Abstract
Active 18S and 25S ribosomal RNAs were produced in trans in yeast, from plasmids containing RNA polymerase II transcription signals and rDNA fragments with unique hybridization tags. Analyses were carried out in cells with temperature-sensitive RNA polymerase I. Functional rRNAs were derived from separate 18S and 5.8/25S rRNA coding units, however, active 25S rRNA could be produced only by cotranscription with 5.8S rRNA. The results demonstrate that the polycistronic organization of the large rDNA operon is not required for successful processing of rRNA or assembly of functional ribosomes. The split operon system should facilitate future efforts to dissect eukaryotic ribosome biogenesis.
The eukaryotic 18S, 5.8S, and 25/28S ribosomal RNAs are generated in the nucleolus from a single large precursor. Processing of this transcript occurs concomitantly with formation of modified nucleotides, binding of ribosomal proteins, and vectoral transport of the developing subunits (1–4). All eukaryotic organisms are thought to process rRNA in a similar way (5, 6). In yeast, the 35S primary transcript is first cleaved at a site in the 5′ external transcribed spacer (5′ ETS) called A0. This is followed by cleavages designated A1 and A2/A3, which define the 5′ end of 18S RNA (A1) and divide the transcript into two intermediates containing the 18S and 5.8/25S RNAs (Fig. 1A). These intermediates undergo further cleavages to generate the mature rRNAs (10, 11).
Figure 1.
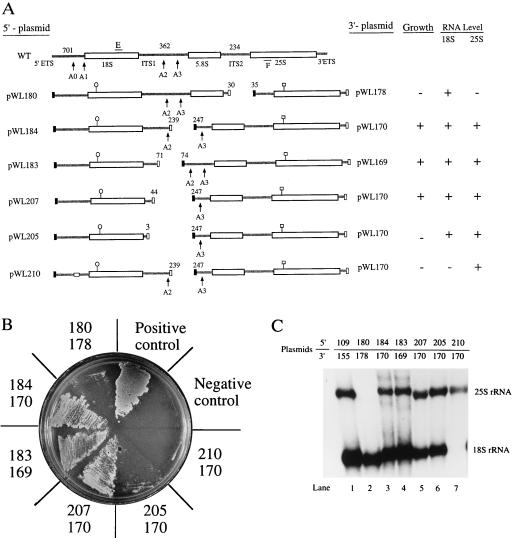
The large rDNA operon of S. cerevisiae can be successfully expressed in two parts. (A) Expression of small and large subunit rRNAs in trans. The yeast rDNA operon was divided at the sites indicated, and each segment was expressed from a different plasmid, under control of GAL7 promoters and terminators (closed and open boxes). The experimental 18S and 25S RNA coding units contained unique hybridization tags (○| and □|) and the plasmids harboring the 5′ and 3′ portions of the operon carried URA3 and TRP1 genes (see Materials and Methods). rDNA function was evaluated in yeast strain NOY504, which contains a ts RNA polymerase I mutation and mutations in URA3 and TRP1 (7). Plasmid pWL210 is identical to pWL184 except that it contains a 6-nt substitution in the 5′ ETS region (open rectangle) shown to base-pair with U3 snoRNA (472AAAGAG477→UCUUCA; ref. 8). Sites of pre-rRNA processing (A0, A1, etc.) are indicated by arrows. Sizes of the ETS and ITS segments are shown above the wild-type operon. E and F refer to sites used to prime PCR amplification. Growth and RNA levels are indicated at the right. The numbers above the experimental constructs refer to the nucleotide positions at which they were divided. Note that the dissections are imperfect, with at least a few nucleotides absent in each case (see also Fig. 2). (B) Phenotypic properties of the split operons. Transformants containing the plasmid pairs shown in A were streaked on galactose plates at 37°C and incubated for 7 days. The positive control contained a pair of plasmids with intact rDNA operons (pWL109 and pWL155), and the negative control contained vectors without rDNA inserts. Growth phenotypes are summarized in A. +, good growth (colonies evident in 7 days); −, no growth. (C) Northern hybridization analysis of rRNA expressed in trans. Cells were first grown in glucose medium at 25°C and then diluted into galactose medium. RNA was extracted after incubation at 37°C for 7 h (9). Northern assays were carried out using probes specific for unique hybridization tags in 18S and 25S RNAs. Plasmids pWL109 and pWL155 were used as a positive control. The levels of 18S and 25S RNAs are tabulated in A; + indicates wild-type level, and − indicates an undetectable level.
