Abstract
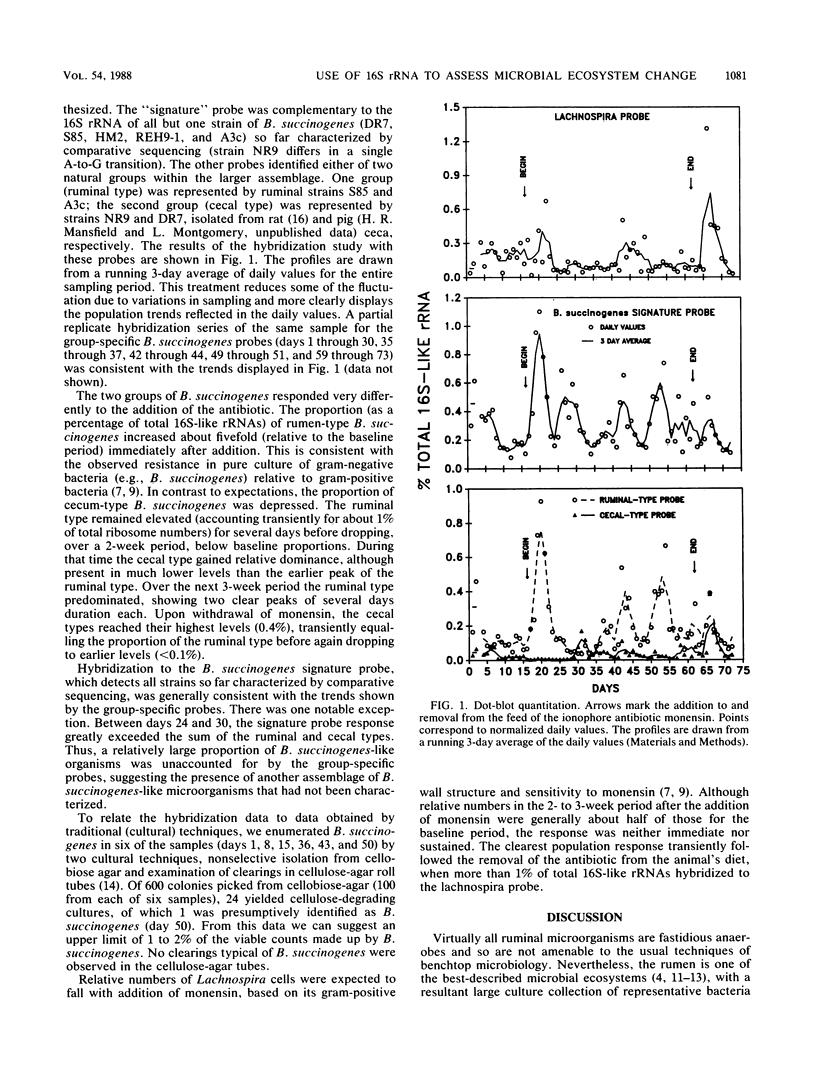
To address the long-standing need for more precise descriptions of natural microbial ecosystems, 16S rRNAs were used to track certain species and phylogenetically coherent groups of microorganisms in their natural setting without culturing. Species- and group-specific 16S rRNA-targeted oligonucleotide hybridization probes were developed to enumerate various strains of Bacteroides succinogenes and Lachnospira multiparus-like organisms in the bovine rumen before, during, and after perturbation of that ecosystem by the addition of the ionophore antibiotic monensin. Based on probe hybridization, relative numbers of L. multiparus-like organisms were depressed about 2-fold during monensin addition and demonstrated a transient 5- to 10-fold increase immediately after removal of the antibiotic from the diet. The most pronounced population changes were observed among different strains of B. succinogenes, as evaluated by three hybridization probes. One probe hybridized to all strains, whereas the other two identified genetically distinct groups represented by strains isolated from the rumen and from the ceca of nonruminants. The rumen-type strains predominated on most days (ca. 0.2 to 0.8% of total ribosome numbers). Their proportion transiently increased about fivefold immediately after the addition of monensin to the feed and then transiently fell below the average premonensin level. During this time (ca. 2 weeks after monensin addition) the cecal type predominated (ca. 0.1 to 0.2%). Cultural enumeration of B. succinogenes on nonselective agar and by observing clearings in cellulose agar media were largely unsuccessful due to the low number of organisms present and the predominance of other cellulolytic species.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergen W. G., Bates D. B. Ionophores: their effect on production efficiency and mode of action. J Anim Sci. 1984 Jun;58(6):1465–1483. doi: 10.2527/jas1984.5861465x. [DOI] [PubMed] [Google Scholar]
- Bryant M. P., Robinson I. M. Effects of diet, time after feeding, and position sampled on numbers of viable bacteria in the bovine rumen. J Dairy Sci. 1968 Dec;51(12):1950–1955. doi: 10.3168/jds.S0022-0302(68)87320-5. [DOI] [PubMed] [Google Scholar]
