Abstract
A new self-sustained terminal protein-primed DNA amplification system has been used to describe in vitro evolutionary changes affecting maintenance of the genome size of bacteriophage φ29. These changes involve generation and efficient amplification of short palindromic molecules containing an inverted duplication of one of the original DNA ends. A template-switching mechanism is proposed to account for the appearance of these molecules. After their formation, they would replicate by means of hairpin intermediates. Relevant kinetic information about this DNA replication system has been obtained from the competition between the input full-length φ29 DNA and its derived truncated versions. The physiological relevance of these molecules and the mechanisms to control their formation are discussed.
Keywords: φ29 DNA replication, DNA amplification, strand-displacement, snap-back
Molecular in vitro evolution was first described by Mills et al. (1) using the bacteriophage Qβ RNA replicase. Since then, several experimental designs have been proposed to study the adaptive changes of an initial population of replicating nucleic acid molecules under selective pressure. In conditions of self-sustained replication, in vitro evolution is guided by direct competition among different replicating molecules for a limited amount of resources (nucleotides, primers, enzymes). As a result, the fastest replicating molecules are exponentially enriched in the growing population (1, 2).
Most of the in vitro evolution studies concerning replication processes have been based on Qβ RNA replicase. The RNA-dependent RNA polymerase of bacteriophage Qβ is a very versatile enzyme, able to replicate a large variety of RNA molecules (ref. 3 and references therein). This property makes this enzyme very suitable for evolutionary studies (1, 4, 5). Basic properties of the Qβ RNA replication process have been elucidated by means of this strategy (6–9).
The so-called strand-displacement amplification (10), allowing exponential enrichment of DNA templates, has been also used for in vitro evolution experiments (11, 12). However, this technique requires exogenous activities to regenerate functional primers after each replication round.
Recently, we have reported a new self-sustained, strand-displacement DNA amplification system based on terminal protein (TP)-primed DNA replication (13). This DNA replication strategy has been found in several prokaryotic and eukaryotic viruses and has been proposed for a variety of linear plasmids isolated from bacteria, fungi, and higher plants (reviewed in ref. 14) and more recently for the replication of the DNA ends of Streptomyces (15). In all these cases, the genome is a linear double-stranded DNA (dsDNA) molecule with a TP covalently linked to both 5′ ends and an inverted terminal repeat of different length, having a minimum of 6 bp in the case of φ29, an average of 1 kb in the case of linear plasmids, and a maximum of 24 kb for the chromosomal DNA of Streptomyces griseus (16). The reported TP-primed DNA amplification system (13) was based on the use of four highly purified proteins involved in Bacillus subtilis phage φ29 DNA replication (reviewed in ref. 14). The viral protein p6 dsDNA-binding protein (DBP) forms a nucleoprotein complex with both φ29 DNA ends, which is further recognized by a heterodimer formed between φ29 DNA polymerase and a free molecule of TP. Initiation of DNA replication takes place by covalently linking dAMP to TP, in a reaction catalyzed by φ29 DNA polymerase. Then, elongation proceeds from both DNA ends by a strand-displacement mechanism. DNA synthesis is continuous from both DNA origins, without the need of lagging strand replication. This process generates large amounts of single-stranded DNA (ssDNA), that are bound by the viral protein p5 [φ29 ssDNA-binding protein (SSB)]. Complete replication of both strands originates two dsDNA molecules with TP attached to their 5′ ends. As reported in the original characterization of the φ29 DNA amplification system (13), four DNA replication proteins (DNA polymerase, TP, DBP, and SSB) are required for efficient DNA amplification.
During the development of this system, we observed that certain conditions favored the generation of short DNA molecules. These products were initially interpreted as abortive molecules resulting from impaired DNA elongation. In this paper, we show that these short DNA molecules are amplification-competent and were not present in the initial DNA population. Therefore, we have further investigated the formation of these molecules, as an example of spontaneous in vitro evolution.
MATERIALS AND METHODS
Nucleotides, Proteins, and DNA Templates.
Unlabeled dNTPs and [α-32P]dATP (400 Ci/mmol; 1 Ci = 37 GBq) were obtained from Amersham. Restriction enzymes, proteinase K, DNase I, and S1 nucleases were purchased from Boehringer Mannheim. φ29 DNA polymerase (Mr = 66,520) and TP (Mr = 30,918) were overproduced in Escherichia coli cells and purified essentially as described (17, 18). Protein p6 (Mr = 11,873) and protein p5 (Mr = 13,212), obtained from φ29-infected B. subtilis cells, were purified as described (19, 20). TP-linked φ29 DNA (Mr = 12.5 × 106) from φ29 sus14(1242) virions, isolated as described (21), was used as input template for in vitro amplification experiments.