Results developed in the past few years have revealed that eukaryotic rRNA processing requires not only proteins but also small nucleolar RNA (snoRNAs), functioning as small nucleolar ribonucleoproteins (12). Thus far, four snoRNAs have been found to be essential for rRNA processing in yeast, and a fifth nonessential species is known to influence the kinetics of the early processing events. Nearly 30 other yeast snoRNAs identified are not essential for growth (12); several of these provide sequence specificity for 2′-O-methylation of rRNA (refs. 13, 14; Ni, Balakin, and M.J.F., unpublished data). Three of the five snoRNAs required for processing are phylogenetically conserved, i.e., U3, U14, and MRP. U3, U14, and a yeast-specific snoRNA, snR30, are required for processing of 18S RNA at sites A1 and A2 (15–17). U3 is known to interact with the 5′ ETS segment and with internal regions in 18S RNA, and U14 base pairs with two different regions in 18S RNA (8, 9, 18). The MRP snoRNA is required for cleavage at a site (A3) upstream of 5.8S RNA (19–21). In vertebrates, production of 18S and 5.8/28S rRNA also requires the U22 and U8 snoRNAs, respectively; homologues to these RNAs are not known in yeast (22, 23). The roles of the snoRNAs required for processing are still obscure, but could include catalyzing hydrolysis and organizing pre-rRNA and other components of the processing apparatus. Processing of rRNA is also expected to involve protein-based nucleases, based on the recent discovery of a yeast homologue of bacterial RNase III (24).
It has been proposed that the processing pathways for the eukaryotic 18S and 5.8/25S RNAs may be independent (25). This hypothesis is supported by deletion mutation results with the yeast rDNA operon, which showed that alterations that affect production of 18S RNA do not block production of 25S RNA and vice versa (26, 27). Consistent with the possibility of independent processing pathways, loss of essential snoRNAs impairs the production of either the 18S or the 5.8/25S rRNAs, but not both.
The goal of the present study was to test the hypothesis that processing, and perhaps ribosome assembly, require only cis-acting elements flanking the individual rRNAs, rather than a contiguous multimeric transcript. To this end, we divided the yeast rDNA operon into two segments, introduced these fragments into separate expression vectors, and asked if ribosomes capable of supporting growth could be produced. The results show that functional ribosomes can, in fact, be generated in this manner.
MATERIALS AND METHODS
Cell Culturing.
Yeast strain NOY504 was kindly provided by M. Nomura (University of California, Irvine) (7). Its genotype is: MATα, rpa12::LEU2, leu2-3, 112, ura3-1, trp-1, his3-11, CAN1-100. Cells were cultured in medium containing 0.67% yeast nitrogen base (amino acid-free), 2% vitamin-free casamino acids, and 2% glucose or galactose, supplemented with uracil (200 μM) or tryptophan (400 μM) as needed. Cells and cultures were manipulated as described by Sherman et al. (28).
DNA Manipulation.
