Abstract
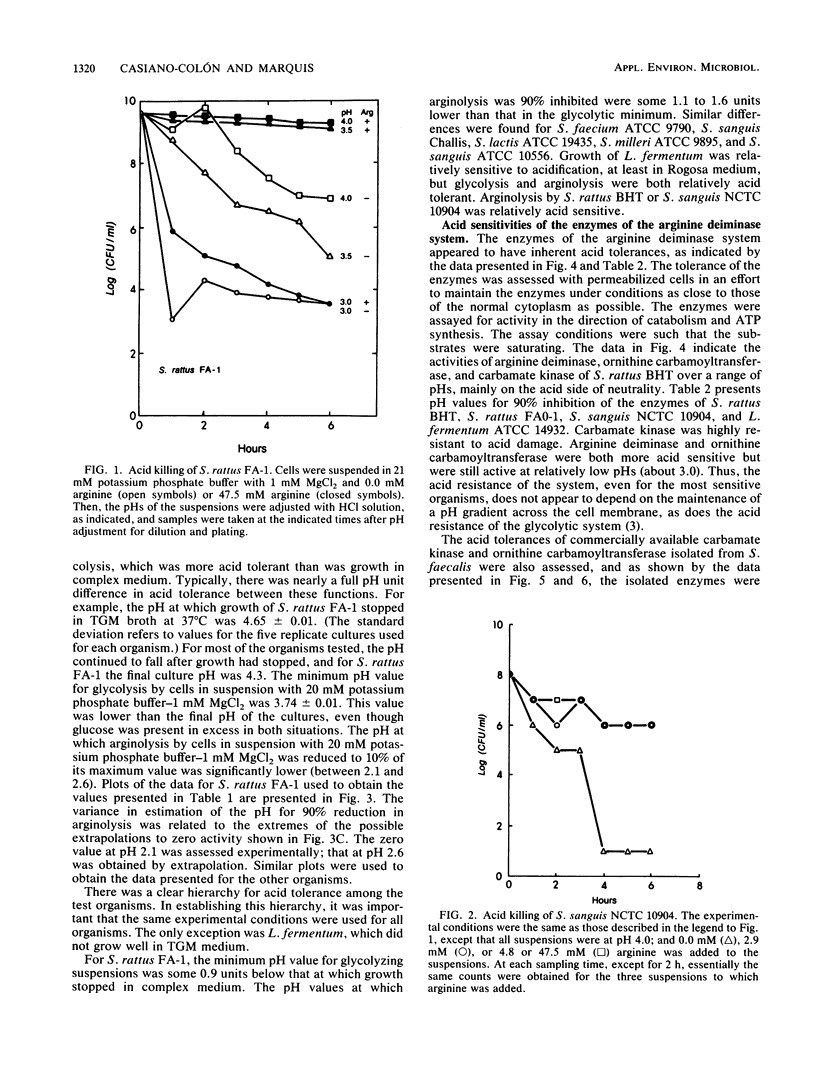
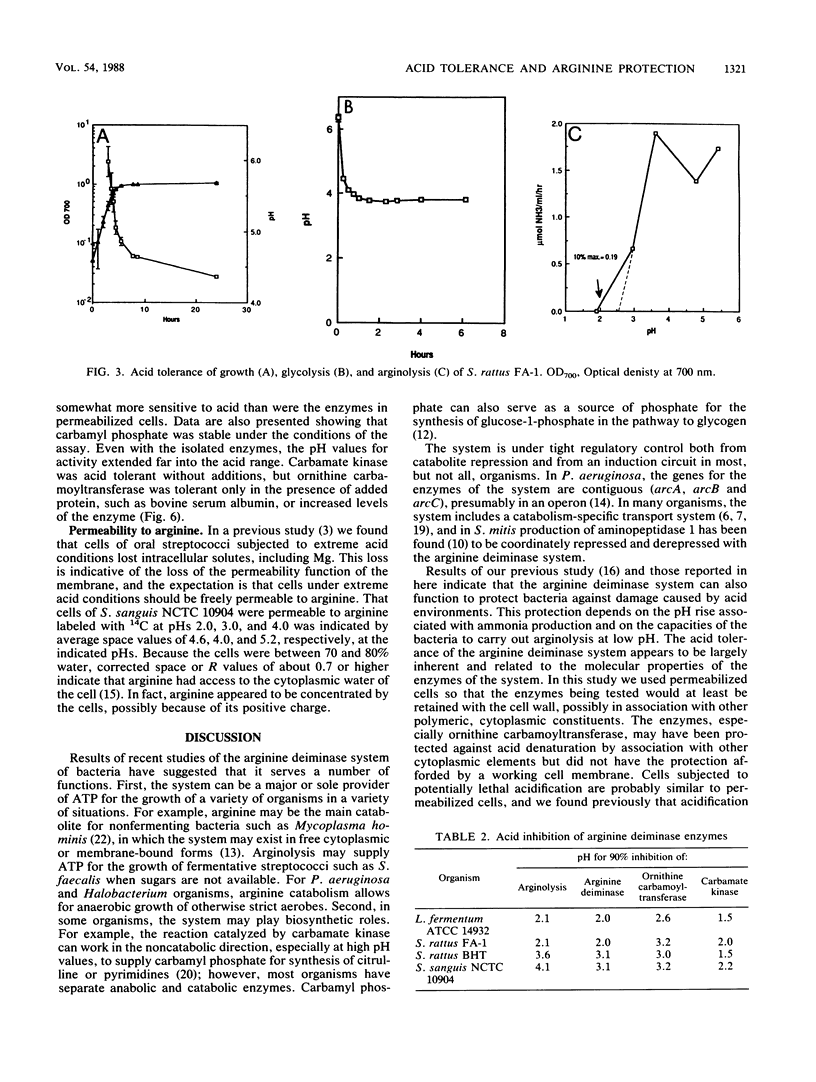
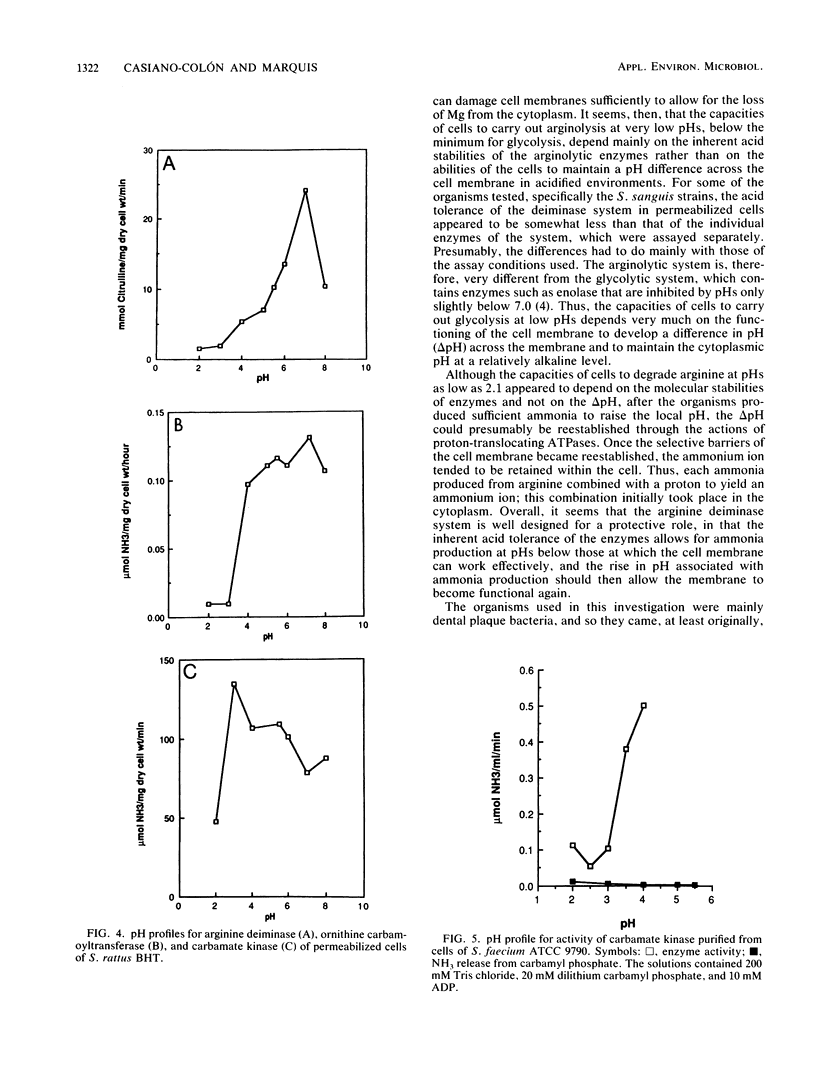
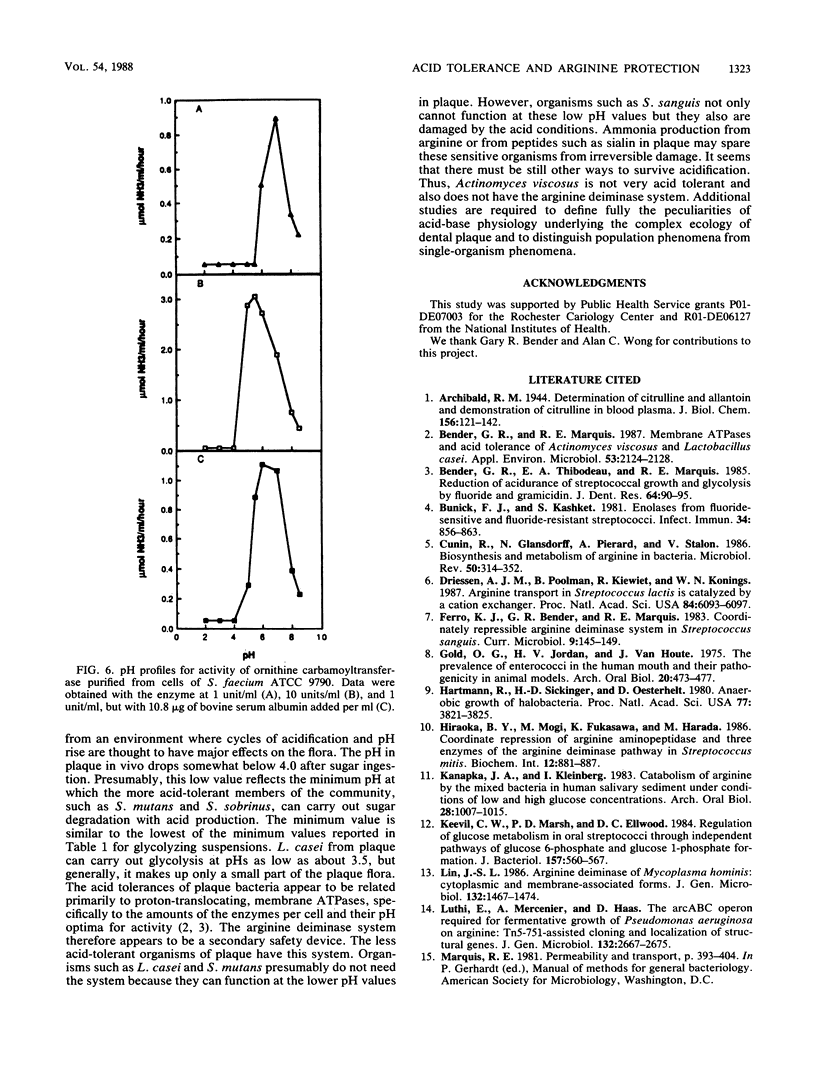
The arginine deiminase system was found to function in protecting bacterial cells against the damaging effects of acid environments. For example, as little as 2.9 mM arginine added to acidified suspensions of Streptococcus sanguis at a pH of 4.0 resulted in ammonia production and protection against killing. The arginine deiminase system was found to have unusual acid tolerance in a variety of lactic acid bacteria. For example, for Streptococcus rattus FA-1, the pH at which arginolysis was reduced to 10% of the maximum was between 2.1 and 2.6, or more than 1 full pH unit below the minimum for glycolysis (pH 3.7), and more than 2 units below the minimum for growth in complex medium (pH 4.7). The acid tolerance of the arginine deiminase system appeared to be primarily molecular and to depend on the tolerance of individual enzymes rather than on the membrane physiology of the bacteria; pH profiles for the activities of arginine deiminase, ornithine carbamoyltransferase, and carbamate kinase in permeabilized cells showed that the enzymes were active at pHs of 3.1 or somewhat lower. Overall, it appeared that ammonia could be produced from arginine at low pH values, even by cells with damaged membranes, and that the ammonia could then protect the cells against acid damage until the environmental pH value rose sufficiently to allow for the reestablishment of a difference in pH (delta pH) across the cell membrane.