Abstract
Cell migration of transformed renal epithelial cells (MDCK-F) depends–in addition to cytoskeletal mechanisms–on the polarized activity of a Ca2+-sensitive K+ channel in the rear part of the cells. However, because of the lack of specific markers for this channel we are not able to determine whether a polarized distribution of the channel protein underlies its functional polarization. To determine whether the migrating MDCK-F cells have retained the ability to target K+ channels to distinct membrane areas we stably transfected the cells with the voltage-dependent K+ channel Kv1.4. Stable expression and insertion into the plasma membrane could be shown by reverse transcription–PCR, genomic PCR, Western blot, and patch-clamp techniques, respectively. The distribution of Kv1.4 was assessed with indirect immunofluorescence by using conventional and confocal microscopy. These experiments revealed that Kv1.4 is expressed only in transfected cells where it elicits the typical voltage-dependent, rapidly inactivating K+ current. The Kv1.4 protein is clustered at the leading edge of protruding lamellipodia of migrating MDCK-F cells. This characteristic distribution of Kv1.4 provides strong evidence that migrating MDCK-F cells are able to insert ion channels into the plasma membrane in an asymmetric way, which reflects the polarization of migrating cells in the plane of movement. These findings suggest that not only epithelial cells and nerve cells, but also migrating cells, can create functionally distinct plasma membrane areas.
Keywords: cell migration, protein sorting, renal epithelial cells
The proper insertion of plasma membrane proteins is a prerequisite for the functional polarization of epithelial cells and thereby for the vectorial transepithelial transport of ions and solutes. Cultured Madin–Darby canine kidney cells (MDCK) are a well characterized epithelial cell model for investigating the mechanisms of membrane protein targeting into apical and basolateral membranes. The “vertical” polarization along the apical-basolateral axis is characterized by different membrane protein and membrane lipid compositions of the apical and basolateral membrane domains. The maintenance of the epithelial polarization is based on cell contact- and tight-junction proteins that form the barrier between the two different membrane domains (1).
In contrast, MDCK cells transformed by prolonged exposure to an alkaline growth medium (MDCK-F cells) have lost this epithelial polarization. MDCK-F cells have the typical phenotype of migrating cells with a wide-spreading lamellipodium and a retracting cell body (2). Hence, instead of being polarized along an apical-basolateral axis, MDCK-F cells are polarized in the direction of migration. This “horizontal” polarization also is reflected by functional differences between the two cell poles. The mechanical force required for migration is generated by different cytoskeletal mechanisms at the cell body (rear part) and at the lamellipodium (front part) of migrating MDCK-F cells. However, this functional polarization is not only restricted to the cytoskeleton but also involves ion transport across the cell membrane and, thereby, the distribution and/or activation of ion channels and transporters. The activity of an endogenous Ca2+-sensitive K+ channel, which is a prerequisite for locomotion, is confined to the rear part of migrating MDCK-F cells (3). These findings imply that migrating MDCK-F cells are still able to differentially sort ion channels like the parent epithelial MDCK cells to generate functionally different plasma membrane domains. This interpretation finds additional support from studies on migrating fibroblasts. These cells have epithelial-like cognate routes from the trans-Golgi network to the plasma membrane (4), and they are able to selectively target a series of membrane proteins such as integrins, Na+/H+ antiporters, and viral glycoproteins (4–6). However, so far it has not been shown whether ion channels are also specifically targeted to one cell pole in migrating cells.
We could not directly determine the cellular distribution of the endogenous Ca2+-activated K+ channel because of the lack of specific antibodies. To address the question of ion channel distribution in migrating cells we thus chose to take advantage of one of the known K+ channels that can be localized by specific antibodies. Therefore, we stably transfected migrating MDCK-F cells with the voltage-dependent K+ channel Kv1.4 (7). We have chosen this K+ channel for the following reasons. (i) Because of its biophysical properties the insertion of functional Kv1.4 K+ channels into the plasma membrane of MDCK-F cells can be easily detected by whole-cell patch-clamp measurements. (ii) The Kv1.4 voltage-activated K+ channel is functionally silent and thus does not influence MDCK-F cell migration. (iii) The Kv1.4 K+ channel is polarized in neuronal cells (8) and exclusively targeted to the basolateral membrane of Kv1.4 transfected MDCK wild-type (WT) cells (9).
