Abstract
Replication of complementary-strand DNA in filamentous phages is initiated by a primer RNA that is synthesized at the minus-strand origin on the viral single-stranded DNA by Escherichia coli RNA polymerase holoenzyme containing the σ70 subunit. We have demonstrated that the affinity of RNA polymerase in vitro to the origin is about 16-fold higher than that to the lacUV5 promoter. We have also shown that the temperature dependence of the primer RNA synthesis is much lower than that of lacUV5 transcription. The high affinity of RNA polymerase to the origin depends on the single strandedness of the “−10 region.” A nucleotide sequence of the nontemplate strand in the −10 region was found to be important for the function, but that of the template strand was not. These observations suggest that σ70 subunit directly interacts with the single-stranded nontemplate strand containing adenine residue(s) at the −10 region of promoter.
Keywords: primer RNA, σ factor, open complex, −10 region, nontemplate strand
In the filamentous bacteriophages (f1, M13, and fd), synthesis of the minus strand is initiated by a RNA primer that is synthesized in the presence of single-stranded DNA binding protein (SSB) by Escherichia coli RNA polymerase holoenzyme containing the σ70 subunit (1, 2). The primer RNA is synthesized at a unique site named the minus-strand origin on the viral single-stranded DNA (ssDNA). The origin contains two hairpin structures named B and C. We have previously reported (1) that the origin and a transcriptional promoter are similar in terms of structure and function. However, the similarity could not be observed in the structure of the “−10 region” in the origin, which is most probably single-stranded (1, 3).
The replication cycle of the phage DNA can be divided into three different stages, which are (i) conversion of viral ssDNA (plus strand) to the double-stranded replicative form (RF) by the synthesis of the minus strand, initiated by RNA polymerase; (ii) multiplication of the RF, which involves, in addition to the minus-strand synthesis, rolling-circle type replication of the plus strand, which is initiated by the phage-encoded gp2 nicking enzyme; and (iii) coating of the single-stranded viral DNA by phage-encoded gp5 (single-strand binding protein) to prevent minus-strand synthesis and to facilitate phage assembly in the membrane (4, 5). The synthesis of the minus strand seems to be quite efficient in the absence of gp5. In vivo, the intermediate single-stranded form during stage ii replication (RF → RF) cannot be detected. Wild-type phage infection does not induce the SOS response, but the infection by a mutant phage defective in the minus-strand origin induces the response (6, 7).
These observations suggest that the affinity of RNA polymerase to the origin in vivo must be higher than to most transcriptional promoters. Otherwise, accumulation of ssDNA would induce the SOS response continuously during the early stage of phage infection. In this report, we show that the affinity of RNA polymerase to the minus-strand origin is much higher than to the lacUV5 promoter in vitro and that the temperature dependence of primer RNA synthesis is much lower than that of transcription from lacUV5 promoter. Two parameters of the −10 region of the origin, its single-stranded structure and a nucleotide sequence containing adenine residue(s) in the nontemplate strand, are essential for the higher affinity to the enzyme.
MATERIALS AND METHODS
Bacteria and Media.
E. coli K38 (HfrC) (8) was generally used for the growth of phage f1. JM109 (supE44, recA1, Δ(lac-proAB)/F′) (9) and DH5 (recA1) (9) were used for the double-origin plasmid assay and for the chloramphenicol acetyltransferase (CAT) assay, respectively. The culture medium used was TY (10).
Enzymes and Chemicals.
E. coli RNA polymerase (holo and core enzymes) and SSB were generous gifts of A. Ishihama and N. Shimamoto, respectively, of the National Institute of Genetics. Restriction enzymes, T4 polynucleotide kinase, and T4 DNA ligase were purchased from Takara. 32P-labeled nucleoside triphosphates and [14C]chloramphenicol were from DuPont/New England Nuclear.
Plasmids and Phages.
H300 is a derivative of wild-type f1 that carries a unique HindIII site at position 5583 [for f1 nucleotide numbers, see Hill and Petersen (11)]. H320 is a derivative of H300 that carries unique XhoI, MluI, and EcoRI sites at positions 5682, 5752, and 5948, respectively. Construction of these restriction sites did not affect the activity of the phage origin (data not shown).