DNA Amplification Assay.
The incubation mixture contained, in 10 μl, 50 mM Tris·HCl, (pH 7.5), 10 mM MgCl2, 20 mM (NH4)2SO4, 1 mM dithiothreitol, 10% glycerol, 0.1 mg/ml bovine serum albumin, 80 μM each dNTP, 10 ng of φ29 DNA polymerase, 25 ng of TP, 10 μg of protein p6 (DBP), and the indicated amount of p5 (SSB). When indicated, [α-32P]dATP (2 μCi) was added to the reaction mixture to label the amplified DNA. The indicated amount of TP-linked φ29 DNA was used as input DNA template. After incubation at 30°C for the indicated times, reactions were stopped by addition of 10 mM EDTA. When indicated, the DNA obtained after one round of amplification was 10-fold diluted and incubated for a next round of amplification, either in the presence or in the absence of a second addition of the indicated replication proteins. Nonincorporated [α-32P]dATP was removed by filtration through Sephadex G-50 spin columns in the presence of 0.1% SDS. When removal of TP from in vitro-amplified DNA was required, samples were incubated with proteinase K (1 μg/μl) in the presence of 0.1% SDS for 1 hr at 37°C. Samples were analyzed by native or alkaline agarose gel electrophoresis (22), followed by autoradiography and/or ethidium bromide staining. DNA synthesis was quantified from the amount of excluded radioactivity (Cerenkov radiation) or by densitometry of the autoradiographs.
Southern Blot.
φ29 DNA (500 ng) was digested with HindIII and resolved in 0.7% agarose gels. Gel transfer was carried out essentially as described (23), using a positively charged nylon membrane (Bio-Rad Zeta-Probe). A mixture of short DNA molecules obtained from in vitro amplification experiments was used as probe for hybridization. As a control, internally labeled full-length φ29 DNA was used in a different hybridization. Band positions were identified by ethidium bromide staining.
RESULTS
Generation of Short DNA Molecules.
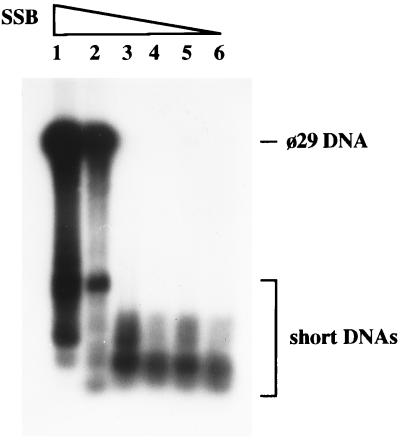
As noticed in the initial characterization of the φ29 DNA amplification system (13), generation of short DNA molecules is dependent on the presence of DNA polymerase, TP, and DBP, but SSB prevents their appearance. In a typical DNA amplification experiment, the relative yield of φ29 DNA and short DNAs is strongly dependent on SSB concentration. As shown in Fig. 1, high concentrations of SSB favored the amplification of mature φ29 DNA, while only short DNAs were generated efficiently at lower SSB concentrations. An interesting observation came from the size analysis of the short DNA products. Several independent amplification experiments were carried out in conditions favoring the appearance of short DNA molecules (0.5 ng of φ29 DNA, in the absence of SSB). After agarose gel electrophoresis under nondenaturing conditions, a quite heterogeneous population of DNA molecules could be observed, their lengths typically varying between 200 and 6000 bp. Although multiple bands were accumulated, they appear to be distinct, and in some cases, only a few bands were observed (two bands being the minimum). Different DNA populations were obtained in different experiments, with no evident preferred length. These results were confirmed under different amplification conditions, varying temperature, ionic strength, and nucleotide concentration (data not shown).
Figure 1.
Effect of SSB on the size of in vitro-amplified DNA. Amplification assays were carried out as described, using 0.5 ng of φ29 TP-DNA as input template. Lanes 1–6 contained 6, 2, 0.7, 0.2, 0.07, and 0.02 μg of SSB, respectively. After 1 hr of incubation at 30°C, reactions were stopped and the size of the amplified DNA was analyzed by alkaline agarose gel electrophoresis followed by autoradiography. The electrophoretic mobility of full-length φ29 DNA and short DNAs is indicated.
The Short DNA Molecules Are in Vitro Replicons.