Our results show that the transfected Kv1.4 K+ channel is localized primarily at specific “hot spots” at the leading edge of migrating MDCK-F cells. These data demonstrate the ability of migrating MDCK-F cells to “horizontally” polarize potassium channel proteins that were shown to be an important prerequisite for directed cell migration.
MATERIALS AND METHODS
Cell Culture.
The experiments were performed with alkali-transformed MDCK-F cells (2), Kv1.4-transfected MDCK-F cells, and Kv1.4-transfected MDCK wild-type cells (kindly provided by Jean Merot, Départément de Biologie Cellulaire et Moléculaire, Gif sur Yvette, France). Cells were cultured under identical conditions at 37°C in HCO3−-buffered MEM, supplemented with 10% fetal calf serum (Biochrom, Berlin) in humidified air with 5% CO2. Kv1.4-transfected cells were always cultured in the presence of 600 μg/ml geneticin (GIBCO/BRL). For immunofluorescence and patch-clamp experiments, cells were seeded on poly-l-lysine-coated coverslips (0.1 g/liter, Serva).
Stable Transfection.
The Kv1.4/pCB6 plasmid was kindly provided by Jean Merot. The ORF of Kv1.4 (GenBank accession no. X16002) was subcloned into the eukaryotic expression vector pCB6 under control of a cytomegalovirus promoter element (10). Transfection was performed with lipofectin according to the manufacturer’s instructions (GIBCO/BRL) by using either 2 μg pCB6/Kv1.4 plasmid/ml MEM− or 2 μg pCB6 plasmid-vector/ml MEM− (mock-transfection). The initial geneticin antibiotical selection of transfected cells was performed for 3 weeks. Geneticin-resistant MDCK-F cells were cloned by single-cell dilution.
PCR Experiments and Western Blotting.
Stable transfection and expression of Kv1.4 in MDCK-F cells was controlled by reverse transcription–PCR (RT-PCR), genomic PCR, and Western Blot analysis. To avoid pCB6/Kv1.4 plasmid DNA contamination, isolated total RNA were treated with DNase (Boehringer Mannheim). Reverse transcription was performed with oligo(dT) cellulose-purified mRNA using Superscript II according to the manufacturer’s instructions (GIBCO/BRL). An additional control reaction was prepared without adding reverse transcriptase (RT−-reaction). RT-PCR was performed in a reaction mixture containing 50 mM KCl, 10 mM Tris⋅HCl, 1.5 mM MgCl2, 0.2 mM of each nucleotide, 20 pmol of each Kv1.4-specific primer, and 2.5 units of Taq polymerase (Boehringer Mannheim). The 5′ primer K1 (sense) 5′-ATGGAGGTGGCAATGGTG-3′ and the 3′ primer K3 (antisense) 5′-CACTGTAGCGGACTGAAC-3′ correspond to the sequence positions 492–511 and 997–1014 of Kv1.4, respectively. The PCR amplification steps (1 min at 94°C, 1 min at 52°C, 1 min at 72°C) were done by using a Progene thermocycler (Techne, Duxfort, England).
Genomic integration of Kv1.4/pCB6 DNA into transfected cells was verified by genomic PCR. Each 500 ng of isolated genomic DNA of Kv1.4-transfected and mock-transfected MDCK-F cells, respectively, was used for PCR amplification. The reaction conditions and the amplification protocol were identical as in the RT-PCR experiments, except for raising the MgCl2 concentration to 2.25 mM and for applying an initial heat denaturation step (94°C for 8 min) before cycling.