Double-origin plasmid pDOWT is a derivative of pBR322 that carries two copies of the HpaII-H fragment of wild-type f1 (from positions 5615 to 5996) inserted in a counterclockwise direction at the EcoRI and BamHI sites, respectively (3). pDO320 is a derivative of pBR322 that carries the f1 HpaII-H fragment inserted at the BamHI site and the HindIII–EcoRI fragment of H320 substituted for the HindIII–EcoRI fragment of pBR322. The mutants in the hairpin C region (m325–m328) were constructed on the plasmid by using synthetic oligonucleotides carrying mutant sequences. The phages carrying these mutations were obtained by ligating the smaller HindIII–EcoRI fragments from the pDO plasmids to the larger HindIII–EcoRI fragment of H320.
pSK+lac1 was constructed as follows. Two complementary 85-mer oligodeoxynucleotides containing the lacUV5 promoter sequence, plac1+ (5′-CGGAATTCTCGAGGCTTTACATTTATGCTTCCGGCTCGTATAATGTGTGGAATTGTGAGCGGATAACAATTTCTGCAGAATTCCG) and plac1−(5′-CGGAATTCTGCAGAAATTGTTATCCGCTCACAATTCCACACATTATACGAGCCGGAAGCATAAATGTAAAGCCTCGAGAATTCCG), were synthesized, annealed, digested with EcoRI and PstI, and inserted between the EcoRI and PstI sites of pBluescript SK(+).
Three plasmids, pKKminus-Ori, pKKOri-10cons, and pKKlacUV5, were constructed by using three couples of oligodeoxynucleotides N14 (5′-pGATCCGATTTAGTGCTTTACGGCACCTCGACCCCAAAAAACTTGATTAGGGA) and N15 (5′-pAGCTTCCCTAATCAAGTTTTTTGGGGTCGAGGTGCCGTAAAGCACTAAATCG), N16 (5′-pGATCCGATTTAGTGCTTTACGGCACCTCGACCCCATATAATTTGATTAGGGA) and N17 (5′-pAGCTTCCCTAATCAAATTATATGGGGTCGAGGTGCCGTAAAGCACTAAATCG), and N20 (5′-pGATCCCCAGGCTTTACACTTTATGCTTCCGGCTCGTATAATGTGTGGAATTA) and N41 (5′-pAGCTTAATTCCACACATTATACGAGCCGGAAGCATAAAGTGTAAAGCCTGGG). They were annealed to each other and inserted between BamHI and HindIII sites of pKK232–8 (12).
Preparation of Template DNA.
Two DNA fragments carrying the lacUV5 promoter sequence, lacUV5–1 (155 bp) and lacUV5–2 (127 bp), were prepared from the recombinant plasmid pSK+lac1 by digestion with ClaI/SacI and ClaI/XbaI, respectively, and purified by PAGE. Another template lacUV5–3 was prepared from pSK+lac1 by PCR using two oligodeoxynucleotides, T7 primer (5′-GTAATACGACTCACTATAGGGC) and T3 primer (5′-AATTAACCCTCACTAAAGGG), and purified by PAGE. The viral single-stranded DNA template was prepared as described (10).
In Vitro Mixed RNA Synthesis.
In vitro mixed RNA synthesis was carried out in a standard reaction (20 μl) that contained 0.1 pmol of template DNA, 0–0.8 pmol of competitor DNA, 480 pmol of E. coli SSB (120 pmol of SSB/0.1 pmol of ssDNA template), 40 mM Hepes·KOH (pH 7.5), 130 mM KCl, bovine serum albumin (0.02 mg/ml), 5 mM MgCl2, 500 μM ATP, 500 μM CTP, 500 μM UTP, 50 μM GTP, and 10 μCi of [α-32P]GTP (800 Ci/mmol; 1 Ci = 37 GBq), 0.1 pmol of RNA polymerase holoenzyme (Eσ70), and heparin (0.2 mg/ml). The reaction was carried out in three steps: the SSB binding step in which template DNA and SSB were mixed at room temperature for 10 min; the preincubation step at 37°C for 10 min after addition of the buffer, salt, and RNA polymerase to the DNA-SSB mixture; and the single-round RNA synthesis step at 37°C for 10 min after addition of NTP substrates, [α-32P]GTP, and heparin. The reaction was stopped by adding 80 μl of a stop solution containing 80 mM NaCl and 10 mM EDTA. Nucleic acid was precipitated with ethanol and analyzed on an 18% polyacrylamide/8 M urea sequencing gel. Autoradiography and measurement of radioactivity in gel bands were carried out with a Fuji image analyzer.
Double-Origin Plasmid Assay.
JM109 cells harboring a double-origin plasmid were grown at 37°C in TY broth to an optical density at 660 nm of 0.3 and divided into two parts. One was infected with f1 at a phage/cell ratio of 50, and the other remained uninfected. Cells were harvested after further incubation at 37°C for 30 min with shaking. Intracellular DNA was extracted and electrophoresed in a 0.7% agarose gel containing 0.5 μg of ethidium bromide per ml.
CAT Assay.