Plasmid pWL109 is a derivative of a URA3 multicopy plasmid (pNOY102, ref. 29) and contains the intact rDNA operon flanked by GAL7 promoter and terminator elements, respectively. The 18S rDNA in this operon contains a hybridization tag at variable region 2 of the coding sequence (9). Plasmid pWL155, a derivative of pNOY199 (an unpublished plasmid construct obtained from M. Nomura), is a TRP1 multicopy vector containing the rDNA allele flanked by the GAL7 promoter and terminator. The experimental rDNA segments were created by a PCR amplification strategy, using a high-fidelity enzyme (pfu polymerase) and primers containing restriction sites. In all cases, at least two independent PCR clones were examined. The sites of separation within the ITS1 segment are shown in Fig. 2. Plasmids pWL180, -184, -183, -207, -205, and -210 were constructed with rDNA fragments created with DNA primers complementary to the individual ITS1 sequence and a primer (E) specific for a site in 18S rDNA. Primer E is 5′-CCAGCAGCCGCGGUAAUU-3′ with a natural SacII site, which is about 560 nucleotides downstream from the 5′ end of 18S RNA. The primers designed to split the operon contained a SalI tail. The resulting 18S rDNA fragments were subcloned into plasmid pWL109 after digestion with SalI and SacII enzymes. Digestion of pWL109 with SalI and SacII removed all of the rDNA insert, except the 5′ ETS and the early part of the 18S rDNA demarcated by the SacII site. For construction of plasmids pWL178, -170, and -169, individual primers specific for the intended split sites were paired with primer F (5′-CACTTTCATTACGCGTATGGGT-3′) containing a natural MluI site for PCR amplification. The primers designed to split the operon contained an XhoI tail. The resulting fragments were subcloned into pWL155 after digestion with XhoI and MluI. The 25S rDNA in pWL155 contained a hybridization tag inserted at the KpnI site of the 5′ coding sequence and a built-in XhoI sequence 29 nucleotides from the start of the rDNA coding sequence. The KpnI site is about 130 nucleotides downstream from the 5′ end of the 25S RNA coding sequence. The tag sequence is 5′-GGAAGCTGCAGCC-3′, which contains a PstI site. Digestion of pWL155 with XhoI and MluI removed all of the rDNA fragment except the portion of the 25S RNA allele demarcated by the MluI site, which is about 700 nucleotides downstream from the 5′ end of the 25S RNA coding sequence. Plasmid pWL210 was derived from pWL184 and contains a 6-nt substitution in the 5′ ETS region known to base-pair with U3 snoRNA (472AAAGAG477→UCUUCA, ref. 8). This same substitution is known to disrupt U3 function and abolish the production of 18S RNA from pre-rRNA (8).
Figure 2.
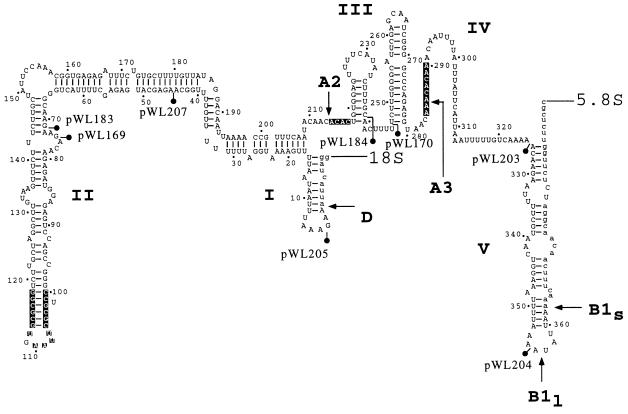
Secondary structure of the yeast ITS1 region. The structure is divided into five domains labeled I-V (adapted from ref. 30). Known processing sites A2, A3, B1L, and B1S are indicated by arrows. Site D corresponds to the 3′ end of mature 18S RNA. B1L and B1S cleavages give rise to long and short forms of 5.8S RNA. The sites used to divide the operon (•|), and the corresponding plasmids are identified. Bold nucleotides are known to be conserved among several yeast species (25).
Northern Analysis.