RT-PCR and genomic PCR amplifications were separated and visualized in 1.5% agarose gels containing ethidium bromide. The specificity of the amplifications was tested by Southern blot hybridizations by using a digoxigenin-ddUTP-labeled internal Kv1.4-specific primer, K2 (5′-GAGAGACTTGCTCACTCCA-3′; position 582–600). The oligonucleotide labeling and chemiluminescence detection of the hybridization were performed according to the manufacturer’s instructions (Boehringer Mannheim).
The Kv1.4 protein expression in MDCK cells was shown by Western blot analysis. Kv1.4-transfected MDCK-F, MDCK-WT, and mock-transfected MDCK-F cells were lysed by 0.1% Triton X-100 in morpholinopropanesulfonic acid-buffered solution, containing 1 mM phenylmethylsulfonyl fluoride. The cell lysate was concentrated with Microcon centrifugation supports (Amicon), separated in 8% polyacrylamide/SDS gels, and blotted onto a nitrocellulose membrane. The Western blot was probed with a Kv1.4-specific antibody and processed with the enhanced chemiluminescence detection system (Amersham). Pictures were digitized by using an HP-ScanJet II CX computer-scanner (Hewlett–Packard).
Patch-Clamp Experiments.
Whole-cell patch-clamp experiments were performed with Kv1.4- and mock-transfected MDCK-F cells (11). The cells were continuously perfused with Hepes-buffered Ringer solution at 37°C (122.5 mM NaCl/5.4 mM KCl/0.8 mM MgCl2/1.2 mM CaCl2/1 mM NaH2PO4/ 5.5 mM glucose/10 mM Hepes). Starting at a holding potential of −80 mV, cells were depolarized in 10-mV steps to +30 mV, with a pulse duration of 200 ms. Blocking of Kv1.4 was performed with 10 mM 4-aminopyridine. Pipettes (Hirschmann Laborgeräte, Eberstadt, Germany) had a resistance of 2–3 MΩ when filled with 140 mM KCl/1 mM MgCl2/10 mM Hepes/1 mM EGTA, pH 7.4. We used a Ca2+-free pipette solution to circumvent the activation of endogenous Ca2+-sensitive K+ channels, which might have obscured Kv1.4-induced currents. Recordings were made with a patch-clamp amplifier (L/M-EPC7; List Electronics, Darmstadt, Germany). Whole-cell currents were filtered at 1.5 kHz with an eight-pole Bessel filter (902 LPF; Frequency Devices, Haverhill, MA), digitized, and analyzed with pclamp software (Axon Instruments, Foster City, CA).
Immunofluorescence Analysis.
Kv1.4- and mock-transfected MDCK-F cells were immunoprobed with a polyclonal antiserum against a fusion protein of Kv1.4 (amino acid position 578–655) and glutathione S-transferase. For indirect immunofluorescence, cells growing on coated coverslips were washed with PBS (containing 1 mM CaCl2 and MgCl2, pH 7.4) and fixed with 3% formaldehyde in PBS for 15 min. Permeabilization was performed with 0.5% Triton X-100 in PBS for 10 min. After washing twice with PBS the cells were blocked with 1% BSA for 30 min. Kv1.4 antibodies were diluted in PBS/BSA, and cells were incubated from 30 min to 3 hr. After washing, cells were probed with Fc-specific Cy3-conjugated F(ab′)2 fragments of anti-rabbit IgG antibodies (Dianova, Hamburg, Germany) for 30 min. All steps were performed at room temperature. Cells were mounted in Mowiol (Hoechst Pharmaceuticals) and examined by using an inverted microscope (Zeiss IM35, Oberkochen, Germany) or a confocal microscope (Bio-Rad MRC600).
RESULTS
Monitoring the Stable Transfection of Kv1.4 in MDCK-F Cells.
