Abstract
The application of a new encoding technology for drug discovery is described. A combinatorial library of mercaptoacyl pyrrolidines has been prepared on a beaded polymeric support. Each polymer bead carries one library constituent in association with an oligomeric “tag,” the structure of which is a record of the specific reagents from which that library member was prepared. After the ligands were solubilized, an array of such beads was screened for angiotensin-converting enzyme inhibitory activity, and the structures of active pyrrolidines were deduced by analysis of the associated tags at sub-picomole levels. Several extremely potent enzyme inhibitors were identified, many from multiple beads. The most potent inhibitor was found to have a Ki of 160 pM, ≈3-fold more active than captopril in the same assay. Direct comparison with iterative deconvolution shows that the encoded screening strategy is a much more efficient means for extracting information from such compound collections, producing more data on a larger number of active structures.
Keywords: combinatorial chemistry, encoded libraries, solid-phase synthesis
Combinatorial library syntheses have become increasingly successful in producing large numbers of pharmaceutically relevant compounds (1–4). Polymer-supported approaches have found particular favor as they simplify product isolation and allow the use of excess reagents to help force reactions to completion. One strategy that has been widely used for the assembly of compound libraries is the “split synthesis” method (5). In this approach, the (solid-supported) starting material is divided into a number of equal portions that are each treated separately with a different reagent. The individual samples are then pooled and mixed before splitting into the desired number of portions for the second chemical step, each portion now containing a mixture of compounds. The split–pool–react cycle can be repeated for each step of library production. Such compound collections can offer an attractive source of molecules for pharmaceutical screening, but the complexity of the mixtures presents challenges in identifying biologically active members of these libraries.
Methods for the deconvolution (or dereplication) of soluble compound pools produced via split synthesis according to some biological assay criterion were first popularized by Houghten et al. (6). The original (and still widely practiced) approach is based upon an iterative process of screening and resynthesis of smaller sublibraries in an attempt to fractionate a mixture into its most active constituent(s). This method requires the synthesis of n sets of pools to identify an active constituent from a library constructed in n steps, typically amounting to three or more library resyntheses. While rather inefficient both in the use of time and reagents, iterative deconvolution analysis has been successfully employed by many workers to identify active components from combinatorial mixtures. Nevertheless, there are a number of caveats that must be appreciated when using this method. The greatest limitation of any assay that compares the activity of pooled compounds is that the activity of a given pool is based on the cumulative activity of all the compounds in the pool—i.e., pools with the same activity may contain many low-affinity compounds or a few high-affinity compounds. Selecting the most active pool for deconvolution does not, therefore, guarantee that the most potent compound in the library will be identified. Theoretical investigations of iterative deconvolutions of RNA oligomer libraries by the ISIS group have suggested that this approach does have a high probability of identifying particularly active (if not always the most active) members of a library. Simulation of a variety of pooling scenarios indicated that the optimal library member is identified under all but the most demanding conditions (i.e., when it is pooled with the most inactive members or when experimental error is large; refs. 7–9).
The use of beaded polymers with the split synthesis strategy results in a solid-supported library where each bead carries (ideally) only one compound (5). This offers the attractive prospect of assaying individual beads for biological activity, with several advantages over the iterative procedure described above: all assays are carried out on single compounds, eliminating the complications resulting from screening mixtures; no assay-specific resynthesis is required; and relatively small amounts of resin are needed, given that a gram of a typical solid support contains ≈1 million beads. The main limitation is the quantity of material that can be prepared on a single bead: most suitable commercially available resins carry sub-nanomole amounts of accessible functionality per bead. While this can be sufficient for determination of biological activity, it is not, in general, enough to subsequently identify unambiguously the active constituent using analytical techniques suitable for the majority of organic molecules (peptides and oligonucleotides are notable exceptions; see ref. 10).
An alternative approach for the identification of active compounds from single beads is to prepare encoded libraries, where the chemical history of each bead is recorded by the cosynthesis of a readily analyzable surrogate marker (or “tag”). Following analysis of the tags carried by a given bead, this code identifies the reagent monomers from which the corresponding compound was assembled. Initially oligonucleotides (11, 12) and peptides (13, 14) were used to encode such libraries, but an expanded interest in the preparation of libraries of nonoligomeric organic compounds necessitated the development of hardier coding chemistries that were capable of withstanding a wider range of chemical procedures (15–19). These tags have been designed to permit decoding by extremely sensitive analytical techniques, such as electron-capture GC (15, 16), fluorescence-based HPLC (17), MS (18), or GC–MS (19), allowing sub-picomole tag detection. This in turn means that the tag need often be present at only a small percentage of the total bead loading, leaving most sites available for the preparation of the compounds under biological investigation. All of these technologies potentially allow a wide variety of chemical libraries to be screened in a single-bead format, with the attendant advantages described above, but without the requirement of developing sensitive analytical methods for each new class of compounds. An interesting extension of this approach allows encoding by radio-frequency irradiation (20, 21).