Cells were first grown in glucose medium at 25°C and then diluted into galactose medium at an OD 660 of 0.8 and incubated at 37°C for 7 h, at which time RNA was extracted. Northern assays were carried out with about 2 μg of RNA using probes specific for the unique hybridization tags in 18S and 25S RNAs (9). The nucleotide sequences for the 18S and 25S probes are 5′-CGCCGAGGATCCAACTAGGGGGCT-3′, 5′-GGGCAGGCTGCAGCTTCCTACCAG-3′, respectively.
RESULTS AND DISCUSSION
Expression Strategy.
In our strategy, the separate rDNA fragments were fused to inducible RNA polymerase II promoters and expressed from different vectors in a test strain (NOY504) with a temperature-sensitive RNA polymerase I mutation (Fig. 1A; ref. 7). To facilitate analysis, unique hybridization tags were incorporated into the 18S and 25S rRNA coding units, in variable sequence regions in each case (Fig. 1). The insertions were found to be phenotypically neutral, as cells that depended on the tagged 18S and 25S RNAs grew as well as cells expressing plasmid-encoded wild-type rRNA (see below).
The rDNA operon was divided into two segments at three different sites. The first site was located 30 nucleotides downstream of the 5.8S coding unit, yielding a 5′ segment encoding both the 18S and 5.8S RNAs and a 3′ segment encoding only 25S RNA (Fig. 1A). The second dissection site was 239 nucleotides beyond the 3′ end of 18S RNA, midway between the A2 and A3 cleavage sites. The third site was located 71 nucleotides beyond the 3′ end of 18S RNA. Separation of the 35S precursor at these latter two sites resulted in a 5′ segment encoding 18S RNA and a 3′ segment encoding both the 5.8S and 25S RNAs. The 5′ segments were cloned separately into a multicopy yeast vector marked with the URA3 gene (pWL180, -184, and -183). The 3′ segments were inserted into a multicopy TRP1 vector (pWL178, -170, and -169). Each fragment was fused to a GAL7 promoter and terminator.
The appropriate plasmid pairs were transformed into the test strain and evaluated for activity. As a positive control, cells were also cotransformed with URA3 and TRP1 plasmids containing intact rDNA operons, also under control of the GAL7 promoter. Cells transformed with empty vectors served as a negative control. Transformants were grown at 25°C on glucose medium lacking uracil and tryptophan for plasmid maintenance. They were then streaked on both galactose and glucose media and incubated at 37°C for 7 days for phenotypic observation. No transformants grew on glucose medium at 37°C, indicating that the GAL7 promoter was repressed, and transcription of chromosomal rDNA was too low to support growth. However, on galactose medium, colonies from the positive control strain appeared 3 days after incubation at 37°C (Fig. 1B).
Active rRNAs Can Be Produced in Trans.
Notably, the two strains in which the rDNA operon was divided into 18S and 5.8S/25S segments showed visible growth 5 days after incubation, with colonies becoming apparent 2 days later (pWL184 + pWL170; pWL183 + pWL169). In contrast, transformants containing the operon divided into 18S/5.8S and 25S segments failed to form visible colonies (pWL180 + pWL178, Fig. 1B). The doubling times for cells containing the plasmid pairs pWL184 + pWL170 and pWL183 + pWL169 were 10.2 and 12.8 h, respectively. This compares with 6.5 h for the positive control (pWL155 + pWL109). To ensure that the split operons were not reassembled into an intact operon in vivo, plasmids were recovered from cells able to grow on galactose, shuttled into Escherichia coli, and then subjected to restriction analysis. The majority of the plasmids were identical to the original forms. For yeast cells containing plasmids pWL184 and pWL170, 16 of 20 E. coli clones harbored the original plasmids. The four remaining clones showed a different restriction pattern, but did not contain intact rDNA operons. These last variants were not able to support growth when reintroduced into the yeast test strain.
Several factors might be responsible for the slower growth associated with the split operons. One possibility is that rRNA production from the GAL7-expression vectors is limited by RNA polymerase II activity. Relative to an intact operon, at least twice as many transcription events would be required to produce the same number of rRNAs in trans. More likely is the possibility that dividing the operon reduces the efficiency of one or more posttranscriptional events.
