Abstract
Methanogens represented about 0.5% of the total bacteria in sediments from a Georgia salt marsh in which Spartina alterniflora is the predominant vegetation. The population of methanogens was composed of at least two groups of nearly equal size. One group was represented by cocci which were able to utilize trimethylamine and were unable to use H2 or acetate. The second group was composed of two subgroups which were able to utilize H2 but were unable to use trimethylamine or acetate. The more common subgroup included rod- or plate-shaped methanogens which could utilize isopropanol in addition to H2 and formate. The second subgroup included Methanococcus maripaludis, which utilized only H2 and formate. Other groups of methanogens were also present, including Methanosarcina sp. which utilized acetate, H2, and methylamines. In addition to the overall variability in the types of methanogens, the numbers of methanogens in sediments also exhibited significant spatial variability both within and between tall- and short-Spartina zones.
Full text
PDF


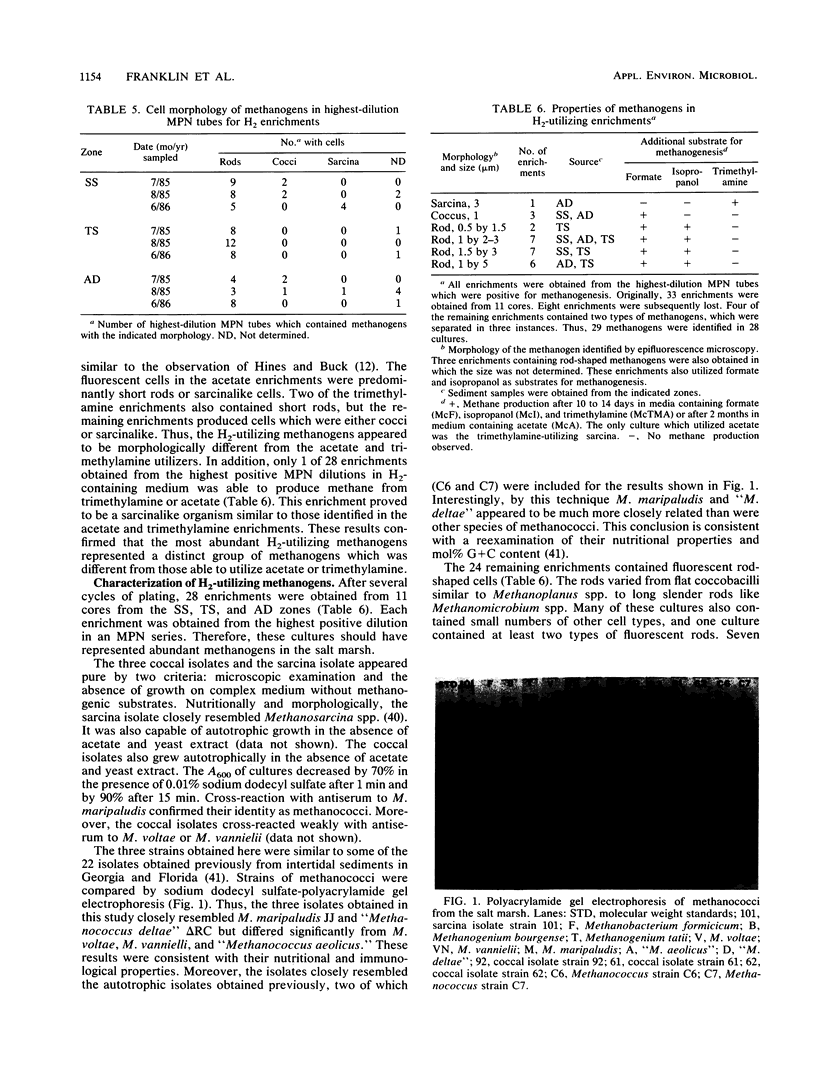



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abram J. W., Nedwell D. B. Hydrogen as a substrate for methanogenesis and sulphate reduction in anaerobic saltmarsh sediment. Arch Microbiol. 1978 Apr 27;117(1):93–97. doi: 10.1007/BF00689357. [DOI] [PubMed] [Google Scholar]
- Abram J. W., Nedwell D. B. Inhibition of methanogenesis by sulphate reducing bacteria competing for transferred hydrogen. Arch Microbiol. 1978 Apr 27;117(1):89–92. doi: 10.1007/BF00689356. [DOI] [PubMed] [Google Scholar]
- Balch W. E., Wolfe R. S. New approach to the cultivation of methanogenic bacteria: 2-mercaptoethanesulfonic acid (HS-CoM)-dependent growth of Methanobacterium ruminantium in a pressureized atmosphere. Appl Environ Microbiol. 1976 Dec;32(6):781–791. doi: 10.1128/aem.32.6.781-791.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G. Bacterial biovolume and biomass estimations. Appl Environ Microbiol. 1985 Jun;49(6):1488–1493. doi: 10.1128/aem.49.6.1488-1493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrain M., Zeikus J. G. Microbial ecophysiology of whey biomethanation: characterization of bacterial trophic populations and prevalent species in continuous culture. Appl Environ Microbiol. 1986 Jan;51(1):188–196. doi: 10.1128/aem.51.1.188-196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway de Macario E., Wolin M. J., Macario A. J. Immunology of archaebacteria that produce methane gas. Science. 1981 Oct 2;214(4516):74–75. doi: 10.1126/science.6792708. [DOI] [PubMed] [Google Scholar]
- Doddema H. J., Vogels G. D. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1978 Nov;36(5):752–754. doi: 10.1128/aem.36.5.752-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNGATE R. E., BRYANT M. P., MAH R. A. THE RUMEN BACTERIA AND PROTOZOA. Annu Rev Microbiol. 1964;18:131–166. doi: 10.1146/annurev.mi.18.100164.001023. [DOI] [PubMed] [Google Scholar]
- Hines M. E., Buck J. D. Distribution of methanogenic and sulfate-reducing bacteria in near-shore marine sediments. Appl Environ Microbiol. 1982 Feb;43(2):447–453. doi: 10.1128/aem.43.2.447-453.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. J., Paynter M. J. Populations of methane-producing bacteria and in vitro methanogenesis in salt marsh and estuarine sediments. Appl Environ Microbiol. 1980 Apr;39(4):864–871. doi: 10.1128/aem.39.4.864-871.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. J., Whitman W. B., Fields R. D., Wolfe R. S. Growth and plating efficiency of methanococci on agar media. Appl Environ Microbiol. 1983 Jul;46(1):220–226. doi: 10.1128/aem.46.1.220-226.