We present here the application of an encoding technology to the screening of a library of highly functionalized pyrrolidines. We have previously described how a potent angiotensin-converting enzyme (ACE) inhibitor was identified from the same compound set by iterative deconvolution (22). Screening of this library against the same biological target offers the first direct comparison of the two approaches.
MATERIALS AND METHODS
Unless otherwise indicated, chemical reagents were purchased from Aldrich. Solvents were from Baxter Diagnostics (McGaw Park, IL) and were the highest quality available.
Encoded Amino Acid Resins.
These were prepared using portions of orthogonally functionalized resin (1.0 g each) as described previously (17) using 9-fluorenylmethoxycarbonyl (Fmoc)-Gly-OH, Fmoc-Phe-OH, Fmoc-Leu-OH, and Fmoc-Ala-OH (all from Bachem). Incorporation of amino acids was monitored by quantitative Fmoc analysis. The amino acid coupling was repeated when this assay indicated a low incorporation.
Encoding Scheme for Pyrrolidine Library.
Three sets of tags (i.e., tags 1 A–D for amino acids, tags 2 A–D for aldehydes, and tags 3 A–E for olefins) were used to code for steps in the library synthesis. Each of the indicated tags comprises a single secondary amine or a mixture of two amines. Tag set 1: A, BB and PP, glycine; B, EB and PP, alanine; C, EB and BB, leucine; and D, PP, phenylalanine. Tag set 2: A, MH and MH′, o-tolualdehyde; B, MH and MD, o-anisaldehyde; C, MD, o-(tert-butyldimethylsilyloxy)-benzaldehyde; and D, MH′ and MD, benzaldehyde. Tag set 3: A, HH, methyl acrylate; B, HH and O′O′, tert-butyl acrylate; C, O′O′, methyl vinyl ketone; D, OO and O′O′, acrylonitrile; and E, HH and OO, methyl methacrylate. (Secondary amine tags are defined as follows: BB, dibutylamine; PP, dipentylamine; HH, dihexylamine; OO, dioctylamine; O′O′, bis-(2-ethylhexyl)amine; MH, methylhexylamine; MH′, methylheptylamine; MD, methyldodecylamine; EB, ethylbutylamine.) To ensure that approximately equal quantities of the amines in any mixture coupled to resin beads, it was necessary to adjust the volume of amines in the mixture in inverse proportion to their chemical reactivity, as determined below.
Determination of Amine Reactivity.
TentaGel-S-NH2 resin (130-μm diameter; 1.0 g, 0.29 mmol·g−1, 0.29 mmol; Rapp Polymere, Tubingen, Germany) was treated with succinic anhydride (0.4 g, 4 mmol, 15 eq) and diisopropylethylamine (0.7 ml, 4 mmol) in N-methyl pyrrolidine (NMP; 10 ml) for 1 h. The resin was activated with an excess of pentafluorophenyl trifluoroacetate and pyridine in NMP (1:1:1; total volume of 6 ml) for 1 h, then split into separate portions and treated with a mixture of two or more secondary amines (total of 0.5 mmol) in NMP (1 ml; amines were obtained from Aldrich, Lancaster Synthesis, and Fluka). After 1 h, the resins were washed well with NMP, 1,4-dioxane, and diethyl ether, and three single beads from each sample hydrolyzed and analyzed as described (17). Relative reactivities were calculated from the observed HPLC peak areas and are expressed relative to HH (= 1.0): MH, 20; MH′, 18; MD, 7; BB, 1.5; EB, 1.7; PP, 1.3; OO, 0.8; and O′O′, 0.008. No notable differences in relative reactivity were detected when amines were applied to differentiated resins containing Fmoc in addition to pentafluorophenyl ester.
Preparation of Encoded Pyrrolidine Library.
