Abstract
In Arabidopsis thaliana, signal transduction of the hormone ethylene involves at least two receptors, ETR1 and ERS, both of which are members of the two-component histidine protein kinase family that is prevalent in prokaryotes. The pathway also contains a negative regulator of ethylene responses, CTR1, which closely resembles members of the Raf protein kinase family. CTR1 is thought to act at or downstream of ETR1 and ERS based on double mutant analysis; however, the signaling mechanisms leading from ethylene perception to the regulation of CTR1 are unknown. By using the yeast two-hybrid assay, we detected a specific interaction between the CTR1 amino-terminal domain and the predicted histidine kinase domain of ETR1 and ERS. We subsequently verified these interactions by using an in vitro protein association assay(s). In addition, we determined that the amino-terminal domain of CTR1 can associate with the predicted receiver domain of ETR1 in vitro. Based on deletion analysis, the portion of CTR1 that interacts with ETR1 roughly aligns with the regulatory region of Raf kinases. These physical associations support the genetic evidence that CTR1 acts in the pathway of ETR1 and ERS and suggest that these interactions could be involved in the regulation of CTR1 activity.
Keywords: hormone, signal transduction, two-component regulators, protein–protein interactions, yeast
Ethylene has numerous effects on plant growth and development, such as fruit-ripening, organ abscission, seed germination, senescence, and the induction of certain defense responses (1). The cloning of genes corresponding to several ethylene-response mutants in Arabidopsis has begun to provide us with insight into the molecular basis of ethylene signal transduction (2–6). In Arabidopsis, there are at least two ethylene receptors, ETR1 and ERS, that are similar to each other. Plants appear to have multiple ethylene receptors; several different ETR1 and ERS homologs have been cloned recently from Arabidopsis (7) and tomato (8, 9). The ETR1 and ERS gene products are predicted to function as histidine protein kinases based on their sequence similarities with the two-component regulator family. The two-component regulators are prevalent in prokaryotes (10) and are starting to be identified in various eukaryotes, including fungi, slime mold, and higher plants (11, 12); one of these, the Arabidopsis CKI1 protein, is potentially a receptor for cytokinin (13). The basic two-component system consists of a histidine autokinase sensor component that directs the activity of a cognate response regulator, which in turn controls downstream signaling (10). After autophosphorylation of the sensor kinase, the phosphoryl group is transferred from the histidine of the sensor kinase to an aspartic acid residue in the receiver domain of the response regulator (10). The ETR1 protein is an example of a hybrid histidine kinase because it contains both a histidine kinase domain and a receiver domain (2). ERS, in contrast, contains only a histidine kinase domain (3). The significance of having an attached receiver domain is unknown, and whether there are separate cognate response regulators for ETR1 and ERS remains to be seen. The amino-terminal portions of ETR1 and ERS, which are 75% identical, do not have significant similarity to any sequences in the current databases. This region of ETR1 and ERS has been shown to bind ethylene reversibly (ref. 14; G. E. Schaller, A. E. Hall, and A. B. Bleecker, personal communication), providing strong evidence that ETR1 and ERS are ethylene receptors. All of the known missense mutations (isolated or introduced) in ETR1 and ERS cause dominant ethylene insensitivity and reside in this amino-terminal region (2, 3).
CTR1 is thought to act at or downstream of both ETR1 and ERS based on double mutant analysis (3, 15). CTR1 is a negative regulator of the ethylene-response pathway because ctr1 null mutants exhibit constitutive ethylene responses even in the absence of ethylene (4). The deduced CTR1 protein sequence is most similar to the Raf family of serine/threonine protein kinases (41% amino acid identity in the kinase domain), suggesting that CTR1 may act in a mitogen-activated protein (MAP) kinase cascade (4). Thus, in the emerging view of ethylene signal transduction in Arabidopsis, ETR1 and ERS are predicted to be histidine autokinases whereas CTR1 is thought to function as a MAP kinase (MAPK) kinase kinase. The signaling mechanisms by which these distinct components might be coupled in the ethylene-response pathway have yet to be elucidated, however. In this study, we used the yeast two-hybrid assay as well as in vitro protein association assays to demonstrate that the presumed regulatory domain of CTR1 can interact directly with the histidine kinase domain of ETR1 and ERS. We also present evidence that CTR1 can associate in vitro with the receiver domain of ETR1. These results provide physical evidence that CTR1 acts in the pathway of both receptors and indicate that ethylene signaling may involve protein–protein interactions between two-component receptors and a Raf-like kinase.
MATERIALS AND METHODS
Yeast Strain, Transformation, and Growth.
Saccharomyces cerevisiae strain L40 was used in these studies. The partial genotype of L40 is MATa his3–200 trp1–901 leu2–3,-112 ade2 LYS2∷(lexAop)4-HIS3 URA3∷(lexAop)8-lacZ GAL4 (16). Yeast transformation was performed by using a lithium acetate-based protocol (17). Standard media were used for growth (18). Glucose was used as the carbon source for transformation, and sucrose was used as the carbon source for the two-hybrid assay.
