring the induction of genes coding for virulence factors (11–13). This response implies that the infectious agent must be able to sense when it is in position to invade. Much of this environmental sensing occurs through two-component signal transduction systems employing a common phosphorylation-dependent mechanism of signal transduction that appears nearly ubiquitous in bacteria (14, 15). Such systems also are found in some lower eukaryotes (16–19) and plants (20). All two-component signaling pathways consist of at least a histidine protein kinase and a response regulator. The kinase autophosphorylates on a histidine residue, and this phosphoryl group is transferred to an aspartate on its cognate response regulator (Fig. 1). The level of signal response is related to the phosphorylation level of the response regulator, and this is controlled by signal-mediated stimulation of the autophosphorylation or phosphatase activities of the kinase. In addition, phosphatases specific for the response regulator may control the system (21). Four features in particular make the two-component family attractive as a potential target for antimicrobials: (i) Significant homology is shared among kinase and response regulator proteins of different genera of bacteria, particularly in those amino acid residues located near active sites (15); (ii) pathogenic bacteria use two-component signal transduction to regulate expression of essential virulence factors that are required for survival inside the host (22–24); (iii) bacteria contain many two-component systems, and some of them are essential for viability (25, 26), and (iv) signal transduction in mammals occurs by a different mechanism.
Figure 1.
Two-component signal transduction in pathogenesis. Signals (external stimuli) residing in the host environment are recognized by the bacterium and stimulate the histidine protein kinase(s) to autophosphorylate (shown in the schematic as the bracketed HPK-P). Each phosphorylated histidine protein kinase (HPK) is mated to a specific response regulator (RR) to which it initiates phosphoryl transfer. Phosphorylation of the response regulator (RR-P) relieves inhibition of its transcriptional activation properties resulting in transcription and expression of the genes that it controls.
Despite the recognition of the essential nature of two-component transduction systems for bacterial survival within the host, few inhibitors of these systems have been described to date. Roychoudhury et al. (27) reported the discovery of inhibitors of the two-component system regulating expression of the alginate capsule in Pseudomonas aeruginosa. One goal of that effort was to identify inhibitors that could be used as adjunct therapy with a bactericidal agent in cystic fibrosis patients by rendering the bacterium susceptible to both the antimicrobial and the defenses of the host. However, these inhibitors did not inhibit bacterial cell growth at concentrations as high as 50 μg/ml when used alone. This report describes the series of inhibitors designed to inhibit two-component systems that are bactericidal against several genera of Gram-positive bacteria.
MATERIALS AND METHODS
Bacteria.
Most bacteria used in this study were obtained from the American Type Culture Collection (ATCC). OC3047 was provided by M. Inouye (Robert Wood Johnson Medical School, Piscataway, NJ). Other bacteria were clinical isolates (OC).
Resistance Emergence.
RWJ-49815 and ciprofloxacin were compared for in vitro selection of high level resistance by serially exposing methicillin-resistant Staphylococcus aureus (MRSA) strains to increasing concentrations of agent. Ten clinical MRSA strains were grown overnight in cation-adjusted Mueller–Hinton broth, and the turbidity of the overnight culture was adjusted to the Kirby–Bauer standard (≈1 × 108 cfu/ml). Samples of the cultures (100 μl) were spread onto the surface of three Mueller–Hinton agar plates containing antibiotic concentrations of twofold below the minimal inhibitory concentration (MIC), at the MIC, and two-fold above the MIC, respectively. After a 48-h incubation at 35°C, discrete colonies from plates containing the highest concentration of RWJ-49815 were grown as described above and were passaged onto agar plates with increasing concentrations of agent. The number of colonies at each concentration was recorded for each passage.
Biochemical Assay.