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiene R. P., Oremland R. S., Catena A., Miller L. G., Capone D. G. Metabolism of reduced methylated sulfur compounds in anaerobic sediments and by a pure culture of an estuarine methanogen. Appl Environ Microbiol. 1986 Nov;52(5):1037–1045. doi: 10.1128/aem.52.5.1037-1045.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl Environ Microbiol. 1984 Oct;48(4):719–725. doi: 10.1128/aem.48.4.719-725.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mackie R. I., Bryant M. P. Metabolic Activity of Fatty Acid-Oxidizing Bacteria and the Contribution of Acetate, Propionate, Butyrate, and CO(2) to Methanogenesis in Cattle Waste at 40 and 60 degrees C. Appl Environ Microbiol. 1981 Jun;41(6):1363–1373. doi: 10.1128/aem.41.6.1363-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink R. W., Dugan P. R. Tentative identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1977 Mar;33(3):713–717. doi: 10.1128/aem.33.3.713-717.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountfort D. O., Asher R. A. Effect of inorganic sulfide on the growth and metabolism of Methanosarcina barkeri strain DM. Appl Environ Microbiol. 1979 Apr;37(4):670–675. doi: 10.1128/aem.37.4.670-675.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountfort D. O., Asher R. A., Mays E. L., Tiedje J. M. Carbon and electron flow in mud and sandflat intertidal sediments at delaware inlet, nelson, new zealand. Appl Environ Microbiol. 1980 Apr;39(4):686–694. doi: 10.1128/aem.39.4.686-694.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann E., Hippe H., Gottschalk G. Betaine: New Oxidant in the Stickland Reaction and Methanogenesis from Betaine and l-Alanine by a Clostridium sporogenes-Methanosarcina barkeri Coculture. Appl Environ Microbiol. 1983 Feb;45(2):474–483. doi: 10.1128/aem.45.2.474-483.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland R. S., Polcin S. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl Environ Microbiol. 1982 Dec;44(6):1270–1276. doi: 10.1128/aem.44.6.1270-1276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paynter M. J., Hungate R. E. Characterization of Methanobacterium mobilis, sp. n., isolated from the bovine rumen. J Bacteriol. 1968 May;95(5):1943–1951. doi: 10.1128/jb.95.5.1943-1951.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E. L., Bakole R. O., Bohlool B. B. Fluorescent-antibody approach to study of rhizobia in soil. J Bacteriol. 1968 Jun;95(6):1987–1992. doi: 10.1128/jb.95.6.1987-1992.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senior E., Lindström E. B., Banat I. M., Nedwell D. B. Sulfate reduction and methanogenesis in the sediment of a saltmarsh on the East coast of the United kingdom. Appl Environ Microbiol. 1982 May;43(5):987–996. doi: 10.1128/aem.43.5.987-996.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Mah R. A. Acetate as sole carbon and energy source for growth of methanosarcina strain 227. Appl Environ Microbiol. 1980 May;39(5):993–999. doi: 10.1128/aem.39.5.993-999.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Baron S. F., Ferry J. G. Methanosarcina acetivorans sp. nov., an Acetotrophic Methane-Producing Bacterium Isolated from Marine Sediments. Appl Environ Microbiol. 1984 May;47(5):971–978. doi: 10.1128/aem.47.5.971-978.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Ferry J. G. Isolation and Characterization of a Methylotrophic Marine Methanogen, Methanococcoides methylutens gen. nov., sp. nov. Appl Environ Microbiol. 1983 Feb;45(2):684–690. doi: 10.1128/aem.45.2.684-690.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F. Growth of methanogenic bacteria in pure culture with 2-propanol and other alcohols as hydrogen donors. Appl Environ Microbiol. 1986 May;51(5):1056–1062. doi: 10.1128/aem.51.5.1056-1062.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]