Abstract
The herpes simplex virus type 1 (HSV-1) genome contains three origins of replication: oriL and two copies of oriS. These origins contain specific sequences, box I and box II, linked by an AT-rich segment, that are recognized by an HSV-1-encoded origin binding protein (UL9 protein) which also possesses DNA helicase activity. Despite its intrinsic helicase activity, the UL9 protein is unable to unwind oriS or the box I element of oriS, either in the presence or absence of the HSV-1-encoded single-strand DNA binding protein, ICP8. However, a complex of the UL9 protein and ICP8 can unwind box I if it contains a 3′ single-stranded tail at least 18 nt in length positioned downstream of box I. These findings suggest a model for the initiation of HSV-1 DNA replication in which a complex consisting of the UL9 protein bound to box I, and ICP8 bound to single-stranded DNA generated at the A+T rich linker, perhaps as a consequence of transcription, unwinds an HSV-1 origin of replication to provide access to the replication machinery with the consequent initiation of viral DNA replication. This mode of unwinding is distinct from that observed for other animal viruses—e.g., simian virus 40 or bovine papilloma virus—in which the initiator protein, T antigen, or E1 protein alone, unwinds elements of the origin sequence, and the single-strand DNA binding protein serves only to keep the separated strands apart.
Keywords: origin-specific unwinding, UL9 protein, ICP8, helicase
Herpes simplex virus type 1 (HSV-1) encodes a 94-kDa origin binding protein, the product of the UL9 gene (1, 2). Studies of HSV-1 DNA replication in vivo have shown the origin binding protein (UL9 protein, also known as OBP) to be essential for viral DNA replication (3, 4). In addition to the UL9 protein, six other viral gene products are required (for a review, see ref. 5). These include a DNA polymerase with its associated processivity enhancing factor, a heterotrimeric helicase-primase or primosome, and a single-strand DNA binding protein (ICP8).
The HSV-1 genome contains three highly homologous origins of replication, oriL and two copies of oriS (5). The UL9 protein that exists in solution as a homodimer (6, 7), specifically and cooperatively binds the two inverted pentanucleotide repeats in boxes I and II of oriS which are separated from each other by an A+T-rich sequence of 18 nt (8, 9). OriL contains a second copy of box I in place of box II (see Fig. 1A). A similar arrangement of binding sites has been observed in the origins of several other herpes viruses including herpes simplex virus type 2 (10), varicella zoster virus (11), equine herpesvirus 1 (12), and human herpes virus type 6 (13). Amino acid sequence comparisons between the HSV-1 UL9 protein and herpes simplex virus type 2, varicella zoster virus, and human herpes virus type 6 homologs show the helicase and DNA binding domains to be highly conserved (14, 15).
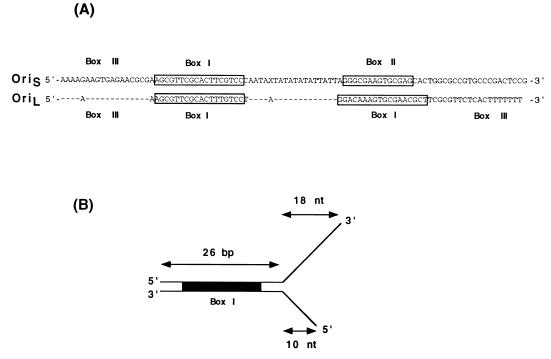
Figure 1.
(A) Nucleotide sequence of HSV-1 replication origins, oriS and oriL. The strong binding sites for the UL9 protein are indicated as boxes I and II; the weak binding site, box III, is also indicated. The X in the oriS sequence indicates a nucleotide absent from oriS but present in oriL. (B) Box I substrate containing two noncomplementary ssDNA tails. Two oligonucleotides, 44-mer (top strand) and 36-mer (bottom strand), were annealed so that a 26-bp duplex region containing the box I sequence and two noncomplementary ssDNA tails was formed.
In addition to its origin binding activity, the UL9 protein exhibits DNA-dependent ATPase and 3′–5′ helicase activities (6, 7, 16, 17). Although the existence of these activities suggests that the UL9 protein binds specifically to elements of the origin (boxes I and II) and unwinds them in a manner analogous to the large T antigen of simian virus 40 (SV40), which also possesses these activities (18, 19), the role of the UL9 protein in unwinding the HSV-1 origins has not yet been clarified. Specific unwinding of the origin is required to provide access to the replication machinery (20). Recent electron microscopic examination of the interaction of UL9 protein with oriS showed that the UL9 protein bends the oriS in the absence of ATP (21), consistent with previous footprinting results (22). Further analysis revealed that the UL9 protein can generate a relatively stable four-stranded structure in the presence of ATP, possibly a manifestation of the origin-specific helicase activity of the UL9 protein (21).
