Abstract
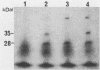
A small cryptic plasmid, pLJ1, was isolated from Lactobacillus helveticus subsp. jugurti and was cloned into Escherichia coli HB101 by using pBR329 as a vector. Plasmid pLJ1 was 3,292 base pairs long and had single restriction endonuclease sites for PvuII, KpnI, AvaII, Acci, HindIII, and EcoRI. In a maxicell system, pLJ1 produced a protein of about 41 kilodaltons.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chassy B. M., Gibson E., Giuffrida A. Evidence for extrachromosomal elements in Lactobacillus. J Bacteriol. 1976 Sep;127(3):1576–1578. doi: 10.1128/jb.127.3.1576-1578.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covarrubias L., Bolivar F. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene. 1982 Jan;17(1):79–89. doi: 10.1016/0378-1119(82)90103-2. [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. H., Savage D. C. Cryptic plasmids in Lactobacillus strains isolated from the murine gastrointestinal tract. Appl Environ Microbiol. 1985 Apr;49(4):1004–1006. doi: 10.1128/aem.49.4.1004-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Rupert C. S. Determination of plasmid molecular weights from ultraviolet sensitivities. Nature. 1978 Mar 30;272(5652):471–472. doi: 10.1038/272471a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley M. B., Fryder V. Plasmids, Lactic Acid Production, and N-Acetyl-d-Glucosamine Fermentation in Lactobacillus helveticus subsp. jugurti. Appl Environ Microbiol. 1978 Apr;35(4):777–781. doi: 10.1128/aem.35.4.777-781.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]