Details of many of the synthetic steps have been described previously (17, 22). Portions of each of the four encoded amino acid resins (0.5 g) were combined. Allyloxycarbonyl (Alloc) was removed from the resulting bead mass using tetrabutylammonium azide (23). The resin was treated with N-Alloc-iminodiacetic anhydride, followed by pentafluorophenyl trifluoroacetate as above, then split into four equal portions. After addition of appropriate mixtures of amines (tag set 2; total ≈2.0 mmol) in NMP (5 ml), the resins were washed thoroughly. Imine formation [with benzaldehyde, o-anisaldehyde, o-tolualdehyde, and o-(tert-butyldimethylsilyloxy)-benzaldehyde in trimethylorthoformate] and, following a further split-pool, silver-catalyzed [2+3] cycloaddition (with methyl acrylate, methyl methacrylate, acrylonitrile, methyl vinyl ketone, and tert-butyl acrylate) proceeded as described (22). The four resin-bound pyrrolidine pools were protected by treatment with Fmoc-Cl (0.26 g, 1.0 mmol) and diisopropylethylamine (0.17 ml, 1.0 mmol) in NMP (5 ml) with shaking at room temperature for 2 h. Two further treatments with Fmoc-Cl in neat pyridine were performed until a constant level of Fmoc-loading was achieved. After an additional Alloc deprotection, amines in tag set 3 were coupled as described above. The resin was pooled and portions (≈10 mg each) acylated individually with each of the three mercaptoacyl chlorides.
Compound Deprotection, Bead Distribution, and Cleavage from the Solid Support.
TentaGel beads from the mercaptoacyl pyrrolidine library were deacetylated collectively using ammonia-saturated methanol for 1 h at room temperature. The beads were washed with 150 mM 2-mercaptoethanol in nitrogen-sparged methanol, then suspended in the same solvent and dispensed into wells of a polystyrene microtiter plate. Microscope-aided visual inspection indicated that ≈80% of the wells held a single bead. Wells with two or more beads were omitted from subsequent analysis. A total of twelve plates was prepared in this manner, each plate containing beads treated with a single mercaptoacyl chloride. Neat trifluoroacetic acid (TFA; 10 μl) was added to each well, and the plate was covered and allowed to stand at room temperature for 1 h. TFA was removed under vacuum and 500 μM 2-mercaptoethanol in ethanol (12 μl) added. The plate was covered and allowed to sit at room temperature overnight.
Assay for ACE Activity.
The in vitro assay for inhibition of rabbit ACE (peptidyl-dipeptidase A; EC 3.4.15.1) was determined by the hydrolysis of hippuryl-His-Leu as described (24). To the plate of cleaved compounds was added nitrogen-sparged 25 mM Hepes buffer (120 μl) containing 0.3 M sodium chloride (pH 8.1). The entire contents were transferred to a Microfluor W (Dynatech) 96-well plate, which had been preblocked with 1% dried nonfat milk in 10 mM Tris/0.15 M NaCl, pH 8.0, overnight. ACE (25 milliunits) was added, and the plates were covered and incubated for 1 h with occasional mixing. The substrate hippuryl-His-Leu was added to each well (to 1 mM final concentration), and the assay allowed to proceed at room temperature for 5 min, with enzymatic activity being determined spectrofluorimetrically using a Fluorolite 1000 plate reader (Dynatech) as described (24). IC50 values for purified inhibitors were determined using varying concentrations of inhibitor (with free thiol content being confirmed by Ellman’s assay) and 1 h preincubation with enzyme. Inhibition curves were constructed using a minimum of seven data points near the IC50, and duplicate experiments were carried out.
Tag Analysis.
The entire contents of desired wells were transferred by pipette into a glass capillary tube sealed at one end. The tube was centrifuged and the presence of the bead was confirmed with the aid of a microscope (×10 objective). The supernatant was removed and the bead was washed with ethanol (2 × 200 μl). After drying briefly under vacuum, the tube was treated with 6 M HCl (100 μl), derivatized with dansyl chloride, and analyzed by HPLC as described previously (17).