E. coli DH5 cells harboring pKK232–8, pKKminus-Ori, pKKOri-10cons, or pKKlacUV5 were grown at 37°C to the midlogarithmic phase. One milliliter of culture was harvested and resuspended in lysozyme (5 mg/ml). After 10 min at 4°C, cells were lyzed by freezing and thawing and centrifuged at 12,000 × g for 15 min at 4°C. CAT activity in the supernatant was determined as described (13) by using [14C]chloramphenicol as substrate.
RESULTS
Competition Experiments of the Minus-Strand Origin and the lacUV5 Promoter for RNA Polymerase.
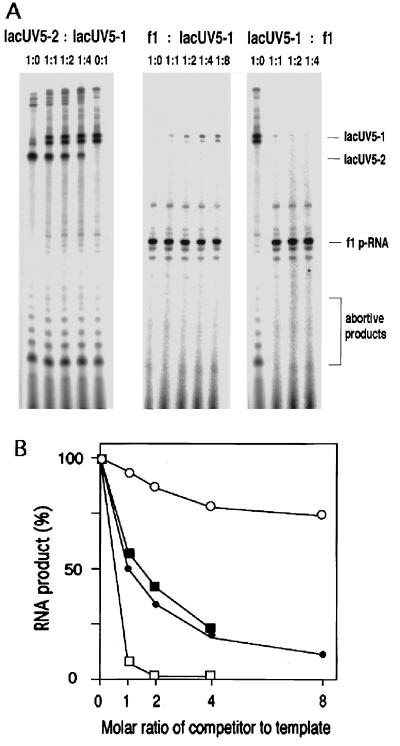
To compare primer RNA synthesis of filamentous phage f1 with general transcription, competition experiments were carried out. Reactions contained 0.1 pmol of RNA polymerase holoenzyme, 0.1 pmol of template DNA, various amounts of competitor DNA, SSB, and heparin. Two DNA fragments containing the lacUV5 promoter sequence, lacUV5–1 (155 bp) and lacUV5–2 (127 bp), were prepared and used as template. RNA synthesis from the lacUV5 promoters was not affected by the addition of E. coli SSB (data not shown). As shown in Fig. 1 A Center and B, primer RNA synthesis from f1 ssDNA was not inhibited by more than 26% even when 0.8 pmol of lacUV5–1 DNA was added as a competitor. Consistent with this result, transcription from 0.1 pmol of lacUV5–1 was drastically decreased to 7% by the addition of 0.1 pmol of f1 DNA (Fig. 1 A Right and B). As a control experiment, we measured transcription from lacUV5–2 in the presence of 0–0.4 pmol of lacUV5–1 fragment as a competitor. The result shown in Fig. 1 A Left and B is consistent with what is expected when the template and competitor have identical affinity to RNA polymerase.
Figure 1.
In vitro mixed RNA synthesis using lacUV5 promoter and f1 minus-strand origin. (A) Electropherogram of RNA products synthesized in the standard reaction that contained 0.1 pmol of template DNA and 0–0.8 pmol of competiter DNA as indicated. (Left) lacUV5–2 as template and lacUV5–1 as competitor. (Center) f1 ssDNA as template and lacUV5–1 as competitor. (Right) lacUV5–1 as template and f1 ssDNA as competitor. Molar ratio of template to competitor is shown above each lane. Position of each RNA product is indicated on the right. (B) Quantification of the data shown in A. Amounts of RNA products were measured by densitometry of autoradiographs using Fuji BAS 2000 system. ○, f1 ssDNA as template and lacUV5–1 as competitor; □, lacUV5–1 as template and f1 ssDNA as competitor; ▪, lacUV5–2 as template and lacUV5–1 as competitor; •, theoretical curve for a template and a competitor of identical affinity.
If in the presence of heparin the amount of RNA synthesized (Px for template x) is proportional to the product of the affinity constant of polymerase to the binding site (Kx and Ky for templates x and y, respectively) and relative concentration of template ([D]x and [D]y), then Px = Kx[D]x/(Kx[D]x + Ky[D]y) and, thus, Kx/Ky = [D]yPx/[D]x(1 − Px).
When this equation was applied to the results shown in Fig. 1B, the average value obtained for Kf1/Kuv5 was approximately 16. Thus, the affinity of RNA polymerase to the f1 minus-strand origin is 16-fold higher than that to the lacUV5 promoter. Although we have not carried out kinetic experiments, the rate of formation of stable complexes with f1 DNA may be about 16-fold higher than with lacUV5 DNA, if exchange between the two stable complexes does not occur at significant rates.
The quantification of primer RNA and lacUV5 transcripts showed that in the absence of competitor DNA the molar yield of the primer RNA was 13% of the amount of f1 ssDNA added as template (0.1 pmol), but the yield of the transcript from lacUV5 was 2.3% of template added (0.1 pmol). In addition, the amount of abortive products synthesized on 0.1 pmol of template in f1 primer RNA synthesis was not greater than 3% of that in the transcription from the lacUV5 promoter (Fig. 1). These results indicate that primer RNA synthesis is much more efficient than transcription.