Autophosphorylation and its inhibition were measured using the KinA kinase of Bacillus subtilis (28). The biochemical assay to measure the inhibition of transfer of phosphate from ATP to KinA contained 0.5 μM KinA in a 150-μM Tris buffer, pH 8.0. The enzyme was incubated with varying concentrations of inhibitor and radiolabeled ATP (37.5 μCi; 1 Ci = 37 GBq) was added to initiate phosphorylation. Phosphorylated products then were separated by using polyacrylamide gel electrophoresis and quantitated with a phosphoimager. The percentage of inhibition was calculated from the intensity of each KinA∼P band after subtraction of the background.
Time Kill Experiments.
In vitro time kill tests were performed to study the bactericidal activity of kinase inhibitors over time. S. aureus OC2089 strain was grown overnight in the Mueller–Hinton broth at 35°C. The overnight culture was diluted in prewarmed Mueller–Hinton broth and incubated in a 35°C-shaking water bath (100 rpm) until the bacteria achieved log phase. This culture was aliquoted into sterile flasks containing RWJ-49815 or levofloxacin at one or four times MIC. These flasks plus a growth control flask without inhibitor were placed into the shaking water bath. Viable cell counts were performed initially and after 2, 4, 6, 24, and 48 h. Plated cultures were incubated at 35°C, and the number of colony-forming units per milliliter was determined.
Taz Assay.
This assay is a secondary whole cell assay measuring the EnvZ/OmpR two-component system that regulates response to osmotic pressure in Escherichia coli. In the modified system, the histidine kinase is a chimeric molecule in which the signal receptor region of the EnvZ has been replaced by the corresponding region of the Tar protein (chemoreceptor for aspartate) (29). As a result, aspartate-stimulated autophosphorylation of the Tar-1:EnvZ (Taz-1) chimera leads to phosphorylation of OmpR and activation of transcription of the ompC promoter, which can be measured by assay of β-galactosidase from a lacZ gene fused to ompC. The E. coli strain OC3047 is a transformant of RU1012 φ(ompC-lacZ) ΔenvZ∷Kmr containing a plasmid encoding for Taz-1. At t = 0 h, aspartate (to 3 mM) and varying concentrations of RWJ-49815 were added to a 96-well microtiter plate containing OC3047 cells growing exponentially in minimal A medium (1) supplemented with 32 μg of polymyxin B nonapeptide/ml. Growth was monitored at 0.5-h intervals by reading the optical density at 600 nm in a microtiter plate reader. To determine levels of β-galactosidase, induction samples were removed at t = 0 and t = 2.25 h, diluted into Z-buffer, lysed by toluene, and assayed in the presence of O-nitrophenyl-β-d-galactoside (30, 31).
RESULTS
Inhibition of KinA Autophosphorylation and Bacterial Growth by RWJ-49815.
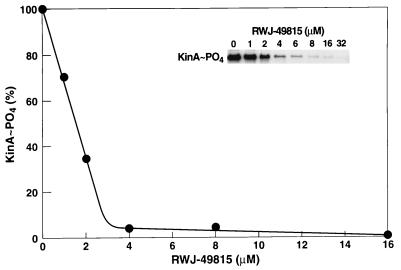
To screen our chemical inventory for inhibitors of two-component systems, we used the KinA kinase and the Spo0F response regulator, which are involved in regulation of sporulation in Bacillus subtilis as a model biochemical assay system (32). Compounds were discovered that selectively inhibited the in vitro autophosphorylation of KinA with [γ−32P] ATP. The percent of inhibition of radiolabel incorporated into KinA, forming KinA-phosphate, was quantified. One compound, RWJ-49815, was found to virtually inhibit the incorporation of phosphate from ATP into KinA at 4.0 μM (Fig. 2), with a one-half maximal inhibitory concentration (IC50) of 1.6 μM. Kinetic analyses revealed that RWJ-49815 was competitive with ATP (data not shown).
Figure 2.
Inhibition of kinase A autophosphorylation by RWJ-49815. Results are expressed as percent of KinA∼P formed in the absence of inhibitor.