Expression of the plasmid-encoded rRNAs was examined by Northern analysis, 7 h after the medium and temperature shifts. The 18S and 25S RNAs produced from the plasmid alleles were distinguished from the chromosomally encoded RNAs with the unique hybridization tags (Fig. 1C). The tagged 18S and 25S RNAs were clearly evident in the positive control strain (pWL109 + pWL155), demonstrating that insertion of the tag sequences did not compromise the stability of these rRNAs. 18S RNA was detected in all strains (Fig. 1C) except the negative control (not shown). Equivalent levels of 18S RNA were observed in cells containing the intact operon and cells harboring 18S rDNA plasmids followed by: (i) 71 nucleotides of 3′ flanking sequence, or (ii) DNA that extends 30 nucleotides beyond the 3′ end of 5.8S RNA (Fig. 1C, lanes 2 and 4). These results indicate that production of 18S RNA requires very little 3′ flanking sequence. Similarly, the level of 25S RNA was the same in strains containing either of the 5.8S/25S transcription units, and these levels were comparable to that of the positive control (Fig. 1C, lanes 1, 3, and 4). In contrast, no 25S RNA was detected in cells in which the 5.8S and 25S RNA coding units were separated (Fig. 1C, lane 2). Taken together, these results indicate that functional small and large subunit rRNAs can be produced in trans from separate transcription units. Active 18S RNA is produced when the 18S rDNA coding unit is separated from the adjoining 5.8/25S rDNA sequences. However, production of functional 25S RNA requires cotranscription of both 5.8S and 25S rRNAs.
To further define the 3′ flanking sequences required for 18S RNA production, we deleted all but 44 and 3 nucleotides, respectively, of the 362-nt internal transcribed spacer-one sequence (ITS1) in an 18S rDNA plasmid (Fig. 1A, detailed in Fig. 2). These constructs were introduced into cells expressing the 5.8S and 25S RNAs from a single plasmid (pWL170). The construct with 44 nucleotides of ITS1 allowed good growth on galactose, whereas the variant with only 3 nucleotides of spacer failed to support growth (Fig. 1B). Interestingly, the mutant phenotype was not due to the absence of 18S RNA, because tagged 18S RNA was observed in these cells (Fig. 1C, compare lanes 5 and 6). The fact that more than 3 nucleotides of spacer flanking the 3′ end of 18S RNA is necessary for growth suggests that ITS1, though dispensable for production of 18S rRNA, has a vital role in the development of active ribosomes. Similarly, it has been shown that deletion of several conserved elements within the second internal transcribed spacer (ITS2; 234 nucleotides) does not compromise 25S RNA production, but does interfere with growth (31). The roles of these internal sequences are not known, but it is reasonable to believe they are involved in some subsequent stage of ribosome maturation.
U3 snoRNA Is Still Required for Processing.
One question prompted by the split operon expression results is whether rRNAs produced in this system bypass the requirement for snoRNAs in processing. Because it is known that cleavages at the A0, A1, and A2 sites require base-pairing of U3 with an element in the 5′ ETS (8), we tested the requirement for U3 when 18S RNA is expressed in trans. Starting with plasmid pWL184, we made a 6-nt substitution mutation in the region shown to base-pair with U3 (pWL210; Fig. 1A). This construct was tested for activity in a strain containing a functional 5.8S/25S rDNA unit (pWL170). The transformants failed to grow on galactose medium at 37°C (Fig. 1B), and a Northern analysis showed that 18S RNA failed to accumulate in this strain (Fig. 1C, lane 7). These results indicate that processing of 18S RNA produced from the split operon also requires base-pairing of U3 with the precursor.
Influence of the ITS Segments.