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender G. R., Marquis R. E. Membrane ATPases and acid tolerance of Actinomyces viscosus and Lactobacillus casei. Appl Environ Microbiol. 1987 Sep;53(9):2124–2128. doi: 10.1128/aem.53.9.2124-2128.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender G. R., Thibodeau E. A., Marquis R. E. Reduction of acidurance of streptococcal growth and glycolysis by fluoride and gramicidin. J Dent Res. 1985 Feb;64(2):90–95. doi: 10.1177/00220345850640021701. [DOI] [PubMed] [Google Scholar]
- Bunick F. J., Kashket S. Enolases from fluoride-sensitive and fluoride-resistant streptococci. Infect Immun. 1981 Dec;34(3):856–863. doi: 10.1128/iai.34.3.856-863.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunin R., Glansdorff N., Piérard A., Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol Rev. 1986 Sep;50(3):314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driessen A. J., Poolman B., Kiewiet R., Konings W. Arginine transport in Streptococcus lactis is catalyzed by a cationic exchanger. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6093–6097. doi: 10.1073/pnas.84.17.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold O. G., Jordan H. V., van Houte J. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch Oral Biol. 1975 Jul;20(7):473–477. doi: 10.1016/0003-9969(75)90236-8. [DOI] [PubMed] [Google Scholar]