In the experiments reported here, we have examined the interaction of the UL9 protein and ICP8 with a duplex box I sequence to which single-stranded DNA (ssDNA) tails are attached. We have found that with this substrate, a complex consisting of the UL9 protein bound to box I and ICP8 bound to the ssDNA tails, can unwind box I, and it does so with a specific directionality.
MATERIALS AND METHODS
Enzymes.
Recombinant UL9 protein was purified from nuclear extracts of Sf21 insect cells as described by Dodson and Lehman (16) with an additional step, Mono-Q chromatography. The purification was monitored by a gel mobility shift assay using 32P-labeled oriS containing DNA and by Western blotting analysis with anti-UL9 antibodies. The purified UL9 protein consisted of a single polypeptide that migrated with a molecular mass of 83 kDa, following SDS/PAGE and silver staining. It contained 3′–5′ helicase, DNA-dependent ATPase, and oriS binding activities. Recombinant ICP8 was purified from whole cell extracts of Sf21 cells according to Boehmer and Lehman (23). The purified ICP8 migrated as a single polypeptide with a molecular mass of 120 kDa, following SDS/PAGE and silver staining. It reacted with anti-ICP8 antibodies, and contained no detectable helicase, nuclease, or ATPase activity.
DNA Substrates.
To prepare the box I substrates, the oligonucleotides 36-mer (5′-CATGCTCGCAGCGGGACGAAGTGCGAACGCTTCGCG-3′) and 44-mer (5′-CGCGAAGCGTTCGCACTTCGTCCCGCCTTCCTGCGCCTTCCTGT-3′) for wild-type box I and 36-mer (5′-CATGCTCGCAGCGGGACGACACCTACGAGCGCCCCG-3′) and 44-mer (5′-CGGGGCGCTCGTAGGTGTCGTCCCGCCTTCCTGCGCCTTCCTGT-3′) for mutant box I (obtained from Operon Technologies, Alameda, CA) were purified by 16% PAGE under denaturing conditions. Bands corresponding to the full-length oligonucleotides were excised and gently homogenized in elution buffer (10 mM Tris·HCl, pH 7.8/0.5 M ammonium acetate/1 mM EDTA/0.1% SDS), and eluted at room temperature overnight. The homogenates were briefly centrifuged in a 0.4 μm filter unit to remove the gel, and the oligonucleotides were precipitated with ethanol. The 44-mer was 5′ end-labeled with T4 polynucleotide kinase and [γ-32P]ATP, and mixed with a 5-fold excess of the 36-mer in 50 mM Tris·HCl (pH 7.5) and 200 mM NaCl. The oligonucleotides were then incubated at 90°C for 10 min, at 65°C for 10 min, and at room temperature for 3 h. The duplex oligonucleotides were electrophoresed through 15% polyacrylamide in 1× TBE (45.5 mM Tris base/44.5 mM boric acid/1 mM EDTA) at 10 V/cm. Bands corresponding to the box I substrate were excised from the gel and eluted as described above. The purity of the substrates was critical since free ssDNA or excess salt inhibited helicase activity.
Box I substrate with only a 3′ single-stranded tail was prepared by annealing the 5′ end-labeled 26-mer (5′-GCGGGACGAAGTGCGAACGCTTCGCG-3′) with the wild-type 44-mer; box I substrate with only a 5′ single-stranded tail was prepared by annealing the 5′ end-labeled 26-mer (5′-CGCGAAGCGTTCGCACTTCGTCCCGC-3′) with the 44-mer (5′-CATGCTCGCACGTCCTTCGCGGGACGAAGTGCGAACGCTTCGCG-3′); box I substrate with ssDNA tails in the opposite orientation was prepared by annealing the 5′ end-labeled 36-mer (5′-CATGCTCGCACGCGAAGCGTTCGCACTTCGTCCCGC-3′) with the 44-mer (5′-GCGGGACGAAGTGCGAACGCTTCGCGCTTCCTGCGCCTTCCTGT-3′).