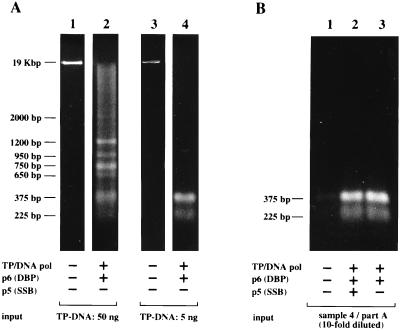
In complete absence of SSB, the short DNA molecules accumulate enough to be clearly detected by ethidium bromide staining, even when a relatively high amount (50 ng) of TP–DNA was used as input template for the amplification reaction (Fig. 2A, lanes 1 and 2). This accumulation suggested that the short DNA molecules are being amplified as competent substrates for multiple rounds of TP-primed DNA amplification. To address this possibility, we carried out serial transfer experiments with different populations of short DNA molecules as the input template. As expected, efficient reamplification was observed after several rounds of transferring reaction products to fresh reaction mixtures, and the length pattern of each DNA population was kept constant throughout several amplification cycles.
Figure 2.
The in vitro-amplified short DNAs replicate by a TP-priming mechanism. (A) Amplification assays were carried out as described, using either 50 ng (lanes 1 and 2) or 5 ng (lanes 3 and 4) of φ29 TP-DNA as input template, in the absence of SSB. After incubation for 90 min at 30°C, half of the reaction was processed as described in Fig. 2, and analyzed by native agarose gel electrophoresis followed by ethidium bromide staining. (B) The amplified DNA corresponding to lane 4 in A was 10-fold diluted and used as new input template for a second amplification assay. Reamplification was allowed for 90 min at 30°C, either with no addition (lane 1) or with the fresh addition of the indicated replication proteins (lanes 2 and 3). The reamplified DNA was treated with proteinase K and analyzed by native agarose gel electrophoresis followed by ethidium bromide staining.
An example of this reamplification experiments is shown in Fig. 2. The short DNAs obtained after a first amplification reaction starting from 5 ng of input TP–DNA (Fig. 2A, lanes 3 and 4) were 10-fold diluted and used as new input for a second round of amplification. Significantly, φ29 DNA polymerase, TP, and p6 were strictly required for reamplification of the short DNA molecules, but the SSB was not needed (see Fig. 2B, lanes 1–3), suggesting that these molecules replicate by a mechanism requiring TP priming at φ29 DNA ends.
Exponential Accumulation of Short DNA Molecules.
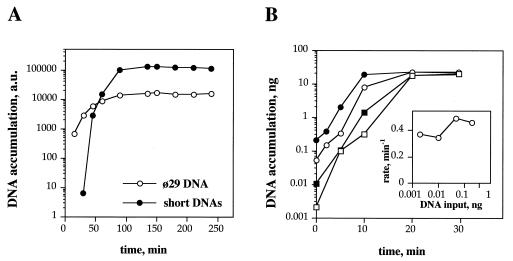
Once we have established the conditions that favor amplification of short DNAs, it was important to know their accumulation kinetics. A DNA amplification experiment was carried out using a low amount of SSB (2 μg), to allow concomitant amplification of φ29 DNA and short DNA molecules. The accumulation of both products was monitored as a function of incubation time. Fig. 3A shows an initial exponential growth for both kinds of molecules, although the accumulation of short DNAs was delayed as compared with φ29 DNA amplification. From the exponential phase of their accumulation time courses, doubling times of 7 min and 1.7 min were estimated for φ29 DNA and the short DNAs, respectively. The fact that both time courses reached saturation at similar times suggests that φ29 DNA and the short DNA molecules are competing for the same replication resources.
Figure 3.
Kinetic analysis of in vitro-amplified DNA. (A) Amplification time course of φ29 DNA and short DNAs. Amplification assays were carried out as described, using 0.5 ng of φ29 TP-DNA as input template, in the presence of 2 μg of SSB. After incubation at 30°C for the indicated times, reactions were stopped and analyzed by 8 M urea/6% PAGE. Quantification was carried out by densitometry of the autoradiograph. The amplified short DNAs were a heterogeneous mixture of bands ranging from 200 to 1000 nt in length. These DNAs were not detectable at the shortest time tested (15 min). (B) Amplification time course of short DNAs. Amplification assays were carried out as described in the absence of SSB. Different amounts of short DNAs were used as template. □, 0.002 ng; ▪, 0.01 ng; ○, 0.05 ng; and •, 0.2 ng. Input DNA template was obtained from a previous amplification assay carried out in the absence of SSB, so only short DNAs were amplified. DNA synthesis was quantified from the amount of DNA-incorporated radioactivity. (Inset) Rates of exponential accumulation of short DNAs using different amounts of input DNA template. Rates were calculated from the linear part of the semilogarithmic plots, according to the equation DNA = DNA0 ∗ e(rate ∗ time).