Plasmid Constructions.
For the two-hybrid assay, protein fusions to the bacterial repressor LexA DNA-binding domain (DB) were made by using the plasmid plexA-NLS, which is a modified form of pBTM116 (16) containing the simian virus 40 nuclear localization signal (a gift of S. Hollenberg, Vollum Institute, Oregon Health Sciences University). Fusions to the Gal4 transcription activation domain (AD) were made by using the plasmids pGAD424 (19) and/or pACTII (20). Restriction fragments of ETR1 (PflMI–AflII; PflMI–XhoI) and ERS (StyI–KpnI) cDNA clones were subcloned into plexA-NLS, and a restriction fragment of CTR1 (DdeI–DdeI) was subcloned into pGAD424 after modification of fragment ends to maintain the reading frame. A fragment encoding the CTR1 kinase domain (residues 538–821) was cloned into pGAD424 after amplification from a cDNA clone by using the PCR (21) and primers designed to incorporate restriction sites in the correct reading frame. Sequences encoding smaller CTR1 regions were cloned similarly into pACTII after amplifying the desired sequences by PCR.
For the in vitro protein association assays, constructs expressing bacterial maltose-binding protein (MBP) fusions were made in vector pMAL-c2 (New England Biolabs). DNA fragments encoding the CTR1 amino-terminal domain (CTR153–568), the ETR1 histidine kinase domain (ETR1293–610), and the ETR1 histidine kinase domain plus receiver domain (ETR1293–729) were subcloned from the two-hybrid vectors above such that the two-hybrid constructs and bacterial constructs expressed the same sequences for each protein. A DNA fragment encoding the ETR1 receiver domain (ETR1604–738) was PCR-amplified from a cDNA clone by using primers designed to incorporate restriction sites in the correct reading frame. For the CKI1 receiver domain (CKI1981–1122), a restriction fragment (EcoRI to 3′ polylinker site XbaI) was subcloned from a full length CKI1 cDNA clone.
Constructs for generating radiolabeled, in vitro-translated CTR1 polypeptides were made in pBluescript SKII (Stratagene). A DNA fragment encoding the CTR1 amino-terminal domain (residues 53–568) first was amplified by PCR using primers designed to incorporate a start and stop codon at the appropriate locations. The CTR1 kinase domain (residues 538–821) was subcloned as a BamHI–XbaI restriction fragment from the two-hybrid pGAD424 construct described above (which already contained an appropriate start and stop codon).
Yeast Two-Hybrid Assay.
Yeast strain L40 was transformed simultaneously with a TRP+ LexA (DB) construct and a LEU+ Gal4 (AD) construct. The ability to drive expression of the yeast HIS3 reporter gene was tested for by growing transformants on selective medium lacking tryptophan, leucine, and histidine. LacZ reporter gene activity in the yeast cells was monitored visually by the 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) filter assay (16) and was quantified by measuring β-galactosidase activity in log-phase liquid cultures as described (22). For the X-Gal filter assay, colonies were lifted onto supported nitrocellulose filters and then cracked open by freezing the filters in liquid nitrogen two-to-three times for 1 min. The thawed filters then were placed on Whatman grade 3MM paper soaked in Z buffer (60 mM Na2HPO4/40 mM NaH2PO4/10 mM KCl/1 mM MgSO4, pH 7.0) plus 0.1% Triton X-100 and 1.5 mg/ml X-Gal. The filters were incubated at room temperature, and the appearance of blue color was monitored over several hours.
To determine the relative levels of fusion proteins expressed in the yeast cells, yeast cultures were grown to late log-phase in medium lacking tryptophan and leucine, and then the cells were harvested by pelleting. Crude extracts were made by boiling the cells in protein sample buffer followed by vortex mixing in the presence of 425- to 600-μm glass beads (Sigma). Aliquots of equivalent cell numbers were loaded onto 7.5% or 10% SDS/PAGE gels and were separated by electrophoresis. Immunoblot analysis was performed as described (22) by using mAbs to either LexA or the transcription activation domain of Gal4 (residues 768–881) as the primary antibody. The secondary antibody used was horseradish peroxidase-conjugated goat antiserum to mouse Igs (Amersham), and immune complexes were detected by using the enhanced chemiluminescence kit (Amersham).
Protein Association Assay in Yeast Extracts.