Box I substrates with differing lengths of 3′ ssDNA tails were prepared by annealing the 5′ end-labeled 26-mer (5′-CGCGAAGCGTTCGCACTTCGTCCCGC-3′), 30-mer (5′-CGCGAAGCGTTCGCACTTCGTCCCGCCTTC-3′), 34-mer (5′-CGCGAAGCGTTCGCACTTCGTCCCGCCTTCCTGC-3′), or 44-mer (5′-CGCGAAGCGTTCGCACTTCGTCCCGCCTTCCTGCGCCTTCCTGT-3′) with the wild-type 36-mer. The duplex substrates were purified as described above.
DNA Unwinding Assay.
To measure unwinding of box I, reaction mixtures (25 μl) containing 50 mM Hepes·KOH (pH 7.8), 100 mM NaCl, 0.5 mM dithiothreitol, 10% glycerol, 10 μg bovine serum albumin, 2 pmol of box I substrate, and 2 pmol of UL9 protein were assembled on ice. ATP (4 mM) was added to the reaction mixtures which were then incubated at 37°C for 60 min. The reactions were stopped by the addition of 6.5 μl of stop solution (100 mM EDTA/1% SDS/20 μg of proteinase K), and the mixtures were incubated for an additional 10 min at 37°C, followed by electrophoresis through a 15% polyacrylamide gel at 10 V/cm. The gel was then dried on Whatman DE81, and quantitated with a PhosphorImager (Molecular Dynamics).
RESULTS
Our initial attempts to demonstrate the specific unwinding of oriS by the UL9 protein were patterned after similar studies with the SV40 large T antigen. In the case of the T antigen, a double hexamer binds, and by virtue of its helicase activity, unwinds the SV40 origin of replication (18, 19). In vitro, the reaction requires a single-strand DNA binding protein (SSB); either the mammalian RP-A or the Escherichia coli SSB, and ATP (24). Similar experiments with the HSV-1 UL9 protein using both linear duplexes containing oriS or plasmids into which oriS had been inserted failed to demonstrate the specific unwinding of sequences within oriS either alone or in the presence of the HSV-1 encoded SSB, ICP8, or heterologous SSBs (unpublished data).
UL9 Protein Binds Box I But Fails to Unwind It.
Failure to detect unwinding of oriS by the UL9 protein prompted us to examine more closely the interaction of the UL9 protein with the high affinity box I sequence alone. Box I is present in both oriS and oriL and consists of 18 nt residues, including the core binding site for the UL9 protein (5) (Fig. 1A). A 32P-labeled, fully duplex box I sequence was incubated with stoichiometric amounts of UL9 protein and ATP in the presence or absence of ICP8. Although hydrolysis of the ATP was observed, the UL9 protein was unable to unwind the box I duplex (data not shown).
A Complex of UL9 Protein and ICP8 Can Unwind Box I with Attached Single Strands.
To examine the possibility that a single-stranded region might be required to promote the unwinding of box I, a box I duplex was synthesized to which ssDNA strands were attached downstream of the box I sequence (Fig. 1B). We reasoned that these single strands might mimic the easily unwound A+T-rich sequence that links boxes I and II (Fig. 1A). Binding of the UL9 protein to box I to which ssDNA tails are attached (box I substrate) was verified by a gel mobility shift analysis (data not shown).
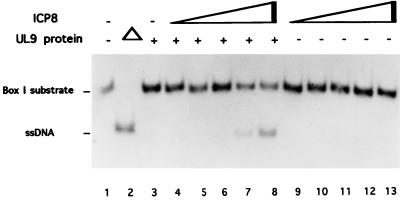
Despite the presence of ssDNA tails, no unwinding of box I by the UL9 protein alone could be detected (Fig. 2). However, upon addition of ICP8, unwinding of the box I substrate by the UL9 protein was observed (Fig. 2). No unwinding was detected with ICP8 alone, which under certain conditions can display helix destabilizing activity (23).
Figure 2.
Unwinding of box I substrate by UL9 protein and ICP8. The UL9 protein and increasing amounts of ICP8 were incubated with the box I substrate (2 pmol) for 60 min at 37°C. Lanes: 1, no protein; 2, boiled substrate; 3, UL9 protein alone; 4–8, 0.001, 0.01, 0.1, 1, and 10 pmol of ICP8 in the presence of the UL9 protein (2 pmol); 9–13, ICP8 in the absence of the UL9 protein.