Temperature Dependence of Priming Reaction and Transcription.
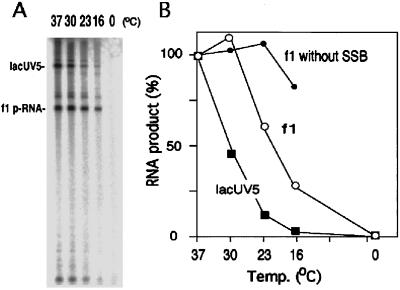
A major structural difference between the minus-strand origin and transcriptional promoters is that the origin is partially single-stranded. We have previously proposed that when the nucleotide sequence of the minus-strand origin is depicted by horizontally aligning the two hairpins as if they were parts of a mostly double-stranded linear molecule (see Fig. 3A Top), the nucleotides specifically required for origin function are found in the −10 and −35 regions (1). In this model, the −10 region is probably single-stranded, and the strand connecting the −35 region and the RNA start site is the nontemplate strand. During initiation of transcription at promoters, the binary complex of RNA polymerase and promoter DNA undergoes a change from a closed complex to an open complex, in which the enzyme is bound to DNA much more tightly. During this transition, a region of about 17 bp around the −10 region is unwound, which leads to the exposure of the template strand to facilitate the polymerization (14). Open complex formation is dependent on temperature and salt concentration (15). We therefore compared primer RNA synthesis and transcription from the lacUV5 promoter for their dependence on reaction temperature in a mixed RNA synthesis system in vitro. The results shown in Fig. 2 indicate that the temperature dependence of primer RNA synthesis is lower than that for lacUV5 transcription. Primer synthesis at 23°C and 16°C was 61% and 27%, respectively, of that at 37°C. On the other hand, lacUV5 transcription at 23°C and 16°C was 12% and 2.1%, respectively, of that at 37°C (Fig. 2). The nonspecific polymerization activity of RNA polymerase on the single-stranded template without SSB at 16°C was 86% of that at 37°C. These results suggest that the energy required to form the open complex in primer RNA synthesis is less than that in lacUV5 transcription.
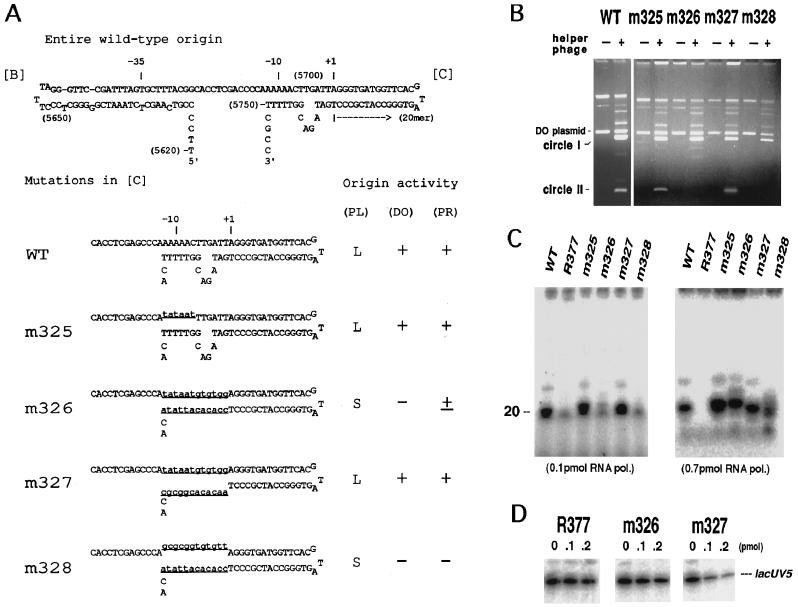
Figure 3.