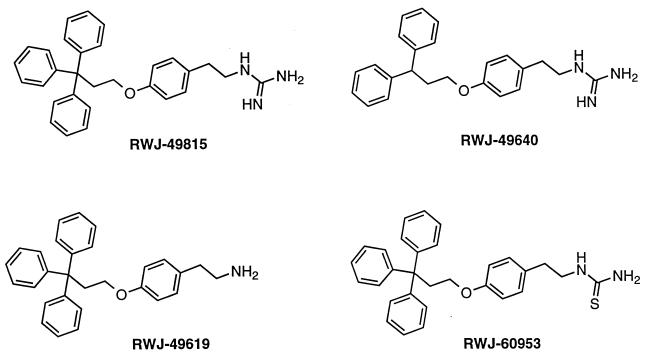
RWJ-49815 is a substituted phenethylguanidine, one of a series of hydrophobic tyramine derivatives (Fig. 3). The compound had antibacterial activity in Gram-positive bacteria with MIC values of 1–2 μg/ml (Table 1). Most importantly, RWJ-49815 inhibited growth of MRSA, vancomycin-resistant Enterococcus faecium, and penicillin-resistant Streptococcus pneumoniae. These pathogens are particularly difficult to treat clinically and can be lethal when untreated. The growth of drug susceptible strains, like S. aureus strain 29213, was inhibited by RWJ-49815 at similar concentrations as resistant strains, like methicillin-resistant S. aureus strain OC2089 (Table 1). RWJ-49815 did not inhibit the Gram-negative species E. coli, P. aeruginosa, or Klebsiella pneumoniae (MICs of >128 μg/ml). However, polymyxin B nonapeptide-treated E. coli (33) were highly sensitive to RWJ-49815 (MICs of 1–2 μg/ml, data not shown), suggesting that the outer membrane of Gram-negative bacteria is a barrier to penetration of the compound into the cells.
Figure 3.
Structures of RWJ-49815 and analogs.
Table 1.
Minimal inhibitory concentrations of RWJ-49815 and analogs against Gram-positive bacteria
| Minimal inhibitory concentrations,* μg/ml
| ||||
|---|---|---|---|---|
| Test organism | RWJ-49815† | RWJ-49619 | RWJ-49640 | RWJ-60953 |
| S. aureus ATTC 29213 | 1 | 4 | 4 | 16 |
| S. aureus ATTC 6538 | 2 | 2 | 1 | >32 |
| S. aureus OC667 (MR) | 2 | 2 | 2 | >32 |
| S. aureus OC2089 (MR) | 2 | 2 | 1 | >32 |
| S. epidermidis OC2603 | 2 | 2 | 2 | >32 |
| E. faecalis ATCC 29212 | 2 | 4 | 8 | >32 |
| E. faecalis OC3041 | 2 | 4 | 8 | >32 |
| E. faecium OC3312 (VanR) | 1 | 4 | 8 | >32 |
| S. pneumoniae OC3570 (PenS) | 2 | 16 | 8 | >32 |
| S. pneumoniae OC3561 (PenI) | 2 | 8 | 4 | >32 |
| S. pneumoniae OC3035 (PenR) | 1 | 8 | 4 | >32 |
Stock solutions of the compounds were prepared in dimethyl sulfoxide at 2.56 mg/ml and MICs were determined with the broth micro-dilution method as described by the National Committee for Clinical Laboratory Standards (1994) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically.
Molecular weights of compounds: RWJ-49815, 449; RWJ-49619, 407; RWJ-49640, 373; RWJ-60953, 466. MR, methicillin-resistant; VanR, vancomycin-resistant; PenS, penicillin-sensitive; PenI, intermediate penicillin-resistant; PenR, penicillin-resistant.