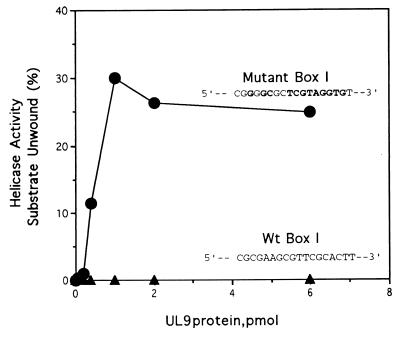
Although the UL9 protein alone was unable to unwind the box I substrate, mutation of the box I sequence to prevent binding of the UL9 protein (25) resulted in significant unwinding (Fig. 3). The ability of the UL9 protein to unwind the mutant duplex in the absence of ICP8 is presumably a manifestation of the nonspecific helicase activity associated with the UL9 protein, which requires the loading site provided by the attached 3′ single strand (17).
Figure 3.
Unwinding activity of the wild-type (Wt) and mutant box I substrates by UL9 protein. The core binding site for UL9 protein was mutated to prevent specific binding (mutations shown in boldface type) (7). Increasing amounts of the UL9 protein (as indicated) were incubated with the box I substrate (2 pmol) for 60 min at 37°C. The reaction mixtures were electrophoresed through a 15% polyacrylamide gel, and the ssDNA products of the reaction were quantitated with a PhosphorImager.
An obvious role for ICP8 in this reaction is to bind to the single strands generated as a consequence of the unwinding of box I by the UL9 protein, thereby preventing their reannealing. This has, in fact, been shown to be the case for the SV40 large T antigen; indeed, a variety of heterologous SSBs (e.g., ICP8 and E. coli SSB) can substitute for RP-A, the mammalian SSB (24, 26). In contrast, unwinding of the box I substrate by the UL9 protein required the homologous SSB, ICP8. Previous experiments had shown that the nonspecific helicase activity of the UL9 protein is stimulated by ICP8 as a consequence of the physical interaction between the two proteins (17, 27). These observations suggest that the unwinding of the box I substrate by the UL9 protein and ICP8 requires a similar interaction.
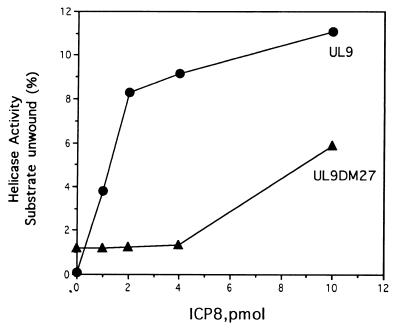
Further support for the specific interaction between ICP8 and the UL9 protein was provided by a mutant UL9 protein (UL9DM27) that lacks 27 amino acids at the C terminus (28) and interacts weakly with ICP8 in solution (29). The mutant protein binds oriS normally (28) and exhibits helicase and DNA-dependent ATPase activities (29). As shown in Fig. 4, the ability of the mutant UL9 protein to unwind the box I substrate in the presence of ICP8 was severely reduced. This finding suggests that an interaction between the C-terminal portion of the UL9 protein and ICP8 is needed to promote the unwinding of box I. In contrast to the wild-type UL9 protein, the mutant protein showed a low level of helicase activity in the absence of ICP8. This effect may be related to its high intrinsic helicase activity (29). These findings demonstrate that a specific complex of UL9 protein and ICP8 can unwind box I to which ssDNA is attached. Because the fully duplex box I substrate lacking tails is not unwound by the UL9 protein–ICP8 complex, we infer that the unwinding of box I is mediated by a complex consisting of UL9 protein and ICP8 bound to the ssDNA. Thus, unlike the case of SV40 large T antigen where unwinding of the origin requires an SSB solely to maintain the separation of the single strands produced by its helicase action, essentially a passive role, ICP8 is an active participant in the unwinding process.
Figure 4.
Requirement of the C-terminal portion of the UL9 protein to unwind box I substrate in the presence of ICP8. The wild-type UL9 protein and mutant UL9 protein (UL9DM27) (2 pmol) each were incubated with increasing amounts of ICP8 and box I substrate (2 pmol) for 60 min at 37°C. Following 15% PAGE, the ssDNA products of the reaction were quantitated with a PhosphorImager.
Unwinding of Box I by the UL9 Protein–ICP8–ssDNA Complex Specifically Requires a 3′ ssDNA Tail Downstream of Box I.