- Hartmann R., Sickinger H. D., Oesterhelt D. Anaerobic growth of halobacteria. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3821–3825. doi: 10.1073/pnas.77.7.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka B. Y., Mogi M., Fukasawa K., Harada M. Coordinate repression of arginine aminopeptidase and three enzymes of the arginine deiminase pathway in Streptococcus mitis. Biochem Int. 1986 Jun;12(6):881–887. [PubMed] [Google Scholar]
- Kanapka J. A., Kleinberg I. Catabolism of arginine by the mixed bacteria in human salivary sediment under conditions of low and high glucose concentration. Arch Oral Biol. 1983;28(11):1007–1015. doi: 10.1016/0003-9969(83)90055-9. [DOI] [PubMed] [Google Scholar]
- Keevil C. W., Marsh P. D., Ellwood D. C. Regulation of glucose metabolism in oral streptococci through independent pathways of glucose 6-phosphate and glucose 1-phosphate formation. J Bacteriol. 1984 Feb;157(2):560–567. doi: 10.1128/jb.157.2.560-567.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. S. Arginine deiminase of Mycoplasma hominis: cytoplasmic and membrane-associated forms. J Gen Microbiol. 1986 Jun;132(6):1467–1474. doi: 10.1099/00221287-132-6-1467. [DOI] [PubMed] [Google Scholar]
- Lüthi E., Mercenier A., Haas D. The arcABC operon required for fermentative growth of Pseudomonas aeruginosa on arginine: Tn5-751-assisted cloning and localization of structural genes. J Gen Microbiol. 1986 Oct;132(10):2667–2675. doi: 10.1099/00221287-132-10-2667. [DOI] [PubMed] [Google Scholar]