RESULTS AND DISCUSSION
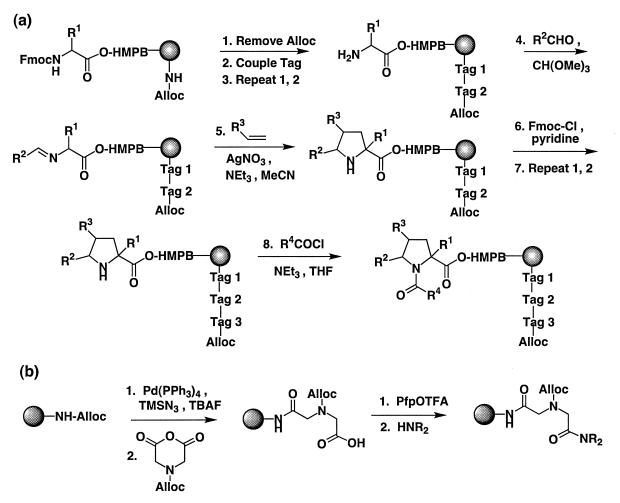
We have previously shown that encoding chemistry based on the assembly of a tertiary amide oligomer is compatible with solid-phase pyrrolidine synthesis (17). Ligand synthesis was carried out on an acid-cleavable linker (4-(4-hydroxymethyl-3-methoxyphenoxy)-butyric acid, HMPB), using building blocks protected with base-labile (Fmoc) and acid-labile (e.g., tert-butyl) groups where appropriate (Fig. 1). The encoding oligomer was attached directly to the polymer resin without an intervening cleavable linker. Tag building blocks were incorporated as N-Alloc derivatives (25) to maintain orthogonal protection of sites for tag and ligand addition. The quality of pyrrolidines prepared in the encoded format was equal to that obtained from unencoded solid-phase synthesis (17).
Figure 1.
(a) Encoded pyrrolidine synthesis on TentaGel resin. Ratio of Fmoc:Alloc protected amines is ≈9:1. (b) Tag addition chemistry (i.e., steps 1 and 2 in a, above).
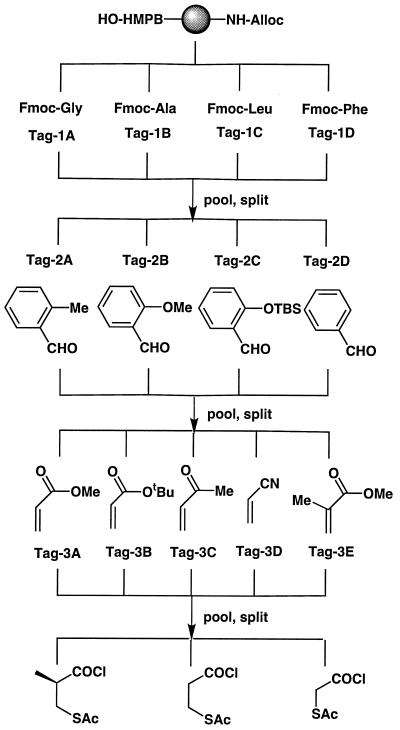
Library assembly (Fig. 2) proceeded in an identical fashion to the synthesis of discrete compounds with the important exception that all resin portions were pooled, randomized, and redivided after each set of building block/tag addition steps. The use of a binary coding strategy (15) allowed nine amines to represent the 240 library members (excluding diastereomers; see below). The amines that encode each set of building blocks were chosen to possess similar reactivity (see Materials and Methods). The final step of library assembly—i.e., acylation with each of the three mercaptoacyl chlorides, was not encoded; rather, the library was stored as three separate, fully encoded sublibraries of 80 compounds each.
Figure 2.
Assembly of encoded mercaptoacyl pyrrolidine library.
The stated pool sizes do not take into account the presence of multiple stereoisomers of each compound, which arise because the key [2+3] cycloaddition proceeds via (predominantly) endo approach of the olefin to either face of a planar, prochiral, resin-bound azomethine ylide intermediate. The major products are a pair of enantiomers in which the substituents at the 4 and 5 positions of the pyrrolidine ring are in a syn relationship with the 2-carboxy group (ref. 22; (diastereomers result from subsequent acylation with the chiral 2-S-mercaptoisobutyryl chloride). Thus the library probably contains more than 500 distinct compounds; however, it is not possible to identify the different diastereomers by encoding, since each bead carries a mixture of such components.
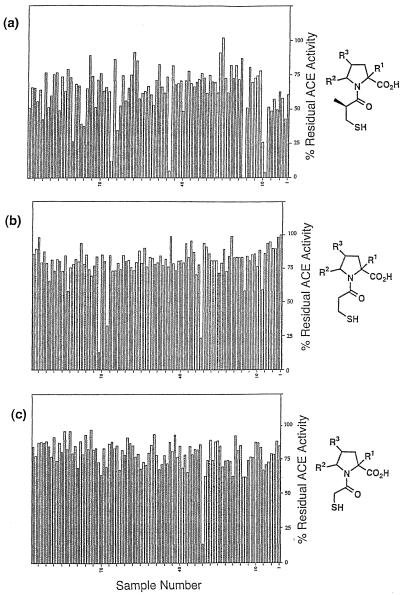
To minimize chemical manipulation of single beads, the S-acetyl protecting group was removed from several milligrams of each of the three sublibraries before distribution into individual wells of 96-well polypropylene plates. A total of 12 plates was prepared, each plate containing beads treated with a single mercaptoacyl chloride. Following cleavage from the solid support compounds were assayed for ACE inhibition at an approximate concentration of 50 nM using a discontinuous fluorogenic assay (24). Data from one plate for each acid chloride are shown in Fig. 3. A few beads gave essentially complete enzyme inhibition, with a small number of additional beads showing moderate to good activity. The majority of wells showed no inhibition. There was generally greater inhibition from the mercaptoisobutyrate beads than from the corresponding acetate or propionate samples, in agreement with our expectations based on assay of pools of these compounds at the initial step of deconvolution analysis (22).