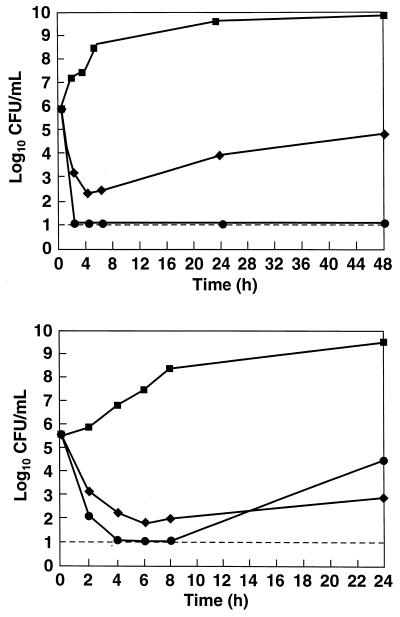
The antibacterial activity of RWJ-49815 is bactericidal (Fig. 4A) against methicillin-resistant S. aureus OC2089. Killing is rapid at one to four times the MIC, and regrowth is suppressed for at least 48 h at one to four times the MIC. Although there is some regrowth of bacteria at 48 h for a single MIC drug treatment, the compound is still inhibitory with ≈1.5-logs killing after 48 h. In fact, the activity of RWJ-49815 compared favorably with the bactericidal quinolone levofloxacin, against methicillin-resistant S. aureus OC2878 at two times the MIC (Fig. 4B).
Figure 4.
Antibacterial activity of RWJ-49815 against MRSA. (A) Time kill curve of RWJ-49815 against MRSA OC2089 at one times and four times MIC. (▪) Drug-free control; (♦) RWJ-49815 one times MIC; (•) RWJ-49815 four times MIC. (B) Time kill curve RWJ-49815 against MRSA OC2878 with a control drug, levofloxacin. The dotted line indicates the limit of detection of bacterial counts. (▪) Drug-free control; (♦) Levofloxacin two times MIC; (•) RWJ-49815 two times MIC.
Inhibitory Activity of RWJ-49815 Derivatives.
Several hydrophobic tyramine derivatives of RWJ-49815 have been synthesized to better define the structural basis for inhibition (Fig. 3). Analogs containing the diphenyl moiety in place of the triphenylmethyl moiety exhibited a 10-fold lower potency than RWJ-49815 in the KinA assay (RWJ-49640; IC50 = 20 μM), and antibacterial activity against enterococci and streptococci pathogens was decreased (Table 1). Compounds in which the highly basic guanidine functionality was replaced by a primary amine also were slightly less potent in the enzyme assay (RWJ-49619; IC50 of 14 μM), with a zero to eightfold increase in MIC values (Table 1). However, replacement of the guanidine moiety with nonbasic functionalities, such as thiourea, yielded analogs (RWJ-60953) that essentially were devoid of activity in the biochemical assay (IC50 of >500 μM) and antibacterial activity (Table 1), thereby establishing the importance of the basic group.
