Abstract
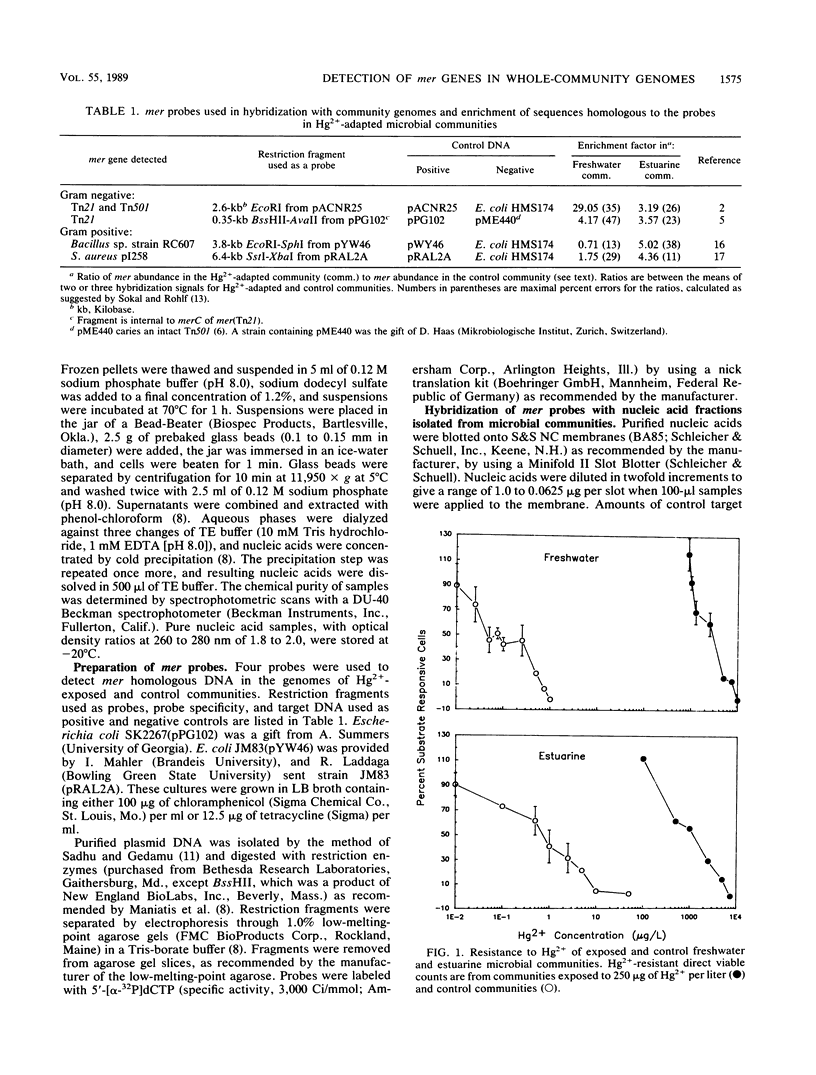
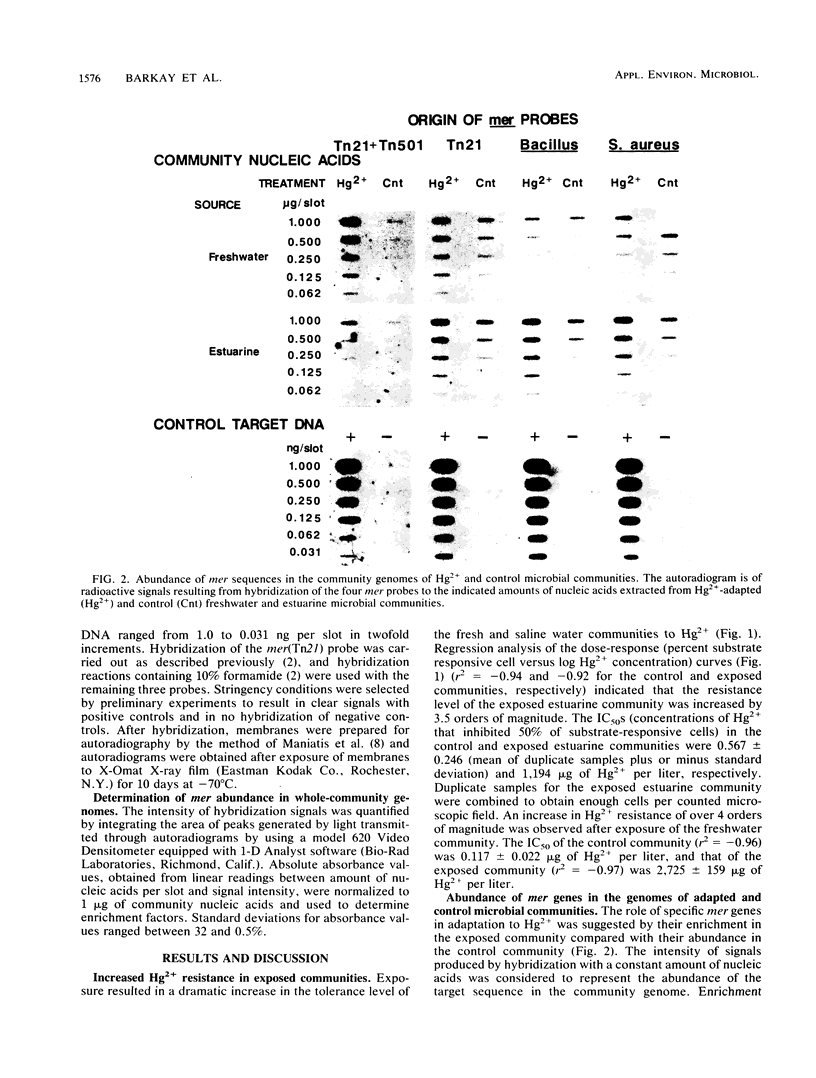
Nucleic acids extracted from microbial biomass without prior culturing were hybridized with probes representing four mer operons to detect genes encoding adaptation to Hg2+ in whole-community genomes. A 29-fold enrichment in sequences similar to the mer genes of transposon Tn501 occurred during adaptation in a freshwater community. In an estuarine community, all four mer genes were only slightly enriched (by three- to fivefold), suggesting that additional, yet uncharacterized, mer genes encoded adaptation to Hg2+.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barkay T. Adaptation of aquatic microbial communities to hg stress. Appl Environ Microbiol. 1987 Dec;53(12):2725–2732. doi: 10.1128/aem.53.12.2725-2732.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Fouts D. L., Olson B. H. Preparation of a DNA gene probe for detection of mercury resistance genes in gram-negative bacterial communities. Appl Environ Microbiol. 1985 Mar;49(3):686–692. doi: 10.1128/aem.49.3.686-692.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkay T., Liebert C., Gillman M. Environmental significance of the potential for mer(Tn21)-mediated reduction of Hg2+ to Hg0 in natural waters. Appl Environ Microbiol. 1989 May;55(5):1196–1202. doi: 10.1128/aem.55.5.1196-1202.