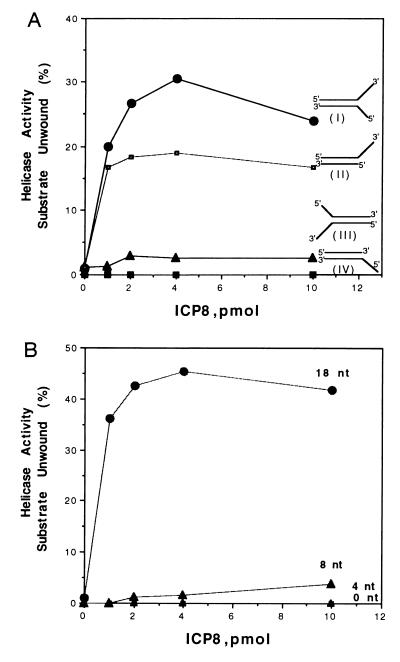
To investigate further the role of the single strands in the unwinding of box I, a series of box I substrates differing only in the length and orientation of their single-stranded tails was synthesized and examined for their ability to be unwound by the UL9 protein–ICP8 complex. These included, in addition to box I with the 3′ and 5′ ssDNA tails used in the previous experiments, box I with only a 3′ ssDNA tail and box I with a 5′ ssDNA tail. (Fig. 5A). As shown in Fig. 5A, only the box I substrate with a 3′ ssDNA tail downstream of box I could be unwound by the UL9 protein–ICP8 complex. Optimal unwinding was observed at a stoichiometry of 1 mol of ICP8 per mol of 3′ ssDNA tail. The UL9 protein alone was unable to unwind any of the box I substrates in the absence of ICP8.
Figure 5.
(A) Requirement of 3′ ssDNA tail to unwind box I substrate by UL9 protein–ICP8 complex. (I) Box I substrate with 3′ and 5′ ssDNA tails; (II) box I with a 18-nt 3′ ssDNA tail; (III) box I with ssDNA tails in opposite orientation; (IV) box I with a 18-nt 5′ ssDNA tail. UL9 protein (2 pmol) was incubated with the box I substrates, and the indicated amounts of ICP8 at 37°C for 60 min. (B) Effect of 3′ ssDNA tail length on unwinding of box I by UL9 protein–ICP8 complex. Following 15% PAGE, the ssDNA products were quantitated with a PhosphorImager.
Because of the importance of the 3′ ssDNA tail, we examined the influence of 3′ ssDNA tail length on the ability of the UL9 protein–ICP8 complex to unwind box I. Of the substrates tested, only box I with a 18-nt-long ssDNA tail was efficiently unwound; there was a low level of unwinding of box I with an 8-nt ssDNA tail (Fig. 5B). Box I with a 4-nt ssDNA tail was inert, as was box I lacking a 3′ tail (Fig. 5B). Estimates of the binding site size of ICP8 for ssDNA range from 12 to 18 nt (23, 30, 31), suggesting that binding of at least one ICP8 monomer to the ssDNA tail is required to form an active complex with the UL9 protein. These results provide further support for the idea that the unwinding of box I requires a complex consisting of the UL9 protein and ICP8 bound to ssDNA.
Unwinding of Box I Requires Specific Orientation of the UL9 Protein–ICP8 Complex.
The nucleotide sequence of the two UL9 binding sites (boxes I and II) in oriS are inverted relative to each other, and joined by an 18-nt A+T-rich linker (Fig. 1A). Inasmuch as a 3′ ssDNA tail, at least 18 nt in length attached to box I is required by the UL9 protein–ICP8 complex, we wished to determine whether a specific orientation of the UL9 protein when bound to box I was required to unwind the box I substrate. We therefore prepared box I with the ssDNA tails upstream of box I. Despite the fact that the 18-nt 3′ ssDNA tail can serve as an ICP8 binding site, unwinding of box I was significantly reduced (Fig. 5A, substrate III). These findings suggest that to unwind box I the UL9 protein must bind in such a way that its C terminus can interact directly with ICP8 bound to the single strand at the 3′ end of the box I sequence.
DISCUSSION
Although the UL9 protein encoded by HSV-1 binds tightly and specifically to the HSV-1 origins of replication, oriL and oriS, and possesses DNA helicase activity, the unwinding of sequences within these origins either in the presence or absence of an SSB has not been observed. In contrast, the SV40 large T antigen, the initiator protein for SV40 DNA replication, can bind, and by virtue of its intrinsic helicase activity, unwind the SV40 origin (18, 19). In vitro, an SSB, either the homologous RP-A or the heterologous E. coli SSB, is required to maintain the separated strands.