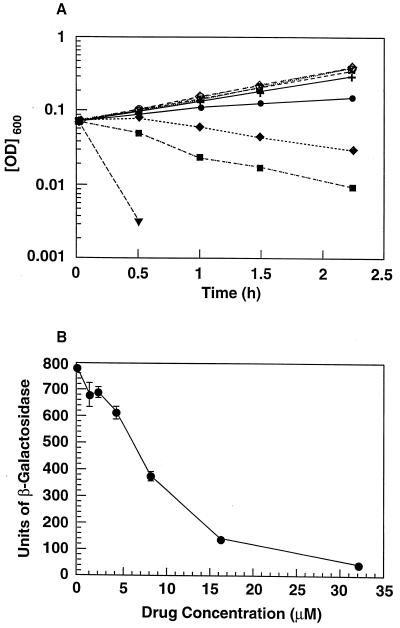
Two lines of evidence suggest that the observed growth inhibition by hydrophobic tyramine derivatives (as well as those of related compounds in the same series) is a primary consequence of their inhibition of one or more two-component systems. First, there was a direct correlation between the activity of these compounds against the kinase A target and their ability to inhibit growth of S. aureus (Fig. 5). Second, in a whole cell E. coli assay, RWJ-49815 showed a concentration-dependent inhibition of Taz-1 that extended to concentrations of the drug at which no effect on growth rate is apparent (Fig. 6). The Taz-1 system is a modification of the EnvZ/OmpR signal transduction pair, which normally is involved in the response of E. coli to changes in osmolarity by modifying the levels of the porins OmpC and OmpF. In this system, the kinase is a chimeric molecule in which the signal receptor region of EnvZ has been replaced with the corresponding region of the Tar protein (chemoreceptor for aspartate) (29, 30). As a result, aspartate stimulates Taz-1 to phosphorylate OmpR and the signal transduction effects can be monitored through the changes in expression of an ompC-lacZ gene fusion. RWJ-49815 inhibited both bacterial growth and OmpR-dependent inducibility of the reporter gene in a concentration-dependent manner (Fig. 6). However, the inhibitory effects of RWJ-49815 on inducibility of the reporter gene were stronger and precede those of the inhibitory effect of drugs on growth rate. Whereas the effects on growth rate first become apparent at 8 μM (Fig. 6A), there was already a small but distinct inhibition of the OmpR-dependent inducibility of the reporter gene at RWJ-49815 concentrations ≤4 μM (Fig. 6B). The effects of RWJ-49815 on OmpR clearly preceded those on growth rate and suggested a causal relationship.
Figure 5.
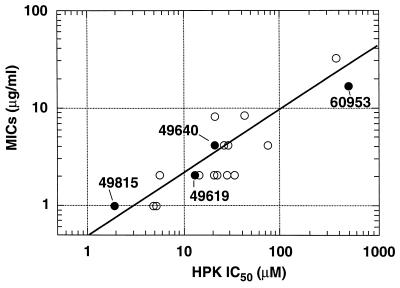
Correlation of antibacterial activity of RWJ-49815 with enzyme inhibition. The 18 tyramine derivatives (○), including the four identified by structures in this report (•), are plotted on log scales, comparing the target-based inhibition of activity (IC50s) vs. the MICs against MRSA strain OC2089.
Figure 6.
(A) Concentration-dependent effect of RWJ-49815 on the growth rate of E. coli strain OC3047 harboring the Taz-1 two-component system and reporter gene. Concentrations of RWJ-49815: (○) μM; (⋄) 1 μM; (□) 2 μM; (x) 4 μM; (+) 8 μM; (•) 16 μM; (♦) 32 μM; (▪) 64 μM; and (▾) 128 μM. Bacteria incubated with concentrations of RWJ-49815 up to 4 μM had mean generation times of 56 min that were indistinguishable from the no-drug control. Bacteria incubated with 8 μM RWJ-49815 had a doubling time of 67 min. (B) Effect of RWJ-49815 on the Taz-1 induction of β-galactosidase.
Resistance Emergence to RWJ-49815.
In vitro resistance emergence experiments have been conducted in which 10 independent clinical strains of methicillin-resistant S. aureus grown in broth overnight were spread onto the surface of agar plates containing twofold serial dilutions of test agents [RWJ-49815 and ciprofloxacin (a quinolone)]. Viable bacteria selected from colonies on agar plates containing the highest drug concentration were passaged on solid medium, which contained increasing concentrations of RWJ-49815 or ciprofloxacin. After five passages, both the number of colonies and the fold increase in MIC for selection was less for RWJ-49815 (zero to 16-fold increase) than for ciprofloxacin (eight to 256-fold).