To evaluate the kinetic behavior of these short DNAs under conditions in which φ29 DNA is not present, we took advantage of their suitability as substrates for DNA amplification. A population of short DNAs was generated in the absence of φ29 SSB, so no amplification of φ29 DNA was observed. Different amounts of these molecules were subsequently used as input for DNA amplification experiments. The resulting time courses are shown in Fig. 3B. Exponential growth of the input DNA was observed before saturation was reached. Both, exponential rates and saturation levels were fairly independent from the amount of input DNA (see Fig. 3B Inset). Duplication times ≈1.7 min were calculated from the exponential rates.
Nature of the Short DNA Molecules.
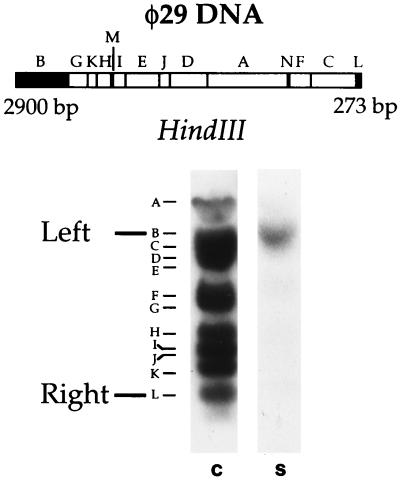
The sequence relationship between φ29 DNA and the short DNA molecules was addressed by means of Southern blot, restriction digestion, and DNA sequencing. A population of internally labeled short DNAs was hybridized to HindIII-digested φ29 DNA blotted onto the membrane. As shown in Fig. 4, a clear hybridization signal was observed with a 2900-bp HindIII fragment containing the left φ29 DNA end. No signal corresponding to the right end was observed. For a rapid examination of different populations of short DNAs, a restriction analysis was carried out. DNA obtained from several amplification experiments was assayed with a battery of 20 restriction enzymes whose target sites are located at the right or left end or at internal regions of φ29 DNA (data not shown). All the short DNAs tested were shown to have target sites corresponding either to the left or to the right φ29 DNA end. However, of ≈30 DNA molecules, only one was cut by restriction enzymes corresponding to the right end. No DNA with restriction sites of internal regions of φ29 DNA was found. Furthermore, several restriction enzymes without target sites in φ29 DNA were unable to cut any short DNA.
Figure 4.
Nature of the in vitro-amplified short DNAs. (Upper) HindIII restriction map of φ29 DNA. The fragments corresponding to the DNA ends are highlighted and their size is indicated. (Lower) Southern blot analysis of short DNAs. Amplification assay was carried out as described, using 0.5 ng of φ29 TP-DNA as input template, in the absence of SSB, so only short DNAs were amplified. This sample was blotted against HindIII-digested φ29 DNA (S), as described. The position of the fragments corresponding to the φ29 DNA ends is indicated in the control lane (C), in which internally labeled full-length φ29 DNA was used as probe.
These experiments strongly suggest that the short amplified DNAs are minireplicons derived from the φ29 DNA template, preferentially having sequences corresponding to its left portion. The ends of these short DNAs were shown to be identical to either the right or the left end of φ29 DNA, by DNA sequencing using oligonucleotides specific for φ29 DNA ends. In agreement with the restriction analysis, sequences corresponding to both φ29 DNA ends were not found on individual DNA molecules. Full sequence of these DNAs could not be obtained due to strong stop sites found by several DNA polymerases when reading the central region of the molecules (data not shown).
Structural Properties of the Short DNA Molecules.
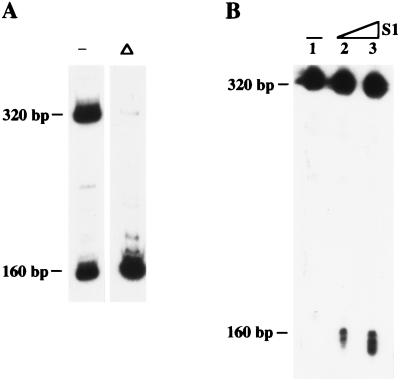
To unravel the replication mechanism of these short DNAs, we carried out several experiments to determine their structural properties. Fig. 5A shows a population of two DNAs (160 bp and 320 bp, approximately, as resolved by native electrophoresis) obtained by in vitro DNA amplification in the absence of SSB. Both DNAs correspond to the left end of φ29 DNA. The presence of TP in these molecules was deduced from gel mobility-shifts after treatment with proteinase K (data not shown). On the other hand, based on the exhaustive restriction analysis mentioned above and DNase I digestion experiments (data not shown), we concluded that these two molecules are, for the most part, dsDNA.