Constructs expressing MBP fusions were maintained in Escherichia coli strain BL21-DE3 and were induced with 0.3 mM isopropyl β-d-thiogalactopyranoside for 2.5 hr. Extracts were prepared by freezing and sonication. The fusion proteins were purified on amylose-containing beads as described by the manufacturer (New England Biolabs), and samples were visualized on SDS/PAGE gels stained with Coomassie blue. Cultures of yeast strain L40 carrying either plexA-NLS constructs or pGAD424 constructs were grown in selective medium to late log-phase, and then extracts were made by disrupting the cells by vortex mixing with glass beads in the presence of immunoprecipitation buffer (0.6 M sorbitol/50 mM Tris, pH 7.5/140 mM NaCl/5 mM EDTA/10 mg/ml BSA/0.06% Triton) (23). To the yeast extracts, we added equivalent amounts of either MBP or MBP fusion attached to the amylose resin. The protein-containing resin was mixed by rotation with the yeast extract for 1 hr at 4°C and then pelleted. The pellet was washed twice with immunoprecipitation buffer and then three times in immunoprecipitation buffer lacking BSA. Samples were suspended in protein sample buffer, boiled, and then separated on 7.5% SDS/PAGE gels. Immunoblot analysis was performed as described above. Before blocking with nonfat milk, the filters were stained with Ponceau S (Sigma) both to visualize the total protein population (24) and to ensure that similar amounts of MBP and/or MBP fusions were present.
In Vitro Protein Association Assay.
Constructs expressing MBP fusions were maintained in E. coli strain BL21-DE3 carrying the pLysS vector (Novagen). Fusion proteins were induced as described above, and extracts were prepared by subjecting the cells to two freeze/thaw cycles followed by a brief sonication. As above, the fusion proteins were purified on amylose-containing beads, and samples were visualized on SDS/PAGE gels stained with Coomassie blue. Radiolabeled test proteins were synthesized with the TnT T7 Quick Coupled Transcription/Translation System (Promega) by using [35S]-methionine as the radiolabel. Assays were performed by mixing 5 μg of MBP fusion attached to amylose beads with either 5 μl or 25 μl of radiolabeled test protein in the presence of bead-binding buffer (50 mM KH2PO4/150 mM KCl/1 mM MgCl2/10% glycerol/10 mg/ml BSA/0.5% Triton X-100). Samples were rotated for 1.5 hr at 4°C, were pelleted, and were washed three times with bead-binding buffer lacking BSA. The samples were suspended in protein sample buffer, were boiled, and then were separated on 8.5% SDS/PAGE gels. After fixation, gels were soaked in Amplify (Amersham), and the test proteins were visualized by autoradiography.
RESULTS
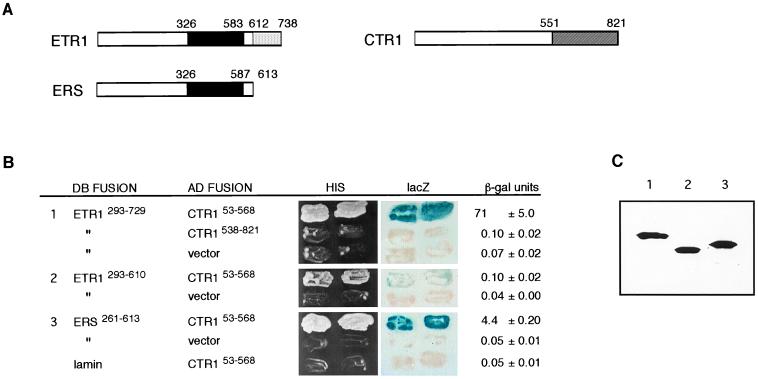
To test whether the Arabidopsis ETR1 and CTR1 proteins can associate physically, we used a LexA version of the yeast two-hybrid assay (25). The following protein fusions were expressed simultaneously in the yeast L40 reporter strain: a fusion of the bacterial repressor LexA DB with either the ETR1 histidine kinase domain (DB–ETR1293–610) or the ETR1 histidine kinase domain plus receiver domain (DB–ETR1293–729) and a fusion of the yeast Gal4 AD to almost the entire amino-terminal domain of CTR1 (AD–CTR153–568). The ETR1 and CTR1 protein domains are depicted in Fig. 1A. The detection of protein interactions was based on the expression of two chromosomally located reporter genes: HIS3, which confers growth in the absence of histidine, and lacZ, which produces β-galactosidase. We found that yeast transformants containing either of the DB–ETR1 constructs together with the AD–CTR153–568 construct were His+ (histidine prototrophs) and produced blue color in the presence of the β-galactosidase substrate X-Gal, indicating that the histidine kinase region of ETR1 can interact with the amino-terminal domain of CTR1 (Fig. 1B). The interaction was specific because transformants expressing the DB–ETR1 fusions and either AD alone or AD fused to the CTR1 serine/threonine kinase domain (AD–CTR1538–821) were His− (histidine auxotrophs) (for example, Fig. 1B). In addition, transformants expressing the DB–ETR1 fusions and AD fused to the S. cerevisiae Gβ protein Ste4p (22) (which was included as a negative control) were also His− (data not shown). Similarly, there was no detectable interaction among AD–CTR153–568 and either a human lamin negative control (16) (Fig. 1B), the Arabidopsis protein phosphatase TOPPZ (ref. 26 and data not shown), or the amino-terminal region of ETR1 (residues 1–293 fused to the amino terminus of LexA) (data not shown). An interaction test using the reciprocal two-hybrid fusions of ETR1 and CTR1 could not be performed because yeast transformants expressing DB–CTR153–568 alone were found to exhibit reporter gene activity.