DISCUSSION
It is now well established that two-component signal transduction systems are essential components of the virulence mechanisms of a wide variety of bacterial pathogens (22). Mutation of the virulence genes including the two-component sensing mechanism has serious consequences for the establishment and progression of an infection. The proteins of two-component systems are highly homologous to each other across Gram-positive and Gram-negative species, and they represent the only common feature of the virulence mechanisms of a variety of pathogens. Because of these features, such signal transduction systems may be viewed as an attractive target for anti-infective intervention (34). Because most virulence mechanisms and two-component systems are not found to be essential for growth in laboratory media, a contrary viewpoint has remained rooted in the minds of some putative experts. However, recently, research in Caulobacter crescentus has established that two-component signal transduction systems are involved in the regulation of DNA replication and the cell cycle and are essential for viability (25, 26). Growth essential systems also have been found in Bacillus subtilis and other Gram-positive species (C. Fabret and J.A.H., unpublished data). Genome sequencing of B. subtilis has revealed at least 35 two-component systems (35), and E. coli probably contains about the same number (36). A broad-spectrum inhibitor of these systems is likely to have viability and virulence consequences. Genome sequencing of pathogens such as Haemophilus influenzae, Borrelia burgdorferi, or Mycoplasma genitalium, however, revealed few, if any, genes coding for two-component-signaling systems (37–39). The small genome and limited metabolic capacities of the latter two organisms suggests that they have little need or ability to respond to their environment by regulating gene expression. Regardless of the reasons, two-component system inhibitors therefore are not likely to be bactericidal to this limited subgroup of pathogens.
The compounds described in this communication were bactericidal to Gram-positive microorganisms, even pathogens that are particularly difficult to treat clinically. They were ineffective on Gram-negative microorganisms unless the organisms were treated with a membrane-permeabilizing compound suggesting that the outer membrane of this class of bacteria presents a barrier to penetration of the compound. Moreover, treated E. coli were killed very effectively by RWJ-49815 indicating that the targets of its action are present in Gram-negative bacteria. The actual target(s) of the compound in either class of microorganisms has not been firmly established. Because the efficacy of growth inhibition by RWJ-48915 and its analogs correlates directly with their ability to inhibit kinase A activity, the target of RWJ-49815 action is thought to be a two-component kinase. However, it cannot be excluded that in vitro kinase A activity assays the efficacy of inhibition at a site on kinase A shared with another nonkinase protein whose inhibition would prevent growth. The resistance emergence studies presented here could be interpreted as indicating that RWJ-49815 inhibits more than one essential target. Multiple targets could diminish resistance emergence from target modification but not from nontarget-based mechanisms of resistance such as inhibitor exclusion.
RWJ-49815 is a potent inhibitor of the autophosphorylation reaction of kinase A. The probable target for this inhibitor is some feature of the ATP-binding site of the kinase because inhibition is competitive with respect to ATP. This site is one of the two major common features of all of the two-component kinases, and the motifs that constitute this site are unique from most other known kinases. Therefore, it is not surprising that a broad-spectrum inhibitor of autophosphorylation in two-component kinases such as RWJ-49815 would be directed toward the site. The previously described compounds with antikinase activity that are structurally different from RWJ-49815 also are directed toward the ATP-binding region (27). This type of ATP-binding site also is found in the serine protein kinases of antisigma factors in B. subtilis (40). Serine protein kinases unlike two-component kinases do not use a histidine phosphate intermediate. A family of serine protein kinases with similar characteristics are found in mitochondria in which they phosphorylate α-ketoacid dehydrogenase complexes to regulate their activity (41, 42). Development of RWJ-49815 and its derivatives as therapeutics will require consideration of the consequences of potential inhibition of these mitochondrial enzymes.
RWJ-49815 and its derivatives are the first series of bacterial two-component system inhibitors with bactericidal activity against Gram-positive pathogens. Their spectrum of activity includes methicillin-resistant S. aureus, vancomycin-resistant enterococci, and penicillin-resistant S. pneumoniae. Although RWJ-49815 is bactericidal, even inhibitors of two-component systems that are not bactericidal may prove to be valuable adjunct therapeutic agents. For example, high-level vancomycin resistance (VanA phenotype) in enterococci is regulated by the VanR–VanS two-component system, the inhibition of which should render the microorganism again sensitive to vancomycin (43). Other examples of two-component system-dependent antibiotic resistance are known (44). It seems likely that inhibition of these signal transduction mechanisms will find many and varied uses as part of the arsenal against infectious disease.