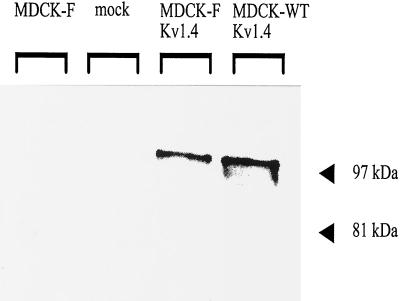
The mRNA expression and the genomic integration of Kv1.4 in transfected MDCK-F cells was performed by RT-PCR and genomic PCR amplifications, respectively. Kv1.4-specific oligonucleotides yielded a single PCR product, which had the expected size of 522 bp. The specificity of this fragment was proven further by internal hybridization with a third Kv1.4-specific oligonucleotide (Fig. 1, lanes 3 and 7). In contrast, mock-transfected and nontransfected MDCK-F cells did not express Kv1.4 (Fig. 1, lanes 1, 2, 6, and 8). The same results were obtained in SDS/PAGE and Western blot experiments (Fig. 2). The Kv1.4-specific antibody recognized a 99-kDa protein band only in whole-cell lysates of Kv1.4-transfected MDCK-F cells. Kv1.4-transfected MDCK-WT cells served as a positive control (9). This 99-kDa band corresponded in size to the functional posttranslationally processed and glycosylated Kv1.4 α-subunit protein (12, 13). In contrast to the findings of Le Maout et al. (9), we were not able to detect an additional band with a molecular mass of around 80 kDa corresponding to the unglycosylated Kv1.4 protein. This could be because we did not use Na+-butyrate for enhanced Kv1.4 expression. The application of this expression stimulus led to strong Kv1.4 protein synthesis, which could be monitored by immunofluorescence (data not shown). Taken together, these data clearly showed that we successfully transfected MDCK-F cells with the neuronal K+ channel Kv1.4.
Figure 1.
(A) RT-PCR and genomic PCR amplification products of transfected MDCK-F cells, separated in an ethidium bromide-stained agarose gel. Loading: M, marker (506 and 1,018 bp). Lanes: 1–5, RT-PCR of MDCK-F cells; 1, nontransfected cells; 2, mock-transfected cells; 3, Kv1.4-transfected cells; 4, H2O control; 5, RT− control; 6–8, genomic PCR of MDCK-F cells; 6, nontransfected cells; 7, Kv1.4-transfected cells; 8, mock-transfected cells. (B) Southern blot hybridization with a digoxigenin-ddUTP-labeled Kv1.4-specific oligonucleotide. Only the stably transfected cell clones in lanes 3 and 7 exhibited the Kv1.4-specific, single 522-bp band in RT-PCR and genomic PCR experiments, as shown in A.
Figure 2.
Western blot detection of Kv1.4 expression in transfected MDCK-F cells. The blotted cell protein lysates were probed with an anti-Kv1.4 polyclonal antibody and detected with the enhanced chemiluminescence system (Amersham). Loading: lane 1, nontransfected MDCK-F cells; lane 2, mock-transfected MDCK-F cells; lane 3, Kv1.4-transfected MDCK-F cells; lane 4, Kv1.4-transfected MDCK-wild-type cells as a positive control for Kv1.4 expression. The protein band of approximately 99 kDa representing Kv1.4 was detected only in Kv1.4-transfected cells.
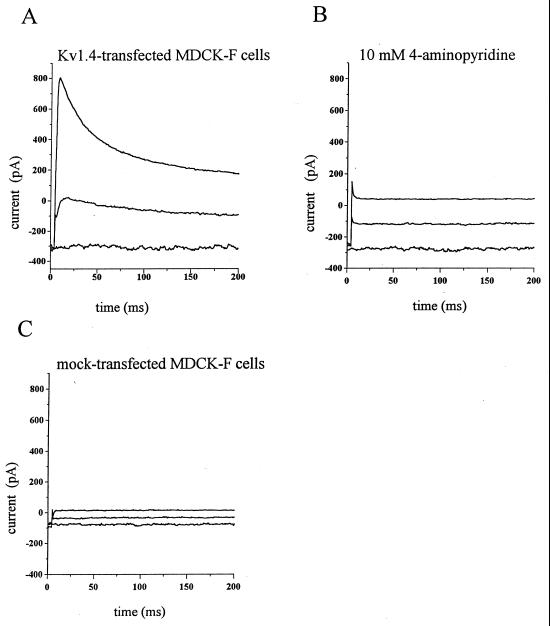
The membrane insertion of Kv1.4 was proven by patch-clamp experiments in the whole-cell configuration. Expression of the Kv1.4 potassium channel in Xenopus oocytes generated a characteristic, fast-inactivating outward current that was activated by membrane depolarization (7). Because of the absence of such a current in mock-transfected MDCK-F cells, functional Kv1.4 was easily recognized. Starting from a holding potential of −80 mV we depolarized the membrane potential of MDCK-F cells in 10-mV steps. Depolarization pulses lasted 200 ms. The original traces in Fig. 3A show the typical Kv1.4-mediated fast inactivating current. The channel activated between −60 and −50 mV followed by rapid inactivation (Fig. 3A). Blocking Kv1.4 channel with the K+ channel inhibitor 4-aminopyridine (10 mM) abolished this current response (Fig. 3B). The current block was reversible (data not shown). As expected, mock-transfected MDCK-F cells did not express this voltage-activated K+ current (Fig. 3C). Thus, the electrophysiological data confirmed our molecular analysis for functional Kv1.4 expression and membrane insertion in transfected MDCK-F cells.