Figure 3.
ACE inhibition by mercaptoacyl pyrrolidines cleaved from single beads. Data is from one 96-well plate each of pyrrolidines acylated with mercaptoisobutyryl (a), mercaptopropionyl (b), and mercaptoacetyl (c) groups.
The most active beads were removed from the various plates for decoding. After washing, the beads were air-dried and treated with concentrated hydrochloric acid, and the liberated amines were dansylated and and analyzed as described previously (17). The building blocks predicted by the observed amines from selected beads are shown in Table 1. While only a small subset of building blocks were identified on the most active beads (i.e., those giving >70% inhibition), random selection of less active mercaptoisobutyryl beads indicated that the building blocks were evenly distributed in the library.
Table 1.
ACE inhibitory activity and component building block identities deduced from decoding analysis
| Acylating group | % inhibited | Amino acid | Aldehyde | Olefin | Compound |
|---|---|---|---|---|---|
| Mercaptoisobutyryl | 97 | Gly | Bz | MA | 1 |
| 97 | Gly | Bz | MA | 1 | |
| 96 | Gly | Bz | MA | 1 | |
| 95 | Gly | Tol | MA | 5 | |
| 88 | Gly | Bz | MMA | 4 | |
| 88 | Gly | Bz | MA | 1 | |
| 87 | Gly | Bz | MMA | 4 | |
| 82 | Gly | Tol | MA | 5 | |
| 81 | Gly | Anis | MA | 6 | |
| 78 | Gly | Bz | MA | 1 | |
| 76 | Gly | Tol | MA | 5 | |
| 68 | Gly | Bz | MMA | 4 | |
| 63 | Phe | TBS | MVK | ||
| 57 | Phe | TBS | TBA | ||
| 50 | Ala | Bz | MMA | ||
| 39 | Leu | Bz | TBA | ||
| 30 | Phe | Bz | MMA | ||
| 35 | Phe | TBS | MMA | ||
| 24 | Leu | Tol | AN | ||
| 18 | Ala | TBS | MMA | ||
| 4 | Phe | Anis | MA | ||
| Mercaptopropiornyl | 91 | Gly | Bz | MA | 2 |
| 91 | Gly | Bz | MA | 2 | |
| 87 | Gly | Bz | MA | 2 | |
| 83 | Gly | Bz | MA | 2 | |
| 78 | Gly | Bz | MMA | 7 | |
| 76 | Gly | Bz | MMA | 7 | |
| 74 | Gly | Bz | MMA | 7 | |
| Mercaptoacetyl | 87 | Gly | Bz | MA | 3 |
| 77 | Gly | Bz | MA | 3 |
Beads showing greatest activity (at a single concentration of ≈50 nM) from each of the three mercaptoacyl sublibraries were selected for decoding, along with representative less active beads from the mercaptoisobutyryl pool. Bz, benzaldehyde; Tol, o-tolualdehyde; Anis, o-anisaldehyde; TBS, o-(tert-butyldimethyl-silyloxy)-benzaldehyde; MA, methyl acrylate; MMA, methyl methacrylate; TBA, tert-butyl acrylate; MVK, methyl vinyl ketone; AN, acrylonitrile.
Since each of the three sublibraries comprised 80 building block combinations, on average, every 96-well plate should contain one representative of each. The most inhibitory compounds were generally identified at close to the expected rate—e.g., from the four plates prepared from mercaptopropionyl chloride we found exactly four beads with tags corresponding to compound 2. For weaker compounds this was less often true; for instance, compound 6 was represented by a single example. Replicates of these less potent compounds were, perhaps, less clearly distinguished from the background of lower activity compounds.
Where the same compound was identified from multiple beads, a reasonably consistent level of enzyme inhibition was obtained from the single assay point determinations (see Table 1). While the most active compound in the entire library (1) was found on the three beads giving the greatest inhibition (≈97%), it was also found on two beads showing significantly less potent activity (88 and 78% inhibition). It is likely that this variation reflects deviations in the amount of compound obtained from different beads, with bead size variability having a predominant influence.