- Chen M., Wolin M. J. Effect of monensin and lasalocid-sodium on the growth of methanogenic and rumen saccharolytic bacteria. Appl Environ Microbiol. 1979 Jul;38(1):72–77. doi: 10.1128/aem.38.1.72-77.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford Mark. California field test goes forward. Science. 1987 May 1;236(4801):511–511. doi: 10.1126/science.11643987. [DOI] [PubMed] [Google Scholar]
- Dennis S. M., Nagaraja T. G., Bartley E. E. Effects of lasalocid or monensin on lactate-producing or -using rumen bacteria. J Anim Sci. 1981 Feb;52(2):418–426. doi: 10.2527/jas1981.522418x. [DOI] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Hobson P. N., Wallace R. J. Microbial ecology and activities in the rumen: Part II. Crit Rev Microbiol. 1982 May;9(4):253–320. doi: 10.3109/10408418209104492. [DOI] [PubMed] [Google Scholar]
- Hobson P. N., Wallace R. J. Microbial ecology and activities in the rumen: part 1. Crit Rev Microbiol. 1982 Apr;9(3):165–225. doi: 10.3109/10408418209104490. [DOI] [PubMed] [Google Scholar]
- Macy J. M., Farrand J. R., Montgomery L. Cellulolytic and non-cellulolytic bacteria in rat gastrointestinal tracts. Appl Environ Microbiol. 1982 Dec;44(6):1428–1434. doi: 10.1128/aem.44.6.1428-1434.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery L., Macy J. M. Characterization of rat cecum cellulolytic bacteria. Appl Environ Microbiol. 1982 Dec;44(6):1435–1443. doi: 10.1128/aem.44.6.1435-1443.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl D. A., Lane D. J., Olsen G. J., Pace N. R. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl Environ Microbiol. 1985 Jun;49(6):1379–1384. doi: 10.1128/aem.49.6.1379-1384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson M. J., Hirose T., Miyake T., Kawashima E. H., Itakura K. The use of synthetic oligonucleotides as hybridization probes. II. Hybridization of oligonucleotides of mixed sequence to rabbit beta-globin DNA. Nucleic Acids Res. 1981 Feb 25;9(4):879–894. doi: 10.1093/nar/9.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstone H. D., Davis C. L., Bryant M. P. Effect of monensin on breakdown of protein by ruminal microorganisms in vitro. J Anim Sci. 1981 Sep;53(3):803–809. doi: 10.2527/jas1981.533803x. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gutell R., Gupta R., Noller H. F. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983 Dec;47(4):621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Macke T. J., Fox G. E. A phylogenetic definition of the major eubacterial taxa. Syst Appl Microbiol. 1985;6:143–151. doi: 10.1016/s0723-2020(85)80047-3. [DOI] [PubMed] [Google Scholar]