Figure 3.
Transient outward K+ currents induced by depolarization of the membrane potential measured in the whole-cell configuration of the patch-clamp technique. The depolarizing pulses started from a holding potential of −80 mV. The elicited currents in A–C correspond to membrane depolarizations to −80 mV, −40 mV, and −10 mV, respectively. (A) Whole-cell current measurement of a Kv1.4-transfected MDCK-F cell under control conditions. The Kv1.4 current was activated at −60 to −50 mV, and it decayed exponentially within 50 ms. (B) The same cell as in A in the presence of 10 mM 4-aminopyridine. (C) No Kv1.4-like current could be recorded in mock-transfected MDCK-F cells.
Immunolocalization of Kv1.4.
To address the question of whether Kv1.4-transfected MDCK-F cells were able to differentially target the Kv1.4 protein we determined the protein’s cellular localization by means of indirect immunofluorescence. The Kv1.4-specific polyclonal antibody used in this study is directed against the intracellular C terminus of Kv1.4 and previously has been employed to demonstrate the polarized expression of Kv1.4 in neurons and in transfected MDCK-WT cells (8, 9).
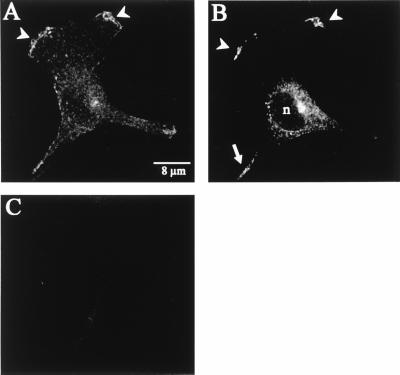
The transfected Kv1.4 K+ channel could be clearly identified in the leading edge of the lamellipodium. The staining disclosed clusters along the circumference of the lamellipodium (Fig. 4A, white arrows). There were apparently “hot spots” of Kv1.4 in areas where the lamellipodium protruded. In addition, Kv1.4 was always present in the perinuclear region. Na+-butyrate-induced overexpression of Kv1.4 led to a strong increment of this signal (data not shown). As expected, we found neither Kv1.4-specific nor unspecific binding of the primary or secondary antibody in mock-transfected MDCK-F cells (Fig. 4B). These experiments with conventional, indirect immunofluorescence suggested that Kv1.4 proteins were preferentially targeted to the leading edge of actively protruding lamellipodia. To confirm this conclusion and to better resolve the perinuclear staining we performed additional experiments with confocal immunofluorescence microscopy. As depicted in Fig. 5 A and B, confocal horizontal sections proved the distinct distribution of Kv1.4 in MDCK-F cells. Fig. 5A corresponded to the lowermost section of a Kv1.4-transfected MDCK-F cell. Only the extreme periphery of the cell nucleus was imaged in this scan section. The presented K+ channel was unequivocally concentrated in the ruffled membrane at the extended part of the leading edge. Kv1.4 was observed neither in the plasma membrane of the cell body (Fig. 5A) nor in ruffles that moved backwards on the lamella toward the cell body (not shown). Fig. 5B underscores this findings and enabled us to ascribe the earlier-mentioned perinuclear staining of Kv1.4 to an intracellular localization. The upper confocal view of the same cell (approximately 400 nm above the first section) clearly disclosed the unstained nucleus (n) and the perinuclear Kv1.4-localization, which was likely to represent the endoplasmic reticulum and/or the Golgi network. Mock-transfected MDCK-F cells showed no unspecific binding of the primary or secondary antibody (Fig. 5C). Sometimes we detected signals in thin cell extensions near the rear part of the cell (Fig. 5B, white arrow). Such cell structures often showed unspecific binding of tested, fluorescently labeled antibodies. Although we never detected Kv1.4 in the plasma membrane of the cell body we could not completely rule out some minor insertion of Kv1.4 at this cell pole.