To define cis-acting elements necessary for 25S RNA production, we examined the roles of the ITS2 and ITS1 segments. One nonproductive construct lacked 35 nucleotides at the 5′ end of ITS2 (pWL178; Fig. 1A). Because it was unclear if the loss of 25S RNA was due to the absence of these 35 nucleotides, we examined the activity of 25S rDNA joined to a complete ITS2 sequence (pWL208, Fig. 3A). This construct was analyzed in conjunction with a plasmid encoding 18S and 5.8S RNAs (pWL180; Fig. 3A). Transformants containing this 25S rDNA allele also failed to support growth (Fig. 3B). Northern analysis showed that 25S RNA did not accumulate, suggesting that the 5′ cis-segment required for production of 25S RNA extends at least up to 5.8S RNA (Fig. 3C, lane 3).
Figure 3.
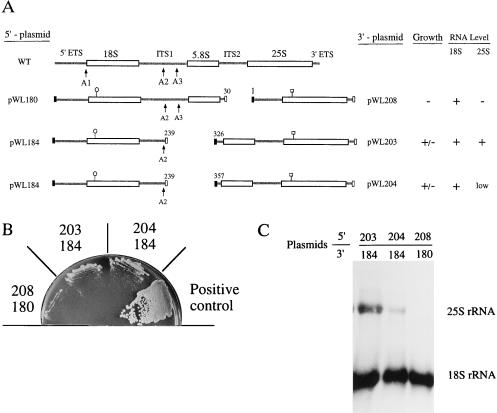
The ITS1 region is required for production of 25S RNA. (A) Structure of ITS1 deletion variants. The strategy used to construct these plasmids is described in Fig. 1A, along with details for construction of plasmids pWL180 and pWL184 (see Materials and Methods). Novel 3′-rDNA plasmids pWL208, -203, and -204 were prepared by subcloning appropriate PCR fragments into pWL155 after double digestion with XhoI and MluI. All rDNAs were expressed with GAL7 promoters and terminators. (B) Growth properties of cells containing the plasmid pairs shown in A. Phenotypes were analyzed on galactose medium, as described in Fig. 1B and are summarized in A. ± indicates extremely slow growth. (C) Northern hybridization analysis of rRNA expressed from the different split operons; details of the assay are provided in Fig. 1C. Levels of 18S and 25S RNA are tabulated in A.
Knowing that 25S RNA can be produced from a construct containing the 5.8S and 25S RNA coding units and the last 116 nucleotides of the (362-nt) ITS1 segment (pWL170), we next examined shorter variants that contained only 37 or 7 nucleotides of the ITS1 segment (Fig. 3A). Both deletions eliminated the A3 cleavage site. RNA produced from the longer construct retains the potential to create a stem structure that forms between the 3′ portion of the ITS1 segment and the 5′ end of 5.8S RNA (domain V, Fig. 2). This stem structure was previously found to be important for maturation of 25S RNA (30). The shorter construct lacks both domains IV and V and retains only 7 upstream nucleotides from a naturally occurring short form of 5.8S RNA produced by wild-type cells. These constructs were analyzed in combination with a fragment containing the remaining 5′ portion of the operon (pWL184, Fig. 3A). Both sets of transformants grew extremely slowly on galactose at 37°C (Fig. 3B). Northern analysis showed that a low level of 25S RNA was produced in cells containing the 5.8S/25S rDNA fragment preceded by only 7 nucleotides, demonstrating that efficient production of 25S RNA requires additional ITS1 DNA (Fig. 3C, lane 2). In contrast, the level of 25S RNA in cells expressing the rDNA segment with 37 nucleotides of ITS1 DNA (Fig. 3C, lane 1) was similar to that of the positive control (not shown). These results showed that the 3′ end of the ITS1 sequence from positions 326 to 362 (Fig. 2) is critical for production of 25S RNA in trans, and that nucleotides 239–325 are important for the production of a functional large ribosomal subunit.