Figure 5.
Structural analysis of in vitro-amplified short DNAs. (A) Native analysis of the DNA population. To analyze the intramolecular renaturability, the in vitro-amplified short DNAs were heat-denatured for 1 min at 100°C and rapidly cooled on ice (Δ). The same sample without heat denaturation was used as control (−). After addition of glycerol gel loading buffer, samples were analyzed in nondenaturing 6% PAGE, followed by autoradiography. (B) Denaturing analysis of the DNA population (lane 1) and location of S1 nuclease-sensitivity sites (lanes 2–3). S1 nuclease digestions of in vitro-amplified short DNAs were carried out for 5 min at 37°C. Lanes 1–3 contained 0, 0.2, and 0.5 units of S1 nuclease, respectively, as defined by the manufacturer. Reactions were directly quenched with formamide gel loading buffer and analyzed in 8 M urea/6% PAGE, followed by autoradiography.
Interestingly, when the same two-DNA population was analyzed in denaturing conditions, only one band was observed (Fig. 5B, lane 1), and its mobility corresponded to the upper band detected in nondenaturing gels (≈320 bp). These results and the 2-fold difference in size between these two fragments clearly suggested that the lower band (≈160 bp) is a self-complementary single strand forming a hairpin and that the longest molecule is a fully palindromic dsDNA, each strand being equivalent to the shortest molecule.
It has been shown that duplex hairpins undergo instantaneous intramolecular renaturation following denaturation, since the complementary strands are covalently linked (24). Since both kind of molecules (hairpin and palindrome) are expected to produce hairpin structures after denaturation and intrastrand renaturation, we studied their renaturability by analyzing their mobility in nondenaturing PAGE after heat denaturation followed by rapid cooling. As shown in Fig. 5A, the population of the two bands (320 bp and 160 bp) was mainly converted in a single band (160 bp) by this treatment. This result confirms both the hairpin structure of the shortest molecule and the palindromic nature of the largest molecule. Moreover, the presence and rough location of the ssDNA loop in the hairpin molecule could be demonstrated by digestion with low concentrations of the single strand-specific S1 nuclease and further analysis in denaturing PAGE. As shown in Fig. 5B, a fraction of the 320-bp band, observed with no S1 treatment (lane 1), could be converted into a band of about half-size (160 bp) after digestion with low concentrations of S1 nuclease (lanes 2–3), indicating the presence of a S1-sensitive site most likely located in the middle of the hairpin molecule.
The proposed structure for both molecules was confirmed by restriction mapping (data not shown). Similar results were obtained with more complex populations of amplified DNA. In all the cases, it was possible to detect the presence of fully palindromic dsDNA molecules with their corresponding hairpin partners (data not shown).
DISCUSSION
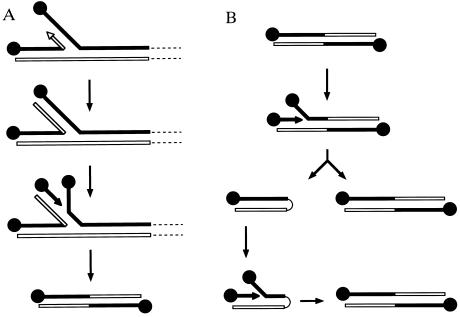
In this paper we have demonstrated that the φ29 DNA replication system can potentially generate in vitro a variety of short DNA molecules, which, in turn, can be efficiently amplified using the φ29 DNA replication machinery. The simplest model that takes into account both the protein requirements for generating these molecules and their structural properties is based on a template-switching mechanism. During strand-displacement DNA synthesis, large amounts of ssDNA are generated. As the displaced strand is not used for lagging strand DNA synthesis, it is available for the polymerase as a competing template. If a template switching event occurs (Fig. 6A), DNA synthesis would continue until the end of the displaced strand is reached, generating a branched molecule with a new DNA replication origin. Initiation on the new origin would lead to replication of the snapped-back strand, producing an inverted duplication of the DNA end. This molecule would contain two identical and fully functional origins of DNA replication. Once formed, these palindromic molecules would replicate according to the mechanism depicted in Fig. 6B. If the starting molecule is short enough (see below for a kinetic estimation of the length limit), initiation at either end very likely would produce a full displacement of the complementary strand before another initiation event can take place at the other end. Due to its self-complementarity, the fully displaced strand could fold back to form a duplex hairpin structure with one DNA replication origin. After initiation on this origin, the initial symmetrical molecule would be regenerated. The fact that both kind of molecules (duplex hairpins and inverted duplications) have been detected in our DNA amplification experiments, strongly supports this model. On the other hand, the difficulties found in sequencing the central part of these molecules with different DNA polymerases are also in agreement with their palindromic structure.