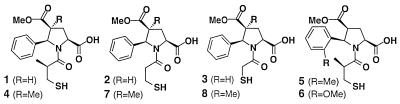
Table 2 shows the structures of the most active compounds identified by this approach. To confirm the relative potencies suggested by the single bead assays, several of the predicted compounds were synthesized by preparative solid-phase methods (i.e., without encoding). Activities of the purified mercaptoacyl (d/l)-pyrrolidine isomer mixtures are in good agreement with the rank order determined from the single bead screening. In two cases (compounds 1 and 4), we have been able to separate the diastereomeric products by preparative HPLC and determine the inhibition constants for the individual isomers. In both instances, the activity resided in a single diastereomer, presumably that having the pyrrolidine ring in the l-configuration (in accordance with literature data; see for example ref. 26).
Table 2.
Structures of most potent ACE inhibitors identified from individual bead assay and activity of purified compounds
| Compound no. | Mean inhibition from single beads, % (n) | IC50 for purified compounds, nM
|
||
|---|---|---|---|---|
| Mixture of isomers | Separated Diastereomers
|
|||
| l-Pyrrolidine | d-Pyrrolidine | |||
| 1 | 91 ± 8 (5) | 0.61 | 0.16 | >100 |
| 2 | 88 ± 5 (4) | 1.3 | ||
| 3 | 82 ± 5 (2) | 13 | ||
| 4 | 81 ± 13 (3) | 4.0 | 0.64 | >100 |
| 5 | 84 ± 8 (3) | |||
| 6 | 81 (1) | |||
| 7 | 76 ± 2 (3) | 32 | ||
| 8 | 69 ± 4 (2) | 400 | ||
Note that the stereochemistry of the pyrrolidine ring is shown in the l-configuration for convenience.
The data presented here allow direct comparison of encoded library screening with our previous study using iterative deconvolution. Compound 1, the most active compound found in this study, is the same compound identified by deconvolution analysis (22). It is gratifying to find that both assay strategies predict the same optimal structure from this library, a compound with more potent in vitro biological activity than captopril (Ki ≈ 500 pM; ref. 27) and many other compounds found over multiple years of traditional medicinal chemistry efforts. This fact illustrates the use of both of these combinatorial approaches in optimizing leads for drug discovery. There are, however, significant practical and strategic differences between the two approaches, as described below.
Deconvolution requires sublibraries to be repeatedly prepared and several rounds of assays to be carried out, whereas an encoded library need only be prepared once and screened once, albeit at the cost of adopting a more complex synthetic scheme. However, this initial investment can be justified, since screening encoded libraries makes very efficient use of materials: all the single-bead screening described above used <30 mg of beads. Since ≈2 g of library was prepared, sufficient material remains for testing against a large number of biological targets.
For the encoded library screening described here, assays are carried out on discrete compounds, whereas deconvolution relies on testing mixtures with the attendant risks both of false positive and false negative results due to the problems associated with cumulative potency (vide supra). The main use of this encoding approach is likely to be found in the screening of large libraries. Here the number of individual assay determinations required to thoroughly sample the library may be impractical. Bead pooling may be adopted in conjunction with a “tiered” or partial compound release strategy, using either (i) two (or more) linkers that are cleavable under chemically orthogonal conditions (28), or (ii) a single linker moiety that demonstrates predictable and convenient cleavage kinetics [e.g., a photolabile linker (29, 30)]. Here, an initial fraction of the compound (e.g., 50%) is cleaved from every bead in a pool of, for example, 100 beads chosen at random from the library. After assay for some desired activity, the beads from active pools are recovered and individually arrayed for a second round of compound cleavage, assay, and finally decoding analysis. In contrast to a conventional deconvolution analysis where the pooled compounds share similar structural features by virtue of containing one or more fixed building block(s), the encoded library approach allows beads to be pooled in a purely stochastic fashion. This should reduce the probability that the activity of any pool is due to the cumulative activity of multiple compounds of modest potency, as any active molecule is unlikely to be pooled with compounds having similar structures.