Figure 4.
Immunofluorescence micrographs of stably transfected MDCK-F cells. (A) Kv1.4 was localized at the leading edge of the protruding lamellipodium and in the cell body of fixed and permeabilized cells. Kv1.4 is not evenly lining the leading edge, but it has a clustered appearance (arrows). (B) In mock-transfected MDCK-F cells there is no unspecific binding of the primary or secondary antibody.
Figure 5.
Confocal microscopy of immunostained Kv1.4-transfected MDCK-F cells. (A) Lowermost scan section. Only the nuclear periphery was imaged in this section. Kv1.4 was strictly localized at the leading edge of the lamellipodium (arrowheads). No accumulation of the Kv1.4 protein could be detected in plasma membrane surrounding the cell body. (B) Upper section of the same cell as in A (approximately 400 nm above). Kv1.4 was clearly detected at the leading-edge membrane (arrowheads), as in A. The intracellular staining was restricted to the endoplasmic reticulum and/or to the Golgi network surrounding the nucleus. The fluorescently labeled cell extension is marked with an arrow. This staining is most likely because of unspecific binding of the secondary antibody. (C) Mock-transfected MDCK-F cells showed no signal.
Our observations of the Kv1.4 distribution indicated a clustered appearance of the transfected K+ channel at the leading edge of actively migrating MDCK-F cells. Therefore, we conclude that the transformed and migrating MDCK-F cells are able to differentially localize the Kv1.4 K+ channel. This indicates that not only epithelial and nerve cells but also migrating cells are able to generate functionally specialized plasma membrane areas.
DISCUSSION
The aim of our study was to examine whether transformed and migrating MDCK-F cells still have the capacity to target ion channels to specific regions within the cell membrane. Previous functional studies have shown that MDCK-F cells are polarized with respect to the activity of an endogenous Ca2+-dependent K+ channel whose activity is higher in the cell body than in the lamellipodium (3, 14). These studies provide strong evidence for a functional link between localized volume regulation mediated by ion channels and transporters and cytoskeletal rearrangements during MDCK-F cell migration. The confinement of the endogenous Ca2+-sensitive K+ channel to the cell body of MDCK-F cells can be due to either preferential activation of the channel at this site, e.g., channel activation by intracellular Ca2+ gradients (15), and/or the polarized localization of the K+ channel protein in the plasma membrane of the cell body. The latter possibility would require that MDCK-F cells are able to specifically sort membrane proteins to the cell body or to the lamellipodium. The role of differential membrane protein targeting for the functional asymmetry of migrating cells has been shown in several studies for integrins, receptors for chemoattractants, and the Na+/H+ exchanger (5, 6, 16). These proteins are sorted into the leading edge of the protruding lamellipodium where they exert important roles in cell crawling. However, to date it had not been determined whether ion channels also can be specifically directed to the cell body or lamellipodia of migrating cells. To address the question of ion channel distribution in migrating cells we stably transfected MDCK-F cells with the well characterized, voltage-activated Kv1.4 K+ channel (7). The permanent expression of Kv1.4 in transfected MDCK-F cells was shown with molecular and electrophysiological experiments. The activation of Kv1.4 upon membrane depolarization and its fast inactivation prove