Figure 1.
Interaction of ETR1 and ERS proteins with the CTR1 protein in the yeast two-hybrid assay. (A) Schematic structures of ETR1, ERS, and CTR1 proteins. Domains are approximated based on sequence homology with two-component proteins and Raf kinases, respectively; the solid black region in ETR1 and ERS indicates the histidine kinase domain, the light-shaded region in ETR1 indicates the receiver domain, and the shaded region of CTR1 indicates the serine/threonine kinase domain. (B) Two-hybrid constructs and results. Regions of ETR1, ERS, and CTR1 that were fused to the DB or AD are indicated by residue numbers. The vector is pGAD424, which produces AD alone. Human lamin was used as a nonspecific control. HIS shows growth of transformants on medium lacking histidine. lacZ shows the X-Gal filter assay of the same transformants (grown in the presence of histidine) after 1 hr at room temperature. β-gal units gives β-galactosidase activity in modified Miller units (22). Three transformants of each were measured, and the average ± the SD is presented. (C) Immunoblot analysis of the DB fusion proteins present in yeast double transformants. The same transformants that were tested in the above assays were analyzed for relative levels of DB fusion proteins. We tested two transformants of each and observed similar levels for each pair; one representative of each is shown. Immunoblot analysis of the AD fusions is shown in Fig. 4.
The interaction of AD–CTR153–568 with DB–ETR1293–729 (which contains the receiver domain) was much stronger than that with DB–ETR1293–610 (which lacks the receiver domain); transformants carrying AD–CTR153–568 and DB–ETR1293–729 grew faster in the absence of histidine, produced blue color sooner in the X-Gal filter assay (5 min vs. 3–4 hr), and contained much higher β-galactosidase activity than transformants carrying AD–CTR153–568 and DB–ETR1293–610 (Fig. 1B). This difference is unlikely to be caused by variation in the amount of DB–ETR1 protein because immunoblotting revealed similar levels of the various DB fusion proteins in the yeast cells (Fig. 1C). One possible explanation is that the ETR1 receiver domain itself might interact with AD–CTR153–568. We were unable to test for this interaction in the two-hybrid assay because, as with DB–CTR153–568, transformants expressing only DB fused with the ETR1 receiver domain exhibited reporter gene activity. To test for an interaction between CTR1 and the ETR1 receiver domain, we used an in vitro assay, which is described later.
Next, we asked whether AD–CTR153–568 can interact similarly with the putative ethylene receptor ERS. ERS has 58% amino acid sequence identity with ETR1 in the histidine kinase domain, but ERS lacks a receiver domain (3) (Fig. 1A). We tested for a two-hybrid interaction between AD–CTR153–568 and a fusion of DB to the ERS histidine kinase domain (DB–ERS261–613) as described for ETR1. Again, the yeast transformants were His+ and blue (Fig. 1B). The ability of the cells to grow in the absence of histidine and to turn blue in the X-Gal filter assay was indistinguishable from that observed with the DB–ETR1293–729 transformants. However, comparison of β-galactosidase activity indicated that the interaction with DB–ERS261–613 is weaker than that with DB–ETR1293–729, perhaps because of the absence of a receiver domain. As before, the level of DB–ERS261–613 fusion protein expressed in the yeast cells was similar to that of the DB–ETR1 fusions (Fig. 1C). CTR1 therefore can associate with both ethylene receptors, ETR1 and ERS, in the yeast two-hybrid assay.
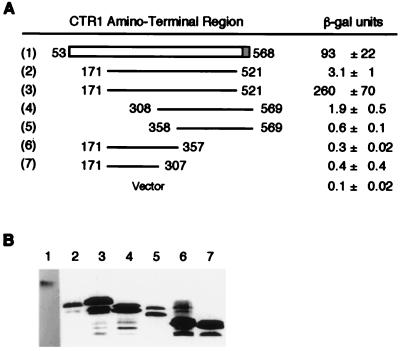
We attempted to define the interacting region of CTR1 by testing truncated versions of AD–CTR153–568 in the two-hybrid assay with DB–ETR1293–729 (Fig. 2). We found that a truncated version (CTR1 residues 171–521) and smaller CTR1 fusions were expressed poorly in the yeast cells, so we switched from using the pGAD424 two-hybrid vector to using pACTII, which has a stronger promoter (19). When highly expressed, the AD–CTR1171–521 fusion protein interacted strongly, and AD–CTR1308–569 interacted weakly with DB–ETR1293–729 based on measurements of β-galactosidase activity (Fig. 2). Little to no interaction was detected with smaller fusions (Fig. 2). Based on the less stringent assay of cell growth in the absence of histidine, AD–CTR1308–569 interacted with DB–ETR1293–729 nearly as well as did AD–CTR1171–521, in that both gave rise to thick patches of yeast cells within 2 days on selective medium lacking histidine. Very little growth was observed for fusions with CTR1 residues 171–357 or 171–307 (data not shown). Taken together, these results suggest that the minimal interacting region of CTR1 has an amino-terminal border between residues 171 and 308 and a carboxyl-terminal border between residues 521 and 569.