The single-bead library assay potentially allows more structure–activity relationship data to be obtained than from a deconvolution. While both approaches identified the same most active compound (1), the second and third most inhibitory compounds found in this work (i.e., 2 and 4) were discarded at the first and second steps of deconvolution, respectively. Clear structure–activity trends are evident from the inhibitors isolated here. For example, the preference for 4-carbomethoxy substitution on the pyrrolidine ring and the systematically deleterious effect of additional 4′-methyl substitution are apparent, as is the striking requirement for the α-unsubstituted proline ring system in the most potent inhibitors. The ability to collect substantial amounts of structure–activity relationship, even at the level of a high-throughput primary screen, may be of considerable use in the development of quantitative models of ligand interactions with biological macromolecules and is a significant advantage of the single-bead screening approach.
A further limitation of iterative deconvolution is that the outcome may be dependent on the synthetic pathway chosen. To avoid the necessity of handling large numbers of separate samples, it is desirable to assay initially the pools obtained by segregating the final set of building blocks in the synthetic sequence. In ideal cases, the pools defined by each of these final reagents will show clear differences in potency, allowing a reasonable selection for the next iterative resynthesis. Often, however, this choice may not be clear-cut, necessitating an arbitrary decision; while this may indicate tolerance by the target for those structural differences in the ligand (with no deleterious effect on the outcome of the analysis), in certain cases it may also be the result of pooling anomalies as described above [causing the best compound(s) to be discarded]. In contrast, encoded libraries allow selection and identification of compounds independent of the synthetic route.
One significant limitation of this encoded bead technology is the amount of material available for screening. For high-fidelity compound synthesis, the maximal solution concentration that can be achieved from individual beads (of the type currently used) in a microtiter assay volume of ≈200 μl is around 1 μM. For many targets, less potent compounds could be leads worth pursuing. This provides an impetus for the development of miniaturized assays to take further advantage of the economy of the bead-based format. Further enhancements would permit multiple assays from a single bead allowing compound retesting, sequential dilution (removing the reliance on a single data point), and determination of selectivity by assay against a panel of targets. We are also actively exploring automation strategies to facilitate segregation and recovery of individual encoded beads for both conventional microtiter plate-based as well as reduced volume assay formats.
In summary, we have applied a new encoding strategy to the screening of a library of highly functionalized pyrrolidines and thereby identified a series of potent inhibitors of ACE. The approach is readily scalable to the production of libraries of tens to hundreds of thousands of compounds and is compatible with a wide variety of organic chemistries. The strategy offers a streamlined approach to the identification of active components from combinatorial libraries and gives useful and substantial information on structure–activity relationships. We look forward to reporting the results of analyzing larger and more diverse libraries against further targets of pharmaceutical relevance.
ABBREVIATIONS
- ACE
angiotensin-converting enzyme
- Fmoc
9-fluorenylmethoxycarbonyl
- NMP
N-methyl pyrrolidine
- BB
dibutylamine
- PP
dipentylamine
- HH
dihexylamine
- OO
dioctylamine
- O′O′
bis-(2-ethylhexyl)amine
- MH
methylhexylamine
- MH′
methylheptylamine
- MD
methyldodecylamine
- EB
ethylbutylamine
- Alloc
allyloxycarbonyl
References
- 1.Gallop M A, Barrett R W, Dower W J, Fodor S P A, Gordon E M. J Med Chem. 1994;37:1233–1251. doi: 10.1021/jm00035a001. [DOI] [PubMed] [Google Scholar]