its functional expression in transfected MDCK-F cells. The striking concentration of Kv1.4 at the leading edge of protruding lamellipodia raises the question of what mechanisms underlie this anisotropy. Comparing the distribution of Kv1.4 in MDCK-F cells with that in transfected MDCK-wild-type cells and in neurons may give us some clues. Kv1.4 is detected mainly in axons and presynaptic nerve terminals and in the basolateral membrane of transfected MDCK-wild-type cells (8, 9). The plus ends of microtubules in axons as well as in MDCK-wild-type cells are oriented toward the respective targeting sites of Kv1.4 (17, 18). Interestingly, the plus ends of microtubules have been shown to be oriented toward the lamellipodium in migrating fibroblasts (19). Thus, it is tempting to speculate that the sorting of Kv1.4 in migrating MDCK-F, in transfected epithelial MDCK cells, and in neurons can be attributed to microtubules and microtubule-associated motor proteins. Our experiments raise the question of whether the targeting of ion channels to the lamellipodium of MDCK-F cells and to the basolateral cell membrane of MDCK-wild-type cells is based on a similar mechanism. Recently, it was shown that microtubule motor proteins play a role in basolateral sorting (20) and that basolateral-targeted proteins are represented mainly in the outgrowing lamellipodium of fibroblasts (21). Therefore, we speculate whether the former basolateral plasma membrane region of polarized MDCK-wild-type cells corresponds functionally to the plasma membrane region of the leading lamellipodium of migrating MDCK-F cells.
The clustering of Kv1.4 in the membrane of the leading edge could also be because of the assembly with other proteins in this area. Studies in transfected COS cells have indicated a clustering of Kv1.4 by coexpression with the “postsynaptic density protein” PSD-95 (22, 23). However, the localization of endogenous proteins with functionally similar protein domains like the zonula occludens proteins (ZO-1 and ZO-2) in MDCK-F cells is not known at present (24). Therefore, investigations focusing on the role of cytoskeletal proteins in ion channel targeting in migrating MDCK-F cells should be performed. First results on the localization of the endogenous Na+/K+ ATPase, which is associated with cell-adhesion and cytoskeletal proteins (25), indicate that, in contrast to Kv1.4, it manifests an unpolarized distribution in MDCK-F cells (unpublished observation).
In summary, we have shown that transformed and migrating MDCK-F cells are able to specifically direct and maintain a neuronal K+ channel to the leading edge of the lamellipodium. Thus, migrating cells appear to be able to create functionally different membrane domains. Such a behavior may have important implications for cell migration because the ion transport across the cell membrane at the cell body and at the lamellipodium has to support different cytoskeletal mechanisms: retraction of the cell body and protrusion of the lamellipodium. These processes may be tentatively translated to “cell shrinkage” and “cell swelling,” both of which require a specific setting, activation, and/or distribution of distinct ion channels and transporters in the plasma membrane.
Acknowledgments
We are grateful to Jean Merot for providing the pCB6/Kv1.4 construct and the stable transfected MDCK wild-type cells and to Dr. S. Silbernagl’s group for many fruitful discussions and the excellent working conditions. This work was supported by the SFB 176 grant (University of Würzburg, Germany) and a postdoctoral fellowship of J.R. by the Deutsche Forschungsgemeinschaft.