Figure 2.
Localization of the region of CTR1 that associates with ETR1. (A) Results of the yeast two-hybrid assay. β-Galactosidase activity [in modified Miller units (22)] was measured in yeast transformants expressing DB–ETR1293–729 plus each of the indicated AD–CTR1 fusion proteins. Fusions 1 and 2 were expressed in plasmid pGAD424; fusions 3–7 were expressed in plasmid pACTII (which has a stronger promoter than pGAD424). Three transformants of each were tested, and the average ± the SD is presented. The background level of activity for transformants carrying the pACTII vector is given. (B) Immunoblot analysis of the levels of AD fusion proteins in the transformants using antibody to Gal4 (AD). Two of each of the transformants tested for β-galactosidase activity were examined and were found to be similar; a representative of each is shown. The exposure for construct 1 was approximately four times longer than for the other constructs.
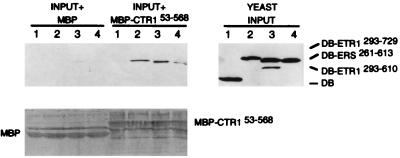
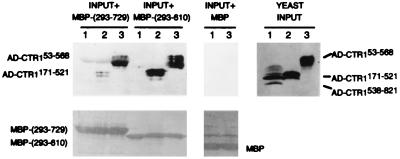
To verify the two-hybrid results, we used two different in vitro protein association assays. In the first assay, we tested whether purified, bacterially expressed MBP fusions could associate with DB or AD fusion proteins present in yeast cell extracts. We found that the purified MBP–CTR153–568 fusion could associate with DB–ETR1293–610, DB–ETR1293–729, and DB–ERS261–613, but not with the DB domain alone (Fig. 3). In the inverse experiment, we found that purified MBP–ETR1293–610 and MBP–ETR1293–729 could associate with AD–CTR153–568 but not with AD–CTR1538–821 (Fig. 4) nor with the AD domain alone (data not shown). Additionally, we confirmed that both MBP–ETR1293–610 and MBP–ETR1293–729 could associate with one of the smaller AD–CTR1 fusions (CTR1 residues 171–521) (Fig. 4). In each case, we verified that MBP alone did not associate with any of the yeast-expressed fusion proteins and that the amount of added MBP fusion was similar for each of the extracts tested (as shown in Figs. 3 and 4). Thus, the interactions we observed in the presence of yeast cell extracts were specific and consistent with the two-hybrid results.
Figure 3.
In vitro association of purified MBP–CTR153–568 with yeast DB–ETR1 and DB–ERS fusions in yeast cell extracts. Bacterially expressed MBP or MBP–CTR153–568 protein was attached to amylose-containing beads, the beads were mixed with yeast extracts 1 through 4, and then the bead-associated proteins were subjected to immunoblot analysis with antibody to LexA (DB). YEAST INPUT is an immunoblot analysis of the yeast extracts before treatment with the beads and indicates the relative amounts of yeast DB fusion proteins present in each extract. The input yeast extracts were: (lane 1) DB alone, (lane 2) DB–ETR1293–729, (lane 3) DB–ERS261–613, and (lane 4) DB–ETR1293–610. For the samples labeled INPUT + MBP, each indicated yeast extract was mixed with MBP attached to beads. For the samples labeled INPUT + MBP–CTR153–568, each indicated yeast extract was mixed with the MBP–CTR153–568 fusion protein attached to beads. The lower panel shows the total protein on the immunoblot filter as detected by Ponceau S.
Figure 4.
In vitro association of purified MBP–ETR1 fusions with yeast AD–CTR1 fusions in yeast cell extracts. Bacterially expressed MBP or the indicated MBP fusion was attached to amylose-containing beads, the beads were mixed with yeast extracts 1 through 3, and then the bead-associated proteins were subjected to immunoblot analysis with antibody to Gal4 (AD). YEAST INPUT is an immunoblot analysis of the yeast extracts before treatment with the beads and indicates the relative amounts of yeast AD fusion proteins present in each extract. The input yeast extracts were: (lane 1) AD–CTR1538–821, (lane 2) AD–CTR1171–521, and (lane 3) AD–CTR153–568. For the samples labeled INPUT + MBP-(293–729), each indicated yeast extract was mixed with the MBP–ETR1293–729 fusion protein attached to beads. For the samples labeled INPUT + MBP-(293–610), the indicated yeast extracts were mixed with the MBP–ETR1293–610 fusion protein attached to beads. For the samples labeled INPUT + MBP, yeast extracts were mixed with MBP attached to beads. The lower panels show the total protein present on the immunoblot filters as detected by Ponceau S. MBP-(293–729) is the MBP–ETR1293–729 fusion, and MBP-(293–610) is the MBP–ETR1293–610 fusion.