Figure 6.
Proposed model for the generation (A) and replication (B) mechanisms of short palindromic TP–DNAs. See Discussion for details.
SSB-mediated inhibition of the generation of short DNAs may be related to its ability to bind the ssDNA generated during strand-displacement DNA synthesis (25). SSB is probably covering the displaced strand as fast as it is produced, therefore reducing the availability of this strand as an alternative template for DNA polymerase. On the other hand, perturbations in the advance of the replication fork are likely to be produced by secondary structures of the displaced ssDNA when SSB is limiting or absent. Then, stalled DNA polymerases would be prone to undergo template switching. According to this model, the absence of SSB would favor the generation of short molecules, but it would have no effect on their replication. This interpretation is in agreement with the null effect of SSB on the reamplification of short DNAs. As schematized in Fig. 6B, once these palindromic molecules are formed, their replication no longer requires the original φ29 DNA molecule, but it is still dependent on the replication proteins DBP, TP, and DNA polymerase. Based on these common requirements, a direct competition between full-length φ29 DNA and its short derivatives is established.
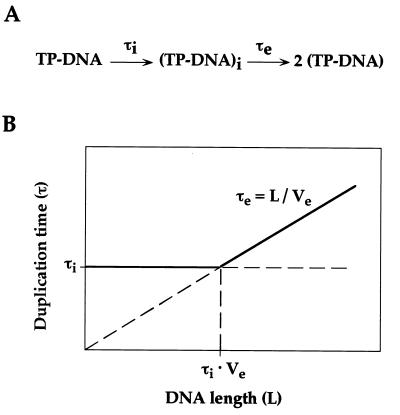
From a kinetic point of view, duplication of TP–DNA requires two distinguishable steps, named protein-primed initiation and full-length DNA elongation (see Fig. 7A). According to this scheme, the slowest of these two steps will determine the duplication time of TP–DNA during exponential amplification. Length reduction is expected to decrease the time required for DNA elongation, that will be equivalent to L/Ve (L being DNA length and Ve being the elongation velocity of DNA polymerase). On the other hand, initiation rate is likely to be independent from DNA length (as far as a minimal length is provided for the replication factors to bind). Taking into account these considerations, a plot of duplication time versus DNA length can be envisioned (see Fig. 7B). For long DNA molecules, duplication time will be determined from their lengths (right part of the plot), shorter molecules replicating faster than longer ones (τe = L/Ve). However, it can be predicted that, below a certain size, the time required for DNA elongation will be shorter than that of the initiation step. Then, duplication time will become length-independent, being limited by the initiation rate (left part of the plot). From Fig. 7B, it can be easily realized that the critical length that separates initiation-limited from elongation-limited DNA duplication equals τi·Ve (τi being the time required for the initiation step).
Figure 7.
(A) Schematic representation of a two-step TP–DNA replication process. Time required for the initiation step of TP-primed DNA replication is indicated as τi. (TP–DNA)i stands for an initiated complex (only one initiation event is considered, for simplicity). Time required for an initiated complex to achieve full-length DNA elongation is indicated as τe. See Discussion for more details. (B) Theoretical dependence between duplication time and DNA length. τi and τe represent the times required for TP-primed initiation and full-length DNA elongation, respectively. L and Ve stand for the length of the replicating molecule and the velocity of DNA polymerase, respectively. See Discussion for more details.
An upper limit of 6 min has been reported for the duplication time of φ29 DNA (13), this value being quite close to the time required for φ29 DNA polymerase to synthesize a full-length DNA strand (≈5 min; refs. 26 and 27). These observations strongly suggest that DNA elongation is the rate-limiting step in the φ29 DNA duplication cycle, under the in vitro amplification conditions. Hence, any TP–DNA molecule shorter than full-length φ29 DNA will have a faster duplication cycle (right part of the plot shown in Fig. 7B). The duplication time of the in vitro amplified short molecules has been estimated as 1.7 min. This value is significantly higher than the expected time required for DNA elongation, considering the elongation velocity of φ29 DNA polymerase (≈4000 nt/min; ref. 27). On the other hand, their duplication time is independent from their length, as demonstrated from the serial transfer experiment (the relative abundance of molecules with different lengths is kept constant through several transfers). These facts imply that protein-primed initiation is rate-limiting for duplication of the short molecules (left part of the plot shown in Fig. 7B). Taking into account the elongation velocity of φ29 DNA polymerase (Ve = 4000 nt/min) and the duplication time imposed by the initiation rate (τi = 1.7 min), a critical length of ≈7000 nt can be calculated. Below this size, duplication velocity reaches its maximal value, being independent from the length of the molecule. This critical size agrees fairly well with the longest molecules observed in our in vitro amplification system (≈6000 nt), indicating that the fastest replicating molecules have been selected. For instance, using these duplication rate values, it can be easily calculated that one short molecule that is generated in a population of 105 φ29 DNA molecules will outnumber its full-length competitors in just 37 min.