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert M. P., Summers A. O. The distribution and divergence of DNA sequences related to the Tn21 and Tn501 mer operons. Plasmid. 1988 Sep;20(2):127–136. doi: 10.1016/0147-619x(88)90015-7. [DOI] [PubMed] [Google Scholar]
- Itoh Y., Watson J. M., Haas D., Leisinger T. Genetic and molecular characterization of the Pseudomonas plasmid pVS1. Plasmid. 1984 May;11(3):206–220. doi: 10.1016/0147-619x(84)90027-1. [DOI] [PubMed] [Google Scholar]
- Sadhu C., Gedamu L. A procedure for the preparation of RNA-free plasmid DNA. Biotechniques. 1988 Jan;6(1):20–21. [PubMed] [Google Scholar]
- Silver S., Misra T. K. Plasmid-mediated heavy metal resistances. Annu Rev Microbiol. 1988;42:717–743. doi: 10.1146/annurev.mi.42.100188.003441. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Mahler I., Levinson H. S., Halvorson H. O. Cloning and expression in Escherichia coli of chromosomal mercury resistance genes from a Bacillus sp. J Bacteriol. 1987 Oct;169(10):4848–4851. doi: 10.1128/jb.169.10.4848-4851.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte W., Green L., Misra T. K., Silver S. Resistance to mercury and to cadmium in chromosomally resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Apr;29(4):663–669. doi: 10.1128/aac.29.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]