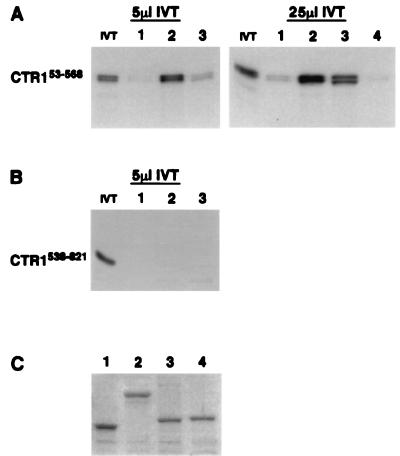
Lastly, we used a second in vitro association assay to demonstrate that CTR1 and ETR1 can interact in the absence of yeast cell extracts. In addition, we determined that the CTR1 amino-terminal domain can associate with the ETR1 receiver domain (an interaction that could not be tested in the two-hybrid assay because of background activation of the reporter genes by either the amino-terminal portion of CTR1 or the receiver domain of ETR1). The assay was performed by mixing in vitro-translated [35S]methionine-labeled CTR1 polypeptides with purified, bacterially expressed MBP fusions attached to amylose-containing beads and then determining whether the radiolabeled proteins were associated with the MBP fusions. To examine the specificity of the interaction, we included MBP fused with the receiver domain (residues 981-1122) of the putative cytokinin receptor CKI1 (13). We found that the radiolabeled CTR1 amino-terminal domain (residues 53–568) could associate with MBP–ETR1293–610 and MBP–ETR1604–738 and only weakly with MBP–CKI1981–1122 and MBP alone (Fig. 5A). Of note, when the amount of added radiolabeled CTR1 protein was increased 5-fold (from 5 to 25 μl), the interaction with MBP–ETR1293–610 and MBP–ETR1604–738 increased whereas the apparent weak association with MBP alone was not enhanced significantly (Fig. 5A). The absence of association with the CKI1 receiver is indicative of the specificity of the CTR1 association with the ETR1 receiver. Furthermore, when we used radiolabeled kinase domain of CTR1 (residues 538–821) as the test protein, there was no substantial interaction with any of the MBP fusions (Fig. 5B). These results indicate that the amino-terminal domain of CTR1 can interact specifically with the ETR1 receiver domain in addition to the ETR1 histidine kinase domain and that endogenous yeast proteins (yeast cell extracts) are not required in these associations.
Figure 5.
In vitro association of radiolabeled CTR1 polypeptides with purified MBP fusions: (i) MBP alone, (ii) MBP–ETR1293–610, (iii) MBP–ETR1604–738, and (iv) MBP-CKI1981–1122. (A) Autoradiograms showing association of the CTR1 amino-terminal domain with MBP fusions 1–4. Bacterially expressed MBP or MBP fusion was attached to amylose-containing beads, and the beads were mixed with 5 or 25 μl of in vitro-translated, radiolabeled CTR1 amino-terminal domain (residues 53–568) (IVT). The bead-associated proteins were separated on SDS/PAGE gels, and the radiolabeled CTR153–568 was visualized by autoradiography. Lane IVT contains 0.1 μl of unassociated radiolabeled CTR153–568. (B) Autoradiogram showing association of the CTR1 kinase domain with MBP fusions 1–3. Bacterially expressed MBP or MBP fusion was attached to amylose-containing beads, and the beads were mixed with 5 μl of IVT. IVT in this case is the radiolabeled in vitro-translated CTR1 kinase domain (residues 538–821). The bead-associated proteins were subjected to SDS/PAGE, and radiolabeled CTR1538–821 was visualized by autoradiography. Lane IVT contains 0.1 μl of unassociated radiolabeled CTR1538–821. The length of exposure is twice that shown in Fig. 5A. (C) Relative amounts of MBP fusions 1–4 used in Fig. 5 A and B separated on SDS/PAGE gels and stained with Coomassie blue.
DISCUSSION
We have detected a physical association between the presumed regulatory domain of the CTR1 protein kinase and the predicted histidine protein kinase domain of the ETR1 and ERS ethylene receptors in both the yeast two-hybrid assay and in vitro. We also observed an in vitro association between the amino-terminal domain of CTR1 and the receiver domain of ETR1. These findings support previous genetic data indicating that CTR1 acts at or downstream of both ETR1 and ERS in the ethylene signal transduction pathway of Arabidopsis. Furthermore, our results suggest that CTR1 might be part of an ethylene receptor complex(es) in Arabidopsis and that the regulation of CTR1 activity by ethylene may involve direct interactions with the two-component receptors.