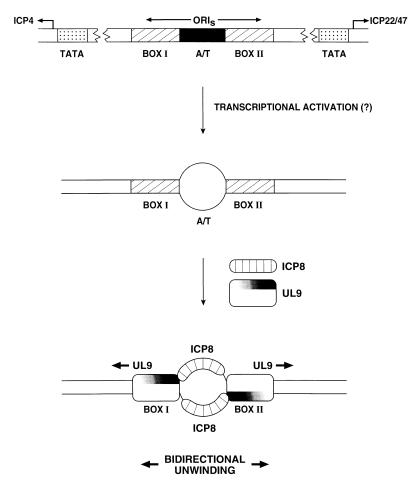
We report here that box I, the high affinity binding site for the HSV-1 UL9 protein within oriS or oriL, can be unwound by the UL9 protein; however, it can only do so if the box I duplex bears an appropriately oriented 3′ ssDNA tail to which at least one ICP8 equivalent is bound. Because a mutant UL9 protein with a C-terminal deletion that interacts weakly with ICP8 shows a correspondingly reduced unwinding activity, we infer that the specific interaction of the C-terminal portion of the UL9 protein with ICP8 is required to potentiate the unwinding of box I. Because the 3′ ssDNA tail must be positioned downstream of box I (Fig. 5A), the unwinding requires a specific orientation of the UL9 protein on box I. These findings lead to a model for the unwinding of an HSV-1 origin of replication by the HSV-1-encoded origin binding protein that differs from that observed for two other viral origins, SV40 and bovine papilloma virus (Fig. 6). In this model, the UL9 protein binds boxes I and/or II with a specific orientation, but is unable to unwind the origin unless ICP8 binds to ssDNA regions generated from the A+T-rich linker as a consequence of limited unwinding. The importance of this linker is emphasized by the finding that deletion of all or part of this sequence inhibits HSV-1 or -2 DNA replication in vivo (10, 32). This limited unwinding may occur prior to or concurrently with UL9 protein binding, and may result from viral immediate early transcription or recruitment of transcription factors to regions flanking the origin (33–35). Transcriptional activation of the initiation of replication has been observed with animal viruses including SV40, bovine papilloma virus, adenovirus, and Epstein–Barr virus as well as E. coli (36, 37). An ICP8 monomer bound to the “top” strand of the unwound A+T-rich sequence can interact with the UL9 protein bound at box I, and the ICP8 monomer bound to the “bottom” strand can interact with UL9 protein bound to box II (or the second box I in oriL) (Fig. 6). Once the two UL9 protein–ICP8 complexes are formed, there is bidirectional unwinding from the two binding sites, and DNA replication can be initiated. In view of the strong homologies in origins of replication and in origin binding proteins, this model could also apply to other human herpesviruses including herpes simplex virus type 2, varicella zoster virus, and human herpes virus type 6.
Figure 6.
Model for bidirectional unwinding of HSV-1 origin of replication (see text for details).
Acknowledgments
We thank Drs. Paul Boehmer and Suk-Hee Lee for UL9DM27 protein and human RP-A, respectively. This work was supported by Grant AI-26538 from the National Institutes of Health.