Origin activity of base-substitution mutants in the −10 region of the minus-strand origin. (A) A possible secondary structure of the wild-type and mutant origins. The curved arrow represents the primer RNA. Numerals in parentheses are f1 nucleotide numbers (11). The altered nucleotides in mutants are shown by underlined lowercase type. Activity of the origins, determined by three methods, plaque size of the phage carrying the mutation, double-origin plasmid assay (3), and primer synthesis in vitro, is qualitatively indicated on the right. (PL), plaque size; L, large; S, small. The results of double-origin plasmid assay (DO) and primer RNA synthesis (PR) are shown in B and C, respectively. For explanation of double-origin plasmid assay, see the text. (B) Results of double-origin plasmid assay for the wild-type, m325, m326, m327, and m328 minus-strand origin. +, Infected with f1; −, uninfected. The positions of solid circles of the parental plasmid (DO plasmid) and its two derivatives, circle I and circle II, are indicated on the left. Intensity of the circle II band is a measure of activity of the mutant origin. (C) In vitro synthesis of primer RNA on mutant origins. Viral ssDNA (0.1 pmol) extracted from mutant phages was used as templates as indicated. RNA synthesized with 0.1 pmol (Left) or 0.7 pmol (Right) of RNA polymerase holoenzyme in a standard reaction was electrophoresed in 12% (Left) or 15% (Right) polyacrylamide gel containing 8 M urea and radioautographed. (D) In vitro mixed RNA synthesis using lacUV5 promoter and mutant origins. The reaction contained 0.1 pmol of lacUV5–3, a DNA fragment carrying lacUV5 promoter, and 0–0.2 pmol of competitor (viral ssDNA) as indicated.
Figure 2.
Effects of temperature on f1 primer RNA synthesis and lacUV5 transcription. (A) Reactions containing 0.1 pmol of f1 ssDNA and 0.1 pmol of lacUV5–2 template with 1.0 pmol of RNA polymerase holoenzyme were carried out. The RNA product was examined by gel electrophoresis and radioautography. (B) Quantification of the data shown in A. In addition, a reaction containing 0.1 pmol of f1 ssDNA template and 1.0 pmol of RNA polymerase in the absense of SSB was carried out to assess temperature dependence of nonspecific polymerization by RNA polymerase (the curve marked f1 without SSB).
Importance of Structure and Base Sequence Around the −10 region of the Origin.
Effect of base substitutions around the −10 region of the minus-strand origin on its function were tested. The mutations were introduced in hairpin C of the origin to give the base sequences shown in Fig. 3A. In m325, the consensus sequence for the promoter −10 region 5′-TATAAT was substituted for the wild-type origin sequence 5′-AAAAAC at nucleotides −12 to −7 on the top strand of the hairpin. In m326, a complete duplex from nucleotide −12 to −1 of lacUV5 promoter was substituted for the corresponding sequence of both top (nontemplate) and bottom (template) strands of the origin. The mutants m327 and m328 were produced from m326 by changing the top and bottom strand sequences (from −12 to −1), respectively, replacing adenosine with cytidine, cytidine with adenosine, guanosine with thymidine, and thymidine with guanosine. The mutations were constructed on a plasmid by using synthetic oligonucleotides and were subsequently transferred to phage.
In vivo activities of the mutant origins were determined by the “double-origin plasmid assay” (3) and by plaque morphology of the phage. Double-origin plasmids were constructed by inserting two copies of an f1 restriction fragment that contained a wild-type plus-strand origin and either a wild-type or a mutant minus-strand origin, in the same orientation into two sites of pBR322. When cells harboring such a plasmid are infected with phage f1, the phage-type rolling-circle replication is initiated from either one of the inserted origins and is terminated at the other. This results in resolution of the plasmid into two circles of different sizes (shown as I and II in Fig. 3B). In the configuration we used, the minus-strand replication of the circle II was dependent on the mutant origin. The results shown in Fig. 3B indicate that circle II carrying m325 or m327 mutation replicated effectively, while that carrying m326 or m328 mutation failed to replicate. Furthermore, mutant phages carrying m325 or m327 formed large plaques, whereas those carrying m326 or m328 mutation formed very small and turbid plaques. These results clearly indicate that the nontemplate strand sequence of the −10 region of lacUV5 promoter plays an important role for the origin function and that the single strandedness in this region is necessary, though not sufficient, for the origin function in vivo.
In vitro activities of the mutant origins were determined by synthesis of primer RNA from viral single-stranded DNA and by competition activity against lacUV5 transcription. A 20-nt primer RNA was produced on wild-type DNA, but not on R377 DNA, which carried a deletion of the entire origin, as described (3, 6). As shown in Fig. 3C, in reactions containing 0.1 pmol of RNA polymerase and 0.1 pmol of single-stranded DNA template, m325 and m327 mutant origins and the wild-type origin produced approximately the same amount of primer, whereas m326 produced less than 5% of the amount. m328 did not produce a significant amount of primer. These results are in full agreement with the results of in vivo assays described above. On the other hand, in reactions containing 0.7 pmol of RNA polymerase, m326 produced the wild-type level of primer RNA (Fig. 3C). The result suggests that m326 has a certain level of origin activity in vitro but has much lower affinity to RNA polymerase than the wild-type origin. Competition experiments against lacUV5 transcription shown in Fig. 3D support this notion. Amounts of the lacUV5 transcript was remarkably reduced by addition of phage DNA carrying m327 origin but not by m326 and R377 viral DNA. Intensity of the bands shown in Fig. 3D was quantified, and average values thus obtained for KR377/Kuv5, Km326/Kuv5, and Km327/Kuv5 were 0.08, 0.16, and 7.2, respectively. The low value for R377 represents the competitive activity of SSB-coated single-stranded phage DNA without the origin and indicates that the competition of transcription by phage DNA (see Fig. 1) is wholly due to the presence of the minus-strand origin.