These last mapping results are in good agreement with results from a previous mutational analysis of ITS1 in an intact operon (30). That study showed that deletion of ITS1 nucleotides 283–294, which eliminates the A3 cleavage site but maintains domain V, had little effect on 25S RNA production. Thus, it seems that the spacer sequence itself rather than the cleavage per se is required for 25S RNA production. Relevant to this, elimination of the A3 cleavage site results in a switch to an alternative processing pathway involving the B1L site (Fig. 2; ref. 32). The hypothesis that spacer nucleotides 326–362 influence pre-rRNA folding is particularly appealing (30). The absence of this part of the spacer in our analysis is predicted to cause improper folding of the 5.8S/25S substrate, and these precursors might be rapidly degraded. We cannot comment on the effects of the ITS1 deletion on 5.8S RNA production, as this RNA was not tagged in our constructs.
Implications for Pre-rRNA Processing.
In Saccharomyces cerevisiae, the primary rRNA precursor is believed to be assembled into a 90S preribosomal RNP complex, which is subsequently converted into two complexes of 43S and 66S (33). The 43S complex contains a 20S precursor to 18S RNA and has been postulated to be transformed into the small ribosomal subunit. The 66S complex contains a 27S rRNA precursor encoding the 5.8S and 25S RNAs, and this complex is viewed as the nascent large ribosomal subunit. The present study demonstrates that cotranscription of eukaryotic rRNAs is not a prerequisite for the formation of functional ribosomal subunits. An early aim of our future work will be to determine if the 90S rRNP maturation complex, which we call the “processome” (34), is formed in the two-part operon system. Interestingly, a few eubacteria and archaebacteria are known to produce the small and large subunit rRNAs from independent transcription units (35–38). This situation is in contrast to most prokaryotic rDNA systems, but could be relevant to the trans expression results reported here. The ability to express active eukaryotic small and large subunit rRNAs from separate genes should facilitate future efforts to dissect the rRNA maturation and subunit assembly processes.
Acknowledgments
We thank M. Nomura for providing rDNA plasmids pNOY102 and pNOY199 and yeast strain NOY504, and T. L. Mason and R. A. Zimmermann for valuable discussions. This work was supported by National Institutes of Health Grant GM19351 to M.J.F.
ABBREVIATIONS
- 5′ ETS
5′ external transcribed spacer
- snoRNA
small nucleolar RNA
- ITS1
internal transcribed spacer-one sequence
- ITS2
second internal transcribed spacer
References
- 1.Warner J R. Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woolford J L, Jr, Warner J R. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Broach J R, Pringle J R, Jones E W, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1991. pp. 587–626. [Google Scholar]
- 3.Woolford Jr J L. Adv Genet. 1991;29:63–118. doi: 10.1016/s0065-2660(08)60107-8. [DOI] [PubMed] [Google Scholar]
- 4.Raue H A, Planta R J. Prog Nucleic Acid Res Mol Biol. 1991;41:81–129. doi: 10.1016/s0079-6603(08)60007-0. [DOI] [PubMed] [Google Scholar]
- 5.Eichler D C, Craig N. Prog Nucleic Acid Res Mol Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- 6.Sollner-Webb B, Tycowski K T, Steitz J A. In: Ribosomal RNA: Evolution, Gene Expression and Function in Protein Synthesis. Zimmermann R A, Dahlberg A E, editors. Boca Raton, FL: CRC; 1996. pp. 469–490. [Google Scholar]
- 7.Nogi Y, Yano R, Dodd J, Carles C, Nomura M. Mol Cell Biol. 1993;13:114–122. doi: 10.1128/mcb.13.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beltrame M, Tollervey D. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang W-Q, Fournier M J. Genes Dev. 1995;9:2433–2443. doi: 10.1101/gad.9.19.2433. [DOI] [PubMed] [Google Scholar]