ABBREVIATIONS
- HSV-1
herpes simplex virus type 1
- SV40
simian virus 40
- SSB
single-strand DNA binding protein
- ssDNA
single-stranded DNA
References
- 1.Elias P, O’Donnell M E, Mocarski E S, Lehman I R. Proc Natl Acad Sci USA. 1986;83:6322–6326. doi: 10.1073/pnas.83.17.6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olivo P D, Nelson N J, Challberg M D. Proc Natl Acad Sci USA. 1988;85:5414–5418. doi: 10.1073/pnas.85.15.5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C A, Nelson N J, McGeoch D J, Challberg M D. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stow N D. J Gen Virol. 1992;73:313–321. doi: 10.1099/0022-1317-73-2-313. [DOI] [PubMed] [Google Scholar]
- 5.Challberg M, Kelly T. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- 6.Bruckner R C, Crute J J, Dodson M S, Lehman I R. J Biol Chem. 1991;266:2669–2674. [PubMed] [Google Scholar]
- 7.Fierer D S, Challberg M S. J Virol. 1992;66:3986–3995. doi: 10.1128/jvi.66.7.3986-3995.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elias P, Gustafsson C M, Hammarsten O. J Biol Chem. 1990;265:17167–17173. [PubMed] [Google Scholar]
- 9.Hazuda D J, Perry H C, McClements W L. J Biol Chem. 1992;267:14309–14315. [PubMed] [Google Scholar]
- 10.Lockshon D, Galloway D A. Mol Cell Biol. 1988;8:4018–4027. doi: 10.1128/mcb.8.10.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, Olivo P D. J Virol. 1994;68:3841–3849. doi: 10.1128/jvi.68.6.3841-3849.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baumann R P, Yalamanchili V R R, O’Callaghan D J. J Virol. 1989;63:1275–1283. doi: 10.1128/jvi.63.3.1275-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewhurst S, Dollard S C, Pellett P E, Dambaugh T R. J Virol. 1993;67:7680–7683. doi: 10.1128/jvi.67.12.7680-7683.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez R, Shao L, Weller S K. J Virol. 1992;66:6735–6746. doi: 10.1128/jvi.66.11.6735-6746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue N, Dambaugh T R, Rapp J C, Pellett P E. J Virol. 1994;68:4126–4136. doi: 10.1128/jvi.68.7.4126-4136.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodson M S, Lehman I R. J Biol Chem. 1993;268:1213–1219. [PubMed] [Google Scholar]
- 17.Boehmer P E, Dodson M S, Lehman I R. J Biol Chem. 1993;268:1220–1225. [PubMed] [Google Scholar]
- 18.Stillman B. Annu Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- 19.Boroweic J A, Dean F B, Bullock P A, Hurwitz J. Cell. 1990;60:181–184. doi: 10.1016/0092-8674(90)90730-3. [DOI] [PubMed] [Google Scholar]
- 20.Bramhill D, Kornberg A. Cell. 1988;54:915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- 21.Makhov A M, Boehmer P E, Lehman I R, Griffith J D. EMBO J. 1996;15:1742–1750. [PMC free article] [PubMed] [Google Scholar]
- 22.Koff A, Schwedes J F, Tegtmeyer P. J Virol. 1991;65:3284–3292. doi: 10.1128/jvi.65.6.3284-3292.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boehmer P E, Lehman I R. J Virol. 1993;67:711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dean F B, Bullock P, Murakami Y, Wobbe C R, Weissbach L, Hurwitz J. Proc Natl Acad Sci USA. 1987;84:16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hazuda D J, Perry H C, Naylor A M, McClements W L. J Biol Chem. 1991;266:24621–24626. [PubMed] [Google Scholar]
- 26.Kenny M K, Lee S-H, Hurwitz J. Proc Natl Acad Sci USA. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boehmer P E, Lehman I R. Proc Natl Acad Sci USA. 1993;90:8444–8448. doi: 10.1073/pnas.90.18.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arbuckle M I, Stow N D. J Gen Virol. 1993;74:1379–1355. doi: 10.1099/0022-1317-74-7-1349. [DOI] [PubMed] [Google Scholar]
- 29.Boehmer P E, Craigie M C, Stow N D, Lehman I R. J Biol Chem. 1994;269:29329–29334. [PubMed] [Google Scholar]
- 30.O’Donnell M E, Elias P, Lehman I R. J Biol Chem. 1987;262:4242–4259. [PubMed] [Google Scholar]
- 31.Gustafsson C M, Falkenberg M, Simonsson S, Valadi H, Elias P. J Biol Chem. 1995;270:19028–19034. doi: 10.1074/jbc.270.32.19028. [DOI] [PubMed] [Google Scholar]
- 32.Gustafsson C M, Hammarsten O, Falkenberg M, Elias P. Proc Natl Acad Sci USA. 1994;91:4629–4633. doi: 10.1073/pnas.91.11.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong S W, Schaffer P A. J Virol. 1991;65:2601–2611. doi: 10.1128/jvi.65.5.2601-2611.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dabrowski C E, Schaffer P A. J Virol. 1991;65:3140–3150. doi: 10.1128/jvi.65.6.3140-3150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voss J H, Roizman B. Proc Natl Acad Sci USA. 1988;85:8454–8458. doi: 10.1073/pnas.85.22.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baker T A, Kornberg A. Cell. 1988;55:113–123. doi: 10.1016/0092-8674(88)90014-1. [DOI] [PubMed] [Google Scholar]
- 37.DePamphilis M L. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]