Table 2.
Resistance emergence of methicillin-resistant S. aureus strains to RWJ-49815 and ciprofloxacin
| RWJ-49815
|
Ciprofloxacin
|
|||
|---|---|---|---|---|
| S. aureus strain | MIC initial, μg/ml | Increase in MIC after passage (fold) | MIC initial, μg/ml | Increase in MIC after passage (fold) |
| OC667 | 4 | 2 | 0.5 | 4 |
| OC2080 | 0.5 | 8 | 0.5 | 8 |
| OC2083 | 2 | 2 | 0.5 | 4 |
| OC2843 | 2 | 2 | 0.5 | 32 |
| OC2878 | 0.5 | 8 | 0.25 | 32 |
| OC2882 | 4 | 2 | 0.5 | 64 |
| OC2997 | 4 | 2 | 0.5 | 32 |
| OC2891 | 4 | 2 | 0.5 | 32 |
| OC2898 | 0.5 | 4 | 0.25 | 64 |
| OC2900 | 0.5 | 4 | 0.25 | 8 |
MIC, minimal inhibitory concentration.
Acknowledgments
We thank M. Inouye for kindly providing the strains used in the Taz-1 assay and H. Krause for technical assistance in part of this work.
ABBREVIATIONS
- MRSA
methicillin-resistant Staphylococcus aureus
- OC
clinical isolate
- MIC
minimal inhibitory concentration
References
- 1.Chu D T, Plattner J J, Katz L. J Med Chem. 1996;39:3853–3874. doi: 10.1021/jm960294s. [DOI] [PubMed] [Google Scholar]
- 2.Eliopoulos G M, Wennersten C B, Gold H S, Moellering R C. Antimicrob Agents Chemother. 1996;40:1745–1747. doi: 10.1128/aac.40.7.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zurenko G E, Yagi B H, Schaadt R D, Allison J W, Kilburn J O, Glickman S E, Hutchinson D K, Barbachyn M R, Brickner S J. Antimicrob Agents Chemother. 1996;40:839–845. doi: 10.1128/aac.40.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mason E O, Lamberth L B, Kaplan S L. Antimicrob Agents Chemother. 1996;40:1039–1040. doi: 10.1128/aac.40.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brickner S J, Hutchinson D K, Barbachyn M R, Manninen P R, Ulanowicz D A, Garmon S A, Grega K C, Hendges S K, Toops D S, Ford C W, et al. J Med Chem. 1996;39:673–679. doi: 10.1021/jm9509556. [DOI] [PubMed] [Google Scholar]
- 6.Sum P-E, Lee V J, Testa R T, Hlavka J J, Ellestad G A, Bloom J D, Gluzman Y, Tally F P. J Med Chem. 1994;37:184–188. doi: 10.1021/jm00027a023. [DOI] [PubMed] [Google Scholar]
- 7.Tally F T, Ellestad G A, Testa R T. J Antimicrob Chemother. 1995;35:449–452. doi: 10.1093/jac/35.4.449. [DOI] [PubMed] [Google Scholar]
- 8.Weiss W J, Jacobus N V, Petersen P J, Testa R T. J Antimicrob Chemother. 1995;36:225–230. doi: 10.1093/jac/36.1.225. [DOI] [PubMed] [Google Scholar]
- 9.Onishi H R, Pelak B A, Gerckens L S, Silver L L, Kahan F M, Chen M, Patchett A A, Galloway S M. Science. 1996;274:980–982. doi: 10.1126/science.274.5289.980. [DOI] [PubMed] [Google Scholar]
- 10.Miller J F, Mekalanos J J, Falkow S. Science. 1989;243:916–922. doi: 10.1126/science.2537530. [DOI] [PubMed] [Google Scholar]
- 11.Fields P I, Groisman E A, Heffron F. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 12.Alpuche-Aranda C M, Swanson J A, Loomis W P, Miller S I. Proc Natl Acad Sci USA. 1992;89:10079–10083. doi: 10.1073/pnas.89.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Véscovi E, Soncini F C, Groisman E A. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 14.Stock J B, Ninfa A J, Stock A M. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parkinson J S, Kofoid E C. Ann Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 16.Maeda T, Wurgler-Murphy S M, Saito H. Nature (London) 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 17.Ota I M, Varshavsky A. Science. 1993;262:566–569. doi: 10.1126/science.8211183. [DOI] [PubMed] [Google Scholar]