Based on sequence analysis, CTR1 is a member of the Raf family of serine/threonine protein kinases identified in Caenorhabditis, Drosophila, and mammals (4). Activation of mammalian Raf is known to involve a number of factors: interaction with the small GTP-binding protein Ras (27, 28), oligomerization on recruitment to the plasma membrane, tyrosine phosphorylation, and/or association with other components (29–34). Also, 14–3-3 proteins play a role in activating Raf, possibly by facilitating dimerization (35–37). From our studies, the minimal interacting region of CTR1 has an amino-terminal border between residues 171 and 308 and a carboxyl-terminal border between residues 521 and 569. Throughout this region, CTR1 displays weak sequence similarity to Raf (4). This region corresponds to the noncatalytic portion of Raf, which is important for Raf kinase activity and is involved in the association of Raf with Ras (27, 28) and 14–3-3 proteins (35, 36). Therefore, this portion of CTR1 may play a regulatory role analogous to that of Raf but involving association with membrane-associated two-component ethylene receptors. Of interest, the tertiary structure of the prototypical bacterial receiver CheY (which is the inferred structure of all receiver domains) is very similar to that of Ras (38, 39), and there are functional similarities at the atomic level between Ras and CheY activation (40). Apart from this, the regulation of CTR1 activity could have other parallels with Raf. ETR1 has been localized to plant membrane fractions and has been shown to exist as a dimer in Arabidopsis (41). Thus, the ethylene receptors conceivably could facilitate membrane association and/or dimerization of CTR1. Assuming a conformational change in the receptor on ethylene binding, the formation or stability of the receptor–CTR1 complex might be conditionally dependent on the presence or absence of ethylene. (We do not know whether the fusion proteins in our experiments resemble an ethylene bound or unbound state.) Another possibility is that the receptors regulate CTR1 by phosphorylation. Finally, although we have shown that the receptor–CTR1 associations can occur in vitro with purified proteins, other components could well be involved in the formation of the receptor–CTR1 complex in Arabidopsis.
The only pathway currently known to contain both a two-component phosphorelay system and a complete MAP kinase cascade is the S. cerevisiae osmolarity-response pathway. [A similar pathway may exist for a stress-activated MAP kinase pathway in Schizosaccharomyces pombe (e.g., ref. 12).] In the yeast omolarity-response pathway, the Ypd1p protein serves as a histidine phosphorelay intermediate between the histidine protein kinase osmolarity sensor Sln1p and the response regulator Ssk1p (42). Through an undefined mechanism, Ssk1p activates a MAP kinase cascade comprised of two MAPKKKs (Ssk2p and Ssk22p), a MAPKK (Pbs2p), and a MAPK (Hog1p) (43). Although two-hybrid interactions have been demonstrated between the sequential components of the two-component phosphorelay portion of this pathway (42), including between the response regulator Ssk1p and the MAPKKKs Ssk2p and Ssk22p (43), it has not been determined whether Sln1p physically interacts with Ssk2p and/or Ssk22p (F. Posas and H. Saito, personal communication). To date, there is no evidence that the ethylene-response pathway in plants contains a phosphorelay system similar to that of the yeast osmolarity-response pathway, which has parallels with several bacterial two-component pathways (11, 44). The protein–protein interactions we have presented in this paper may be indicative of a distinct two-component signaling mechanism.
Acknowledgments
We thank Joe Kieber, Jian Hua, and Tatsuo Kakimoto, respectively, for CTR1, ERS, and CKI1 cDNA clones. We are also grateful to Stan Hollenberg, Erica Golemis, George Sprague, Amy Roth, and John Walker for providing two-hybrid plasmids and strains and Haruo Saito and Anthony Bleecker for sharing unpublished results. We thank Jean–Denis Faure for helpful discussions and Rick Stewart, Zhongchi Liu, and Anthony Bleecker for comments on the manuscript. P.L. was supported in part by National Research Initiative Competitive Grants Program/United States Department of Agriculture Postdoctoral Grant 97–35304-4921, and X.W. was supported in part by the Ministry of Agriculture, The People’s Republic of China. This work was supported by National Research Initiative Competitive Grants Program/United States Department of Agriculture Grant 95–37304-2218 to C.C.