Promoter Activity of the Origin Sequence in Vivo.
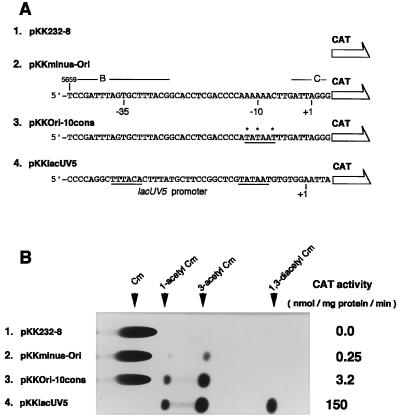
To test whether the nucleotide sequence of the minus-strand origin shows any promoter activity, we constructed a duplex molecule in which one of the strands had a sequence that was identical to the top (nontemplate) strand of the origin (nucleotides 5659 to 5707) (see Fig. 3A). The duplex was inserted into a vector plasmid pKK232–8, which carries a promoter-less cat (CAT) gene and a multicloning site upstream of it (12). Cells carrying the recombinant plasmid pKKminus-Ori thus obtained (Fig. 4A) showed CAT activity of 0.25 nmol per mg of protein per min, whereas the value for a positive control plasmid pKKlacUV5 carrying the lacUV5 promoter sequence without the repressor binding site was 150 (Fig. 4B). The negative control pKK232–8 gave a value 0.0. We also constructed pKKOri-10cons, which carries a sequence identical to pKKminus-Ori except that the −10 region was replaced by the −10 promoter consensus sequence (5′-TATAAT). The CAT activity of the cells carrying this plasmid was 3.2 nmol per mg of protein per min (Fig. 4B). The results indicate that the double-stranded DNA carrying the sequence of nontemplate strand of the origin shows a very weak promoter activity in vivo. If the high affinity of the origin to RNA polymerase compared with the lacUV5 promoter (Fig. 1) is considered, this result suggests importance of single strandedness of the structure around the −10 region in the origin for the strong affinity to RNA polymerase.
Figure 4.
In vivo promoter activity of the minus-strand origin sequence. (A) Nucleotide sequences inserted into pKK232–8 plasmid, which carries a promoter-less cat gene (12). Only the sequences of the nontemplate strand are shown. The insert in pKKminus-Ori carries the f1 wild-type sequence of nucleotides 5659–5707 on the strand shown, which corresponds to the top strand of the minus-strand origin (see Fig. 3A). pKKOri-10cons is identical to pKKminus-Ori except for three base substitutions (marked with asterisks), which yield the consensus sequence of the promoter −10 region. pKKlacUV5 carries a region of the lacUV5 promoter ranging from −46 to +3. (B) CAT activity in E. coli DH5 cells harboring pKK232–8, pKKminus-Ori, pKKOri-10cons, or pKKlacUV5 was determined with [14C]chloramphenicol as substrate. An autoradiograph of a chromatogram is shown. The products of the CAT reaction are indicated on the top. CAT activity was determined from intensity of the spots on the autoradiograph and shown on the right.
DISCUSSION
The high affinity of RNA polymerase to the origin DNA (see Fig. 1) suggests a novel mechanism of promoter recognition by RNA polymerase. The specific binding of the enzyme to the origin and the synthesis of primer RNA are dependent on the presence of σ70 subunit (1, 2). The σ70 subunit is required for promoter recognition by RNA polymerase during transcriptional initiation. The promoter contains conserved hexamer sequences at −10 and −35 regions. Mutations of regions 2.4 and 4.2 of σ70 affect the specific interaction of RNA polymerase with the −10 and −35 regions of the promoter, respectively (16–20). It has also been suggested that σ70 contains two DNA-binding domains that specifically interact with the −10 and −35 sequences (21).