- 10.Raue H A, Planta R J. Gene Expression. 1995;5:71–77. [PMC free article] [PubMed] [Google Scholar]
- 11.Venema J, Tollervey D. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell E S, Fournier M J. Annu Rev Biochem. 1995;35:897–934. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 13.Kiss-Laszlo Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 14.Nicoloso M, Qu L-H, Michot B, Bachellerie J-P. J Mol Biol. 1996;260:178–195. doi: 10.1006/jmbi.1996.0391. [DOI] [PubMed] [Google Scholar]
- 15.Hughes J M, Ares M J. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H V, Zagorski J, Fournier M J. Mol Cell Biol. 1990;10:1145–1152. doi: 10.1128/mcb.10.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrissey J P, Tollervey D. Mol Cell Biol. 1993;13:2469–2477. doi: 10.1128/mcb.13.4.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes J M. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 19.Schmitt M, Clayton D. Mol Cell Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu S, Archer R H, Zengel J M, Lindahl L. Proc Natl Acad Sci USA. 1994;91:659–663. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 22.Tycowski K, Shu M, Steitz J A. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 23.Peculis B A, Steitz J A. Cell. 1993;73:1233–1245. doi: 10.1016/0092-8674(93)90651-6. [DOI] [PubMed] [Google Scholar]
- 24.Elela S A, Igel H, Ares M J. Cell. 1996;85:115–124. doi: 10.1016/s0092-8674(00)81087-9. [DOI] [PubMed] [Google Scholar]
- 25.Van Nues R W, Rientjes J M, van der Sande C A, Zerp S F, Sluiter C, Venema J, Planta R J, Raue H A. Nucleic Acids Res. 1994;22:912–919. doi: 10.1093/nar/22.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musters W, Venema J, van der Linden G, van Heerikhuizen H, Klootwijk J, Planta R J. Mol Cell Biol. 1989;9:551–559. doi: 10.1128/mcb.9.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Nues R W, Venema J, Planta R J, Raue H A. In: The Translational Apparatus: Structure, Function, Regulation, and Evolution. Nierhaus K H, Franceschi F, Subramanian A R, Erdmann V A, Wittmann-Liebold B, editors. New York: Plenum; 1993. pp. 151–162. [Google Scholar]
- 28.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1985. [Google Scholar]
- 29.Nogi Y, Yano R, Nomura M. Proc Natl Acad Sci USA. 1991;88:3962–3966. doi: 10.1073/pnas.88.9.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Nues R W, Venema J, Rientjes J M J, Dirks-Mulder A, Raue H A. Biochem Cell Biol. 1995;73:789–801. doi: 10.1139/o95-087. [DOI] [PubMed] [Google Scholar]
- 31.Van Nues R W, Rientjes J M J, Morre S A, Mollee E, Planta R J, Venema J, Raue H A. J Mol Biol. 1995;250:24–36. doi: 10.1006/jmbi.1995.0355. [DOI] [PubMed] [Google Scholar]
- 32.Henry Y, Wood H, Morrissey J P, Petfalski E, Kearsey S, Tollervey D. EMBO J. 1994;13:2452–2463. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapman J, Retel J, Planta R J. Exp Cell Res. 1975;90:95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- 34.Fournier M J, Maxwell E S. Trends Biochem Sci. 1993;18:131–135. doi: 10.1016/0968-0004(93)90020-n. [DOI] [PubMed] [Google Scholar]
- 35.Tu J, Zillig W. Nucleic Acids Res. 1982;10:7231–7245. doi: 10.1093/nar/10.22.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukunaga M, Mifuchi I. J Bacteriol. 1989;171:5763–5767. doi: 10.1128/jb.171.11.5763-5767.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersson S G E, Zomorodipour A, Winkler H H, Kurland C G. J Bacteriol. 1995;177:4171–4175. doi: 10.1128/jb.177.14.4171-4175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang H, Winkler H H. J Bacteriol. 1993;175:3893–3896. doi: 10.1128/jb.175.12.3893-3896.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]