- Marquis R. E., Bender G. R., Murray D. R., Wong A. Arginine deiminase system and bacterial adaptation to acid environments. Appl Environ Microbiol. 1987 Jan;53(1):198–200. doi: 10.1128/aem.53.1.198-200.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquis R. E., Porterfield N., Matsumura P. Acid-base titration of streptococci and the physical states of intracellular ions. J Bacteriol. 1973 May;114(2):491–498. doi: 10.1128/jb.114.2.491-498.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh P. D., Keevil C. W., Ellwood D. C. Relationship of bioenergetic processes to the pathogenic properties of oral bacteria. J Dent Res. 1984 Mar;63(3):401–406. doi: 10.1177/00220345840630030901. [DOI] [PubMed] [Google Scholar]
- Poolman B., Driessen A. J., Konings W. N. Regulation of arginine-ornithine exchange and the arginine deiminase pathway in Streptococcus lactis. J Bacteriol. 1987 Dec;169(12):5597–5604. doi: 10.1128/jb.169.12.5597-5604.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimke R. T., Berlin C. M., Sweeney E. W., Carroll W. R. The generation of energy by the arginine dihydrolase pathway in Mycoplasma hominis 07. J Biol Chem. 1966 May 25;241(10):2228–2236. [PubMed] [Google Scholar]
- Vander Wauven C., Piérard A., Kley-Raymann M., Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J Bacteriol. 1984 Dec;160(3):928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]