- 18.Posas F, Wurgler-Murphy S M, Maeda T, Witten E A, Thai T C, Saito J. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 19.Alex L A, Borkovich K A, Simon M I. Proc Natl Acad Sci USA. 1996;93:3416–3421. doi: 10.1073/pnas.93.8.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 21.Perego M, Hanstein C G, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 22.Dziejman M, Mekalanos J J. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. , Washington, DC: Am. Soc. Microbiol.; 1995. pp. 305–317. [Google Scholar]
- 23.Groisman E A, Heffron F. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 319–332. [Google Scholar]
- 24.Uhl M A, Miller J F. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 333–349. [Google Scholar]
- 25.Hecht G B, Lane T, Ohta N, Sommer J, Newton A. EMBO J. 1995;14:3915–3924. doi: 10.1002/j.1460-2075.1995.tb00063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quon K C, Marczynski G T, Shapiro L. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 27.Roychoudhury S, Zielinski N A, Ninfa A J, Allen N E, Jungheim L N, Nicas T I, Chakrabarty A M. Proc Natl Acad Sci USA. 1993;90:965–969. doi: 10.1073/pnas.90.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perego M, Cole S P, Burbulys D, Trach K A, Hoch J A. J Bacteriol. 1989;171:6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin T, Inouye M. J Mol Biol. 1993;232:484–492. doi: 10.1006/jmbi.1993.1404. [DOI] [PubMed] [Google Scholar]
- 30.Utsumi R, Brissette R E, Rampersaud A, Forst S A, Oosawa K, Inouye M. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. pp. 352–355. [Google Scholar]
- 32.Burbulys D, Trach K A, Hoch J A. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 33.Chopra I, Hacker K. J Antimicrob Chemother. 1992;29:19–25. doi: 10.1093/jac/29.1.19. [DOI] [PubMed] [Google Scholar]
- 34.Desnottes J. Trends Biotechnol. 1996;14:134–140. doi: 10.1016/0167-7799(96)10015-9. [DOI] [PubMed] [Google Scholar]
- 35.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. Nature (London) 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 36.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 37.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 38.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 39.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, et al. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 40.Min K-T, Hilditch C M, Diederich B, Errington J, Yudkin M D. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 41.Harris R A, Hawes J W, Popov K M, Zhao Y, Shimomura Y, Sato J, Jaskiewicz J, Hurley T D. Adv Enzyme Regul. 1997;37:271–293. doi: 10.1016/s0065-2571(96)00009-x. [DOI] [PubMed] [Google Scholar]
- 42.Harris R A, Popov K M, Zhao Y, Kedishvili N Y, Shimomura Y, Crabb D W. Adv Enzyme Regul. 1995;35:147–162. doi: 10.1016/0065-2571(94)00020-4. [DOI] [PubMed] [Google Scholar]
- 43.Arthur M, Depardieu F, Holman T, Wu Z, Wright G, Walsh C T, Courvalin P. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 387–391. [Google Scholar]
- 44.Salyers A A, Shoemaker N B, Stevens A M. In: Two-Component Signal Transduction. Hoch J A, Silhavy T J, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 393–400. [Google Scholar]