On the basis of footprinting and base-substitution experiments on the minus-strand origin of phage f1, we have previously suggested that a sequence element corresponding to the −35 region of promoters may exist in the hairpin B region (a possible candidate is 5′-TTTACG). Double-stranded structure around this element was required for origin activity (1). It is functionally important for the −10 region to promote DNA melting during open complex formation in transcriptional initiation (14, 15). The sequence around the −10 region of the origin contains an adenine cluster (see Fig. 3A) and is probably in a single-stranded state rather than being annealed to a thymine cluster on the bottom strand. This is supported by the results of base-substitution experiments (Fig. 3 and ref. 1) and by the base sequence of the corresponding region of a rather remotely related phage IKe, in which the complementarity of bases in this region is broken (22). The sequence element AAAAA and the following cytidine on the nontemplate strand of the origin can be replaced by 5′-TATAAT, the consensus sequence of the −10 region of promoters, without losing any origin activity (Fig. 3). The AAAAA sequence, however, cannot be replaced by CCCCC or TTTTT, but the TTTTT sequence on the template strand can be replaced by GGGGG without loss of activity (1). In the mutational study shown in Fig. 3, replacement of the template strand sequence (m327) did not affect origin activity, whereas replacement of the nontemplate strand (m328) destroyed it. These results suggest that adenine residue(s) of the nontemplate strand are important for the origin function, and bases on the template strand in this region are irrelevant. In addition, single strandedness of the −10 region stimulated origin function (compare m326 and m327 in Fig. 3). In fact, the completely double-stranded DNA form that contained the sequence of the nontemplate strand of the origin showed extremely weak activity as a transcriptional promoter (Fig. 4). It is tempting to assume that the single strandedness of the domain surrounding the −10 region of the origin facilitates open complex formation, which thus leads to higher affinity to RNA polymerase than that of transcriptional promoters. This notion is supported by the observation that temperature dependence of primer RNA synthesis was much lower than that of transcription from the lacUV5 promoter (Fig. 2).
The minus-strand origin sequences in a double-stranded form showed extremely weak promoter activity (Fig. 4). This may be due to (i) the sequences of the −35 and −10 regions of the origin that are rather different from the consensus sequences for the −35 and −10 regions of transcriptional promoters and/or (ii) improper distance between the −35 and −10 regions. According to a current model of open complex formation (for review see ref. 23), sequence-specific interaction of the RNA polymerase holoenzyme with the −35 and −10 regions involves twisting of the DNA, which leads to melting of a region around the −10 element. In the wild-type origin, the single strandedness of the −10 region may make the steps that lead to the melting unnecessary. Furthermore, the presence of a single-stranded region between the two elements would allow appropriate phasing between the two elements regardless of the spacer length. The m327 origin that carried single-stranded −10 region showed much higher affinity to RNA polymerase than the m326 origin that carried double-stranded −10 region, whereas both of them contained a single-stranded spacer (see Fig. 3). The reason why the origin depends on single strandedness in the −10 region but promoters do not may be that the single-stranded spacer in the origin would prevent the twisting and the resultant melting around the −10 element, since the DNA backbone in the single-stranded region would rotate freely.
Tripatara and deHaseth (24) have shown that E. coli RNA polymerase can bind to templates containing engineered “bubbles” around the −10 region and can initiate transcription. Crosslinking experiments by Simpson (25) have shown that the σ70 subunit of RNA polymerase is crosslinked to the −3 position of the nontemplate strand. In addition, it was recently reported that RNA polymerase containing σ70 recognizes bases of primarily nontemplate strand of the promoter −10 region (26, 27). Furthermore, by using an oligonucleotide binding assay developed by M. Marr and J. W. Roberts (personal communication), the RNA polymerase holoenzyme formed from a fragment of σ70 containing region 2 and the core enzyme was found to specifically bind a single strand with a sequence of the nontemplate strand of the promoter −10 region (28). These results and our findings strongly support the notion that σ70 subunit of RNA polymerase interacts with the nontemplate strand of the −10 region in a single-stranded state. Adenine residue(s) in the −10 region may be particularly important for direct interaction with the σ70 subunit, probably its region 2.
A mutant origin m326 was inactive in vivo, even though it could produce in vitro a significant amount of primer RNA in the presence of an excess amount of RNA polymerase. The mutant showed much reduced affinity to the enzyme and produced only a small amount of primer RNA at a lower concentration of the enzyme (Fig. 3). Thus, high affinity for RNA polymerase seems to be essential for origin function in vivo. The number of molecules of σ70 subunit present in a cell has been estimated to be 500–700 in the exponential growth phase, and about 1,000 different genes are transcribed in the cell (29). Thus, higher affinity of the origin to RNA polymerase must be necessary to quickly synthesize the minus-strand DNA in phage replication cycle and thus to prevent induction of the SOS response.
Acknowledgments
We thank Akira Ishihama and Nobuo Shimamoto for providing us with RNA polymerase and SSB, respectively; Tapas Kundu and Peter Model for critical reading of the manuscript; and Emiko Suzuki for superb technical assistance. This work was supported in part by grants-in-aid from the Ministry of Education, Science, and Culture of Japan.