A testable prediction of this model is that the kinetic advantage of the short DNAs versus full-length φ29 DNA would be hampered by reducing the initiation rate. Duplication time of φ29 DNA should not be significantly affected, since DNA elongation is its rate-limiting step. Conversely, duplication time of the short DNAs should be increased, approaching that of φ29 DNA. This prediction has been experimentally confirmed by carrying out DNA amplification experiments with subsaturating concentrations of the initiator nucleotide (dATP). The appearance of short DNAs was virtually eliminated in these conditions. However, the final yield of φ29 DNA amplification was also severely decreased (data not shown). This fact is probably related with the φ29 DNA replication strategy. The chances of a replication fork to travel from one DNA end to the opposite one, before another initiation event can take place on it, are increased by reducing the initiation rate. In these conditions, fully displaced strands would likely accumulate without net amplification of the input dsDNA.
In vivo, full displacement of the parental strand is probably avoided by keeping the initiation rate faster than the full-length DNA elongation rate. However, as we have shown, this strategy has the intrinsic risk of favoring amplification of palindromic molecules after a template-switching event. Although these minichromosomes would not be viable, their replication would presumably be detrimental for phage φ29 DNA amplification. According to the results presented in this paper, φ29 SSB seems to be critical to prevent template switching, therefore ensuring a successful phage infection cycle. However, template switching has been shown to participate in the in vivo amplification of inverted repeats in other systems (28–32), and recently, low amounts of palindromic molecules have been detected in B. subtilis cells infected with phage φ29 (V. Murthy, L.B., and M.S., unpublished results). In most cases, amplification of short inverted repeats will give rise to nonviable minichromosomes that will not be stable. However, these molecules might perpetuate through several generations if the host system provides the required replication functions and duplication of the different molecules is coordinated. In this sense, it is very interesting to consider the genus Streptomyces. All the linear chromosomes and plasmids of Streptomyces that have been characterized so far appear to contain terminal inverted repeats and TP (15). Moreover, Streptomyces lividans contains a linear palindromic plasmid whose terminal repeat is identical to the sequence present at the end of the linear chromosome (33). It is very tempting to speculate that these plasmids were generated during replication of the linear chromosome by a template-switching mechanism like the one operating in the in vitro system described here. While the physiological significance of these minichromosomes is presently unclear, the molecular mechanisms responsible for their generation and amplification have a great potential for genome reorganization and evolution.
Acknowledgments
This investigation has been aided by Research Grant 5R01 GM27242-17 from the National Institutes of Health, by Grant PB93-0173 from the Dirección General de Investigación Científica y Técnica, by Grant CHRX-CT 93-0248 from the European Economic Community, and by an Institutional grant from Fundación Ramón Areces. J.M. was recipient of a postdoctoral fellowship from the Comunidad Autónoma de Madrid.
