Abstract
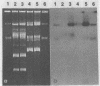
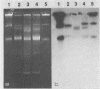
Plasmid pAJ1106 and its deletion derivative, plasmid pAJ2074, conferred lactose-fermenting ability (Lac) and bacteriophage resistance (Hsp) at 30°C to Lac− proteinase (Prt)-negative Lactococcus lactis subsp. lactis and L. lactis subsp. lactis var. diacetylactis recipient strains. An additional plasmid, pAJ331, isolated from the original source strain of pAJ1106, retained Hsp and conjugative ability without Lac. pAJ331 was conjugally transferred to two L. lactis subsp. lactis and one L. lactis subsp. cremoris starter strains. The transconjugants from such crosses acquired resistance to the phages which propagated on the parent recipient strains. Of 10 transconjugant strains carrying pAJ1106 or one of the related plasmids, 8 remained insensitive to phages through five activity test cycles in which cultures were exposed to a large number of industrial phages at incubation temperatures used in lactic casein manufacture. Three of ten strains remained phage insensitive through five cycles of a cheesemaking activity test in which cultures were exposed to approximately 80 different phages through cheesemaking temperatures. Three phages which propagated on transconjugant strains during cheesemaking activity tests were studied in detail. Two were similar (prolate) in morphology and by DNA homology to phages which were shown to be sensitive to the plasmid-encoded phage resistance mechanism. The third phage was a long-tailed, small isometric phage of a type rarely found in New Zealand cheese wheys. The phage resistance mechanism was partially inactivated in most strains at 37°C.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Gautier M., Chopin M. C. Plasmid-Determined Systems for Restriction and Modification Activity and Abortive Infection in Streptococcus cremoris. Appl Environ Microbiol. 1987 May;53(5):923–927. doi: 10.1128/aem.53.5.923-927.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl Environ Microbiol. 1984 Feb;47(2):343–349. doi: 10.1128/aem.47.2.343-349.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Klaenhammer T. R. Bacteriophage Resistance Conferred on Lactic Streptococci by the Conjugative Plasmid pTR2030: Effects on Small Isometric-, Large Isometric-, and Prolate-Headed Phages. Appl Environ Microbiol. 1986 Jun;51(6):1272–1277. doi: 10.1128/aem.51.6.1272-1277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis A. W., Meyer J. Electron microscopic heteroduplex study and restriction endonuclease cleavage analysis of the DNA genomes of three lactic streptococcal bacteriophages. Appl Environ Microbiol. 1986 Mar;51(3):566–571. doi: 10.1128/aem.51.3.566-571.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis Audrey W. Conjugal Transfer in Lactic Streptococci of Plasmid-Encoded Insensitivity to Prolate- and Small Isometric-Headed Bacteriophages. Appl Environ Microbiol. 1988 Mar;54(3):777–783. doi: 10.1128/aem.54.3.777-783.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A. Conjugative 40-megadalton plasmid in Streptococcus lactis subsp. diacetylactis DRC3 is associated with resistance to nisin and bacteriophage. Appl Environ Microbiol. 1984 Jan;47(1):68–74. doi: 10.1128/aem.47.1.68-74.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing W. D., Klaenhammer T. R. Conjugal Transfer of Bacteriophage Resistance Determinants on pTR2030 into Streptococcus cremoris Strains. Appl Environ Microbiol. 1986 Jun;51(6):1264–1271. doi: 10.1128/aem.51.6.1264-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenson L. R., Klaenhammer T. R. Streptococcus cremoris M12R transconjugants carrying the conjugal plasmid pTR2030 are insensitive to attack by lytic bacteriophages. Appl Environ Microbiol. 1985 Oct;50(4):851–858. doi: 10.1128/aem.50.4.851-858.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]