ABBREVIATIONS
- SSB
single-stranded DNA binding protein
- ssDNA
single-stranded DNA
- CAT
chloramphenicol acetyltransferase
References
- 1.Higashitani N, Higashitani A, Guan Z W, Horiuchi K. Genes Cells. 1996;1:829–841. doi: 10.1046/j.1365-2443.1996.d01-279.x. [DOI] [PubMed] [Google Scholar]
- 2.Kaguni J M, Kornberg A. J Biol Chem. 1982;257:5437–5443. [PubMed] [Google Scholar]
- 3.Higashitani N, Higashitani A, Horiuchi K. J Virol. 1993;67:2175–2181. doi: 10.1128/jvi.67.4.2175-2181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horiuchi K, Zinder N D. Proc Natl Acad Sci USA. 1976;73:2341–2345. doi: 10.1073/pnas.73.7.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Model P, Russel M. In: The Bacteriophages. Calendar R, editor. Vol. 2. London: Plenum; 1988. pp. 375–456. [Google Scholar]
- 6.Higashitani N, Higashitani A, Roth A, Horiuchi K. J Bacteriol. 1992;174:1612–1618. doi: 10.1128/jb.174.5.1612-1618.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashitani N, Higashitani A, Horiuchi K. J Bacteriol. 1995;177:3610–3612. doi: 10.1128/jb.177.12.3610-3612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horiuchi K, Zinder N D. Science. 1967;156:1618–1623. doi: 10.1126/science.156.3782.1618. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 10.Zinder N D, Boeke J D. Gene. 1982;19:1–10. doi: 10.1016/0378-1119(82)90183-4. [DOI] [PubMed] [Google Scholar]
- 11.Hill D F, Petersen G B. J Virol. 1982;44:32–46. doi: 10.1128/jvi.44.1.32-46.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosius J, Lupski J R. Methods Enzymol. 1987;153:54–68. doi: 10.1016/0076-6879(87)53047-6. [DOI] [PubMed] [Google Scholar]
- 13.Higashitani A, Nishimura Y, Hara H, Aiba H, Mizuno T, Horiuchi K. Mol Gen Genet. 1993;240:339–347. doi: 10.1007/BF00280384. [DOI] [PubMed] [Google Scholar]
- 14.Gamper H B, Hearst J E. Cell. 1982;29:81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- 15.Chamberlin M J. Annu Rev Biochem. 1974;43:721–775. doi: 10.1146/annurev.bi.43.070174.003445. [DOI] [PubMed] [Google Scholar]
- 16.Siegele D A, Hu J C, Walter W A, Gross C A. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 17.Zuber P, Healy J, Carter H L, III, Cutting S, Moran C P, Jr, Losick R. J Mol Biol. 1989;206:605–614. doi: 10.1016/0022-2836(89)90569-x. [DOI] [PubMed] [Google Scholar]
- 18.Daniels D, Zuber R, Losick R. Proc Natl Acad Sci USA. 1990;87:21–25. doi: 10.1073/pnas.87.20.8075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waldburger C, Gardella T, Wong R, Susskind M M. J Mol Biol. 1990;215:267–276. doi: 10.1016/s0022-2836(05)80345-6. [DOI] [PubMed] [Google Scholar]
- 20.Gardella T, Moyle H, Susskind M M. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- 21.Dombroski A J, Walter W A, Record M T, Jr, Siegele D A, Gross C A. Cell. 1992;70:501–512. doi: 10.1016/0092-8674(92)90174-b. [DOI] [PubMed] [Google Scholar]
- 22.Peeters B P H, Peters R M, Schoenmakers J G G, Konings R N H. J Mol Biol. 1985;181:27–39. doi: 10.1016/0022-2836(85)90322-5. [DOI] [PubMed] [Google Scholar]
- 23.deHaseth P L, Helmann J D. Mol Microbiol. 1995;16:817–824. doi: 10.1111/j.1365-2958.1995.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 24.Tripatara A, deHaseth P L. J Mol Biol. 1993;233:349–358. doi: 10.1006/jmbi.1993.1516. [DOI] [PubMed] [Google Scholar]
- 25.Simpson R B. Cell. 1979;18:277–285. doi: 10.1016/0092-8674(79)90047-3. [DOI] [PubMed] [Google Scholar]
- 26.Ring B Z, Yarnell W S, Roberts J W. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- 27.Roberts C W, Roberts J W. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 28.Severinova E, Severinov K, Fenyoe D, Marr M, Brody E N, Roberts J W, Chait B T, Darst S A. J Mol Biol. 1996;263:637–647. doi: 10.1006/jmbi.1996.0604. [DOI] [PubMed] [Google Scholar]
- 29.Jishage M, Ishihama A. J Bacteriol. 1995;177:6832–6835. doi: 10.1128/jb.177.23.6832-6835.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]