ABBREVIATIONS
- TP
terminal protein
- dsDNA
double-stranded DNA
- DBP
dsDNA-binding protein
- ssDNA
single-stranded DNA
- SSB
ssDNA-binding protein
References
- 1.Mills D R, Petersen R L, Spiegelman S. Proc Natl Acad Sci USA. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eigen M. Naturwissenschaften. 1971;58:465–523. doi: 10.1007/BF00623322. [DOI] [PubMed] [Google Scholar]
- 3.Biebricher C K, Eigen M, Gardiner W C., Jr Biochemistry. 1983;22:2544–2559. doi: 10.1021/bi00279a036. [DOI] [PubMed] [Google Scholar]
- 4.Levisohn R, Spiegelman S. Proc Natl Acad Sci USA. 1968;60:866–872. doi: 10.1073/pnas.60.3.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levisohn R, Spiegelman S. Proc Natl Acad Sci USA. 1969;63:805–811. doi: 10.1073/pnas.63.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biebricher C K, Luce R. Biochemistry. 1993;32:4848–4854. doi: 10.1021/bi00069a021. [DOI] [PubMed] [Google Scholar]
- 7.McCaskill J S, Bauer G J. Proc Natl Acad Sci USA. 1993;90:4191–4195. doi: 10.1073/pnas.90.9.4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biebricher C K, Eigen M, McCaskill J S. J Mol Biol. 1993;231:175–179. doi: 10.1006/jmbi.1993.1271. [DOI] [PubMed] [Google Scholar]
- 9.Rohde N, Daum H, Biebricher C K. J Mol Biol. 1995;249:754–762. doi: 10.1006/jmbi.1995.0334. [DOI] [PubMed] [Google Scholar]
- 10.Walker G T, Little M C, Nadeau J G, Shank D D. Proc Natl Acad Sci USA. 1992;89:392–396. doi: 10.1073/pnas.89.1.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walter N G, Strunk G. Proc Natl Acad Sci USA. 1994;91:7937–7941. doi: 10.1073/pnas.91.17.7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walter N G. J Mol Biol. 1995;254:856–868. doi: 10.1006/jmbi.1995.0661. [DOI] [PubMed] [Google Scholar]
- 13.Blanco L, Lázaro J M, De Vega M, Bonnin A, Salas M. Proc Natl Acad Sci USA. 1994;91:12198–12202. doi: 10.1073/pnas.91.25.12198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salas M. Annu Rev Biochem. 1991;60:39–71. doi: 10.1146/annurev.bi.60.070191.000351. [DOI] [PubMed] [Google Scholar]
- 15.Chen C W. Trends Genet. 1996;12:192–196. doi: 10.1016/0168-9525(96)30014-0. [DOI] [PubMed] [Google Scholar]
- 16.Leahava A, Mizukami T, Kajitani T, Kameoka D, Redenbach M, Shinkawa H, Nimi D, Kinashi H. J Bacteriol. 1995;177:6492–6498. doi: 10.1128/jb.177.22.6492-6498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaballos A, Salas M. Nucleic Acids Res. 1989;17:10353–10366. doi: 10.1093/nar/17.24.10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lázaro J M, Blanco L, Salas M. Methods Enzymol. 1995;262:42–49. doi: 10.1016/0076-6879(95)62007-9. [DOI] [PubMed] [Google Scholar]
- 19.Pastrana R, Lázaro J M, Blanco L, García J A, Méndez E, Salas M. Nucleic Acids Res. 1985;13:3083–3100. doi: 10.1093/nar/13.9.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martín G, Lázaro J M, Méndez E, Salas M. Nucleic Acids Res. 1989;17:3663–3672. doi: 10.1093/nar/17.10.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peñalva M A, Salas M. Proc Natl Acad Sci USA. 1982;79:5522–5526. doi: 10.1073/pnas.79.18.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonell M W, Simon M N, Studier F W. J Mol Biol. 1977;110:119–146. doi: 10.1016/s0022-2836(77)80102-2. [DOI] [PubMed] [Google Scholar]
- 23.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 1. New York: Wiley; 1994. [Google Scholar]
- 24.Goulian M, Lucas Z J, Kornberg A. J Biol Chem. 1968;243:627–638. [PubMed] [Google Scholar]
- 25.Gutiérrez C, Sogo J M, Salas M. J Mol Biol. 1991;222:983–994. doi: 10.1016/0022-2836(91)90589-x. [DOI] [PubMed] [Google Scholar]
- 26.Blanco L, Salas M. Proc Natl Acad Sci USA. 1985;82:6404–6408. doi: 10.1073/pnas.82.19.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soengas M S, Gutiérrez C, Salas M. J Mol Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 28.Nalbantoglu J, Meuth M. Nucleic Acids Res. 1986;14:8361–8371. doi: 10.1093/nar/14.21.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hyrein O, Debatisse M, Buttin G, de Saint Vincent B R. EMBO J. 1988;7:407–417. doi: 10.1002/j.1460-2075.1988.tb02828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouellette M, Hettema E, Wust D, Fase-Fowler F, Borst P. EMBO J. 1991;10:1009–1016. doi: 10.1002/j.1460-2075.1991.tb08035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu C-H, Xu F-Y, Wang K, Pearson A N, Pearson G D. Gene. 1992;110:145–150. doi: 10.1016/0378-1119(92)90641-2. [DOI] [PubMed] [Google Scholar]
- 32.Cohen S, Hassin D, Karby S, Lavi S. Mol Cell Biol. 1994;14:7782–7791. doi: 10.1128/mcb.14.12.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen C W, Yu F-W, Lin Y-S, Kieser H M, Hopwood D A. Mol Microbiol. 1993;7:925–932. doi: 10.1111/j.1365-2958.1993.tb01183.x. [DOI] [PubMed] [Google Scholar]