ABBREVIATION
- RT-PCR
reverse transcription–PCR
References
- 1.Matlin K S, Caplan M J. In: The Kidney: Physiology and Pathophysiology. Seldin D W, Giebisch G, editors. New York: Raven; 1992. pp. 447–473. [Google Scholar]
- 2.Oberleithner H, Westphale H J, Gassner B. Pflügers Arch. 1991;419:418–420. doi: 10.1007/BF00371126. [DOI] [PubMed] [Google Scholar]
- 3.Schwab A, Gabriel K, Finsterwalder F, Folprecht G, Greger R, Kramer A, Oberleithner H. Pflügers Arch. 1995;430:802–807. doi: 10.1007/BF00386179. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimori T, Keller P, Roth M G, Simons K. J Cell Biol. 1996;133:247–256. doi: 10.1083/jcb.133.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawson M A, Maxfield F R. Nature (London) 1995;377:75–79. doi: 10.1038/377075a0. [DOI] [PubMed] [Google Scholar]
- 6.Grinstein S, Woodside M, Waddell T K, Downey G P, Orlowski J, Pouysseguer J, Wong O C P, Foskett J K. EMBO J. 1993;12:5209–5218. doi: 10.1002/j.1460-2075.1993.tb06216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stühmer W, Ruppersberg J P, Schroter K H, Sakmann B, Stocker M, Giese K P, Perschke A, Baumann A, Pongs O. EMBO J. 1989;8:3235–3244. doi: 10.1002/j.1460-2075.1989.tb08483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veh R W, Lichtinghagen R, Sewing S, Wunder F, Grumbach I M, Pongs O. Eur J Neurosci. 1995;7:2189–2205. doi: 10.1111/j.1460-9568.1995.tb00641.x. [DOI] [PubMed] [Google Scholar]
- 9.LeMaout S, Sewing S, Coudrier E, Elalouf J M, Pongs O, Merot J. Mol Membr Biol. 1996;13:143–147. doi: 10.3109/09687689609160590. [DOI] [PubMed] [Google Scholar]
- 10.Brewer C B, Roth M G. J Cell Biol. 1991;114:413–421. doi: 10.1083/jcb.114.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamill O P, Sakmann B. Nature (London) 1981;294:462–464. doi: 10.1038/294462a0. [DOI] [PubMed] [Google Scholar]
- 12.Sheng M, Tsaur M L, Jan Y N, Jan N Y. Neuron. 1992;9:271–284. doi: 10.1016/0896-6273(92)90166-b. [DOI] [PubMed] [Google Scholar]
- 13.Scott V E, Muniz Z M, Sewing S, Lichtinghagen R, Parcej D N, Pongs O, Dolly J O. Biochemistry. 1994;33:1617–1623. doi: 10.1021/bi00173a001. [DOI] [PubMed] [Google Scholar]
- 14.Schwab A, Wojnowski L, Gabriel K, Oberleithner H. J Clin Invest. 1994;93:1631–1636. doi: 10.1172/JCI117144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwab A, Finsterwalder F, Kersting U, Danker T, Oberleithner H. Pflügers Arch. 1997;434:70–76. doi: 10.1007/s004240050364. [DOI] [PubMed] [Google Scholar]
- 16.Sullivan S J, Daukas G, Zigmond S H. J Cell Biol. 1984;99:1461–1467. doi: 10.1083/jcb.99.4.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cid-Arregui A, Parton R G, Simons K, Dotti C G. J Neurosci. 1995;15:4259–4269. doi: 10.1523/JNEUROSCI.15-06-04259.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer E H K, Simons K. J Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vale R D. Curr Opin Cell Biol. 1990;2:15–22. doi: 10.1016/s0955-0674(05)80025-0. [DOI] [PubMed] [Google Scholar]
- 20.Lafont F, Burkhardt J K, Simons K. Nature (London) 1994;372:801–803. doi: 10.1038/372801a0. [DOI] [PubMed] [Google Scholar]
- 21.Peränen J A, Auvinen P, Virta H, Wepf R, Simons K. J Cell Biol. 1996;135:153–167. doi: 10.1083/jcb.135.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim E, Niethammer M, Rothschild A, Jan Y N, Sheng M. Nature (London) 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 23.Kim E, Sheng M. Neuropharmacology. 1996;35:993–1000. doi: 10.1016/0028-3908(96)00093-7. [DOI] [PubMed] [Google Scholar]
- 24.Jesaitis L A, Goodenough D A. J Cell Biol. 1994;124:949–961. doi: 10.1083/jcb.124.6.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson W J, Hammerton R W. J Cell Biol. 1989;108:893–902. doi: 10.1083/jcb.108.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]