ABBREVIATIONS
- DB
DNA-binding domain
- AD
activation domain
- MAP
mitogen-activated protein
- MAPK
MAP kinase
- MBP
maltose-binding protein
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactopyranoside
References
- 1.Abeles F B, Morgan P W, Saltveit M E., Jr . Ethylene in Plant Biology. 2nd Ed. New York: Academic; 1992. [Google Scholar]
- 2.Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- 3.Hua J, Chang C, Sun Q, Meyerowitz E M. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- 4.Kieber J J, Rothenberg M, Roman G, Feldmann K A, Ecker J R. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- 5.Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker J R. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 6.Lehman A, Black R, Ecker J R. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- 7.Hua J, Sakai H, Meyerowitz E M. In: Biology and Biotechnology of the Plant Hormone Ethylene. Kanellis A, Chang C, Kende H, Grierson D, editors. Dordrecht, Netherlands: Kluwer; 1997. pp. 71–76. [Google Scholar]
- 8.Wilkinson J Q, Lanahan M B, Yen H-C, Giovannoni J J, Klee H J. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- 9.Zhou D, Kalaitzis P, Mattoo A K, Tucker M L. Plant Mol Biol. 1996;30:1331–1338. doi: 10.1007/BF00019564. [DOI] [PubMed] [Google Scholar]
- 10.Parkinson J S, Kofoid E C. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 11.Wurgler–Murphy S M, Saito H. Trends Biochem Sci. 1997;22:172–176. doi: 10.1016/s0968-0004(97)01036-0. [DOI] [PubMed] [Google Scholar]
- 12.Shieh J–C, Wilkinson M G, Buck V, Morgan B A, Makino K, Millar J B A. Genes Dev. 1997;11:1008–1022. doi: 10.1101/gad.11.8.1008. [DOI] [PubMed] [Google Scholar]
- 13.Kakimoto T. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 14.Schaller G E, Bleecker A B. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- 15.Roman G, Lubarsky B, Kieber J J, Rothenberg M, Ecker J R. Genetics. 1995;139:1393–1409. doi: 10.1093/genetics/139.3.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vojtek A B, Hollenberg S M, Cooper A J. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 17.Chen D-C, Yang B-C, Kuo T-T. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 18.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 19.Chien C-T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li L, Elledge S J, Peterson C A, Bales E S, Legerski R J. Proc Natl Acad Sci USA. 1994;91:5012–5016. doi: 10.1073/pnas.91.11.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saiki R J, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 22.Clark K L, Dignard D, Thomas D Y, Whiteway M. Mol Cell Biol. 1993;13:1–8. doi: 10.1128/mcb.13.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leeuw T, Fourest–Levin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas D Y, Leberer E. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- 24.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 25.Fields S, Song O. Nature (London) 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 26.Smith R D, Walker J C. Plant Mol Biol. 1993;21:307–316. doi: 10.1007/BF00019946. [DOI] [PubMed] [Google Scholar]
- 27.Daum G, Eisenmann–Tappe I, Fries H W, Troppmair J, Rapp U R. Trends Biochem Sci. 1994;19:474–480. doi: 10.1016/0968-0004(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 28.Avruch J, Zhang X-F, Kyriakis J M. Trends Biochem Sci. 1994;19:279–283. doi: 10.1016/0968-0004(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 29.Leevers S J, Paterson H F, Marshall C J. Nature (London) 1994;369:407–411. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 30.Stokoe D, Macdonald S G, Cadwallader K, Symons M, Hancock J F. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 31.Farrar M A, Alberola-Ila J, Perlmutter R. Nature (London) 1996;383:178–181. doi: 10.1038/383178a0. [DOI] [PubMed] [Google Scholar]
- 32.Luo Z, Tzivion G, Belshaw P J, Vavvas D, Marshall M, Avruch J. Nature (London) 1996;383:181–185. doi: 10.1038/383181a0. [DOI] [PubMed] [Google Scholar]
- 33.Marais R, Light Y, Paterson H F, Marshall C J. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kasid U, Suy S, Dent P, Ray S, Whiteside T L, Sturgill T W. Nature (London) 1996;382:813–816. doi: 10.1038/382813a0. [DOI] [PubMed] [Google Scholar]
- 35.Irie K, Gotoh Y, Yashar B M, Errede B, Nishida E, Matsumoto K. Science. 1994;265:1716–1719. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- 36.Freed E, Symons M, Macdonald S G, McCormick F, Ruggieri R. Science. 1994;265:1713–1716. doi: 10.1126/science.8085158. [DOI] [PubMed] [Google Scholar]
- 37.Fantl W J, Muslin A J, Kikuchi A, Martin J A, MacNicol A M, Gross R W, Williams L T. Nature (London) 1994;371:612–614. doi: 10.1038/371612a0. [DOI] [PubMed] [Google Scholar]
- 38.Artymiuk P J, Rice D W, Mitchell E M, Willett P. Protein Eng. 1990;4:39–43. doi: 10.1093/protein/4.1.39. [DOI] [PubMed] [Google Scholar]
- 39.Chen J M, Lee G, Murphy R B, Brandt–Rauf P W, Pincus M R. Int J Pept Protein Res. 1990;36:1–6. doi: 10.1111/j.1399-3011.1990.tb00077.x. [DOI] [PubMed] [Google Scholar]
- 40.Lukat G S, Lee B H, Mottonen J M, Stock A M, Stock J B. J Biol Chem. 1991;266:8348–8354. [PubMed] [Google Scholar]
- 41.Schaller G E, Ladd A N, Lanahan M B, Spanbauer J M, Bleecker A B. J Biol Chem. 1995;270:12526–12530. doi: 10.1074/jbc.270.21.12526. [DOI] [PubMed] [Google Scholar]
- 42.Posas F, Wurgler–Murphy S M, Maeda T, Witten E A, Thai T C, Saito H. Cell. 1996;86:865–875. doi: 10.1016/s0092-8674(00)80162-2. [DOI] [PubMed] [Google Scholar]
- 43.Maeda T, Takekawa M, Saito H. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 44.Appleby J L, Parkinson J S, Bourret R B. Cell. 1996;86:845–648. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]