- 2.Gordon E M, Barrett R W, Dower W J, Fodor S P A, Gallop M A. J Med Chem. 1994;37:1385–1401. doi: 10.1021/jm00036a001. [DOI] [PubMed] [Google Scholar]
- 3.Terrett N K, Gardner M, Gordon D W, Kobylecki R J, Steele J. Tetrahedron. 1995;51:8135–8173. [Google Scholar]
- 4.Thompson L A, Ellman J A. Chem Rev (Washington, DC) 1996;96:555–600. doi: 10.1021/cr9402081. [DOI] [PubMed] [Google Scholar]
- 5.Furka A, Sebestyen F, Asgedom M, Dibo G. Int J Pept Protein Res. 1991;37:487–493. doi: 10.1111/j.1399-3011.1991.tb00765.x. [DOI] [PubMed] [Google Scholar]
- 6.Houghten R A, Pinilla C, Blondelle S E, Appel J R, Cooley C T, Cuervo J H. Nature (London) 1991;354:84–86. doi: 10.1038/354084a0. [DOI] [PubMed] [Google Scholar]
- 7.Freier S M, Konings D A M, Wyatt J R, Ecker D J. J Med Chem. 1995;38:344–352. doi: 10.1021/jm00002a016. [DOI] [PubMed] [Google Scholar]
- 8.Konings D A M, Wyatt J R, Ecker D J, Freier S M. J Med Chem. 1996;39:2710–2719. doi: 10.1021/jm960168o. [DOI] [PubMed] [Google Scholar]
- 9.Freier S M, Konings D A M, Wyatt J R, Ecker D J. Bioorg Med Chem. 1996;4:717–725. doi: 10.1016/0968-0896(96)00068-5. [DOI] [PubMed] [Google Scholar]
- 10.Lam K S, Hruby V J, Lebl M, Knapp R J, Kazmierski W M, Hersh E M, Salmon S E. Bioorg Med Chem Lett. 1993;3:419–424. [Google Scholar]
- 11.Brenner S, Lerner R A. Proc Natl Acad Sci USA. 1992;89:5181–5183. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Needels M N, Jones D G, Tate E H, Heinkel G L, Kochersperger L M, Dower W J, Barrett R W, Gallop M A. Proc Natl Acad Sci USA. 1993;90:10700–10704. doi: 10.1073/pnas.90.22.10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerr J M, Banville S C, Zuckermann R N. J Am Chem Soc. 1993;115:2529–2531. [Google Scholar]
- 14.Nikolaiev V, Stierandova A, Krchnak V, Seligmann B, Lam K S, Salmon S E, Lebl M. Pept Res. 1993;6:161–170. [PubMed] [Google Scholar]
- 15.Ohlmeyer M H J, Swanson R N, Dillard L W, Reader J C, Asouline G, Kobayashi R, Wigler M, Still W C. Proc Natl Acad Sci USA. 1993;90:10922–10926. doi: 10.1073/pnas.90.23.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nestler H P, Bartlett P A, Still W C. J Org Chem. 1994;59:4723–4724. [Google Scholar]
- 17.Ni Z J, Maclean D, Holmes C P, Ruhland B, Murphy M M, Jacobs J W, Gordon E M, Gallop M A. J Med Chem. 1996;39:1601–1608. doi: 10.1021/jm960043j. [DOI] [PubMed] [Google Scholar]
- 18.Geysen H M, Wagner C D, Bodnar W M, Markworth C J, Parke G J, Schoenen F J, Wagner D S, Kinder D S. Chem Biol. 1996;3:679–688. doi: 10.1016/s1074-5521(96)90136-2. [DOI] [PubMed] [Google Scholar]
- 19.Edwards, P. N., Main, B. G. & Shute, R. E. (1996) U.K. Patent Appl. GB 2,297,551 A.
- 20.Moran E J, Sarshar S, Cargill J F, Shahbaz M M, Lio A, Mjalli A M M, Armstrong R W. J Am Chem Soc. 1995;117:10787–10788. [Google Scholar]
- 21.Nicolaou K C, Xiao X-Y, Parandoosh Z, Senyei A, Nova M P. Angew Chem Int Ed Engl. 1995;34:2289–2291. [Google Scholar]
- 22.Murphy M M, Schullek J R, Gordon E M, Gallop M A. J Am Chem Soc. 1995;117:7029–7030. [Google Scholar]
- 23.Shapiro G, Buechler D. Tetrahedron Lett. 1994;35:5421–5424. [Google Scholar]
- 24.Cheung H S, Cushman D W. Biochim Biophys Acta. 1973;293:451–463. doi: 10.1016/0005-2744(73)90352-5. [DOI] [PubMed] [Google Scholar]
- 25.Loffet A, Zhang H X. Int J Pept Protein Res. 1993;42:346–351. doi: 10.1111/j.1399-3011.1993.tb00504.x. [DOI] [PubMed] [Google Scholar]
- 26.Petrillo E W, Ondetti M A. Med Res Rev. 1982;2:1–41. doi: 10.1002/med.2610020103. [DOI] [PubMed] [Google Scholar]
- 27.Bull H G, Thornberry N A, Cordes M H J, Patchett A A, Cordes E H. J Biol Chem. 1985;260:2952–2962. [PubMed] [Google Scholar]
- 28.Salmon S E, Lam K S, Lebl M, Kandola A, Khattri P S, Wade S, Patek M, Kocis P, Krchnak V, Thorpe D, Felder S. Proc Natl Acad Sci USA. 1993;90:11708–11712. doi: 10.1073/pnas.90.24.11708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Appell K C, Chung T D Y, Ohlmeyer M J H, Sigal N H, Baldwin J J, Chelsky D. J Biomol Screening. 1996;1:27–31. [Google Scholar]
- 30.Holmes C P, Jones D G. J Org Chem. 1995;60:2318–2319. [Google Scholar]