Abstract
CBP (CREB-binding protein) and p300 are versatile coactivators that link transcriptional activators to the basal transcriptional apparatus. In the present study, we identify CBP and p300 as coactivators of the nuclear factor-κB (NF-κB) component p65 (RelA). Consistent with their role as coactivators, both CBP and p300 potentiated p65-activated transcription of E-selectin and VCAM-1–CAT reporter constructs. The N- and C-terminal domains of both CBP/p300 functionally interact with a region of p65 containing the transcriptional activation domain as demonstrated by mammalian two-hybrid assays. Direct physical interactions of CBP/p300 with p65 were demonstrated by glutathione S-transferase fusion protein binding, and coimmunoprecipitation/Western blot studies. The adenovirus E1A 12S protein, which complexes with CBP and p300, inhibited p65-dependent gene expression. Reporter gene expression could be rescued from E1A inhibition by overexpression of CBP or p300. CBP and p300 act as coactivators of p65-driven gene activation and may play an important role in the cytokine-induced expression of various immune and inflammatory genes.
Keywords: nuclear factor-κB, coactivator, E-selectin
Transcriptional regulation, a critical control mechanism in fundamental biologic processes, requires the participation of several classes of proteins: those that bind specific DNA sequences, those that associate with transcriptional regulators through protein–protein interactions, known as transcriptional coactivators or corepressors, and those that perform an architectural function. Collectively, these proteins interact with components of the basal transcriptional apparatus to effect dramatic changes in gene expression (for review, see ref. 1).
Nuclear factor-κB (NF-κB) encompasses an important family of inducible transcriptional activators, critical in the regulation of gene expression in response to injury and inflammatory stimuli. The family members include p50, p52, p65 (RelA), c-Rel, and RelB. In the cell, NF-κB exists as homo- or heterodimers with distinct DNA binding specificities (for review, see refs. 2–7). The family members share a 300-aa Rel homology region that is responsible for protein dimerization, nuclear localization, and DNA binding to κB elements in the enhancer regions of target genes. A heterodimer composed of a p50 subunit bound to a p65 (Rel A) subunit is the common dimer. The p65 subunit, similar to two others in the κB family, RelB and c-Rel, contains two transactivation domains in the C-terminal region of the protein (8). The nuclear localization signal of NF-κB is masked by the binding of an inhibitory protein, IκB, sequestering NF-κB in the cytoplasm in unstimulated cells. Cellular stimulation with inflammatory cytokines, phorbol esters, UV irradiation, or potent oxidants results in the phosphorylation, ubiquitination, and proteosomal degradation of IκB (9–12). This is followed by the rapid translocation of NF-κB to the nucleus where it binds to specific κB elements (13, 14).
Many genes involved in the inflammatory response are induced by NF-κB including the proinflammatory cytokines [interleukin (IL) 1, tumor necrosis factor α (TNF-α), granulocyte–macrophage colony-stimulating factor, IL-2, IL-11, and IL-17], chemokines (IL-8, RANTES, and MCP-1); monocyte chemotactic protein-1 enzymes (inducible nitric oxide synthase and cyclooxygenase II), and the endothelial–leukocyte adhesion molecules [intercellular adhesion molecule 1, vascular cell adhesion molecule 1 (VCAM-1), and E-selectin]. Mice or cell lines derived from mice deficient in p50 (15), p65 (16), RelB (17), c-Rel (18), and IκB-α (19, 20) display various functional immune response defects, emphasizing the critical importance of NF-κB in inflammation.
A second class of proteins important in the initiation of transcription by RNA polymerase II are the coactivators (i.e., proteins that bridge the transcriptional activators and the components of the basal transcriptional apparatus). CREB-binding protein (CBP) is a coactivator that interacts with the cAMP-response element binding protein (CREB), a process dependent on the cAMP-dependent protein kinase A and its phosphorylation of CREB (21, 22). A closely related cofactor, p300, was independently isolated on the basis of its interactions with adenovirus E1A (23). Both CBP and p300 interact with a variety of transcriptional activators, including CREB/ATF (22, 24, 25), c-Jun (25, 26), c-Myb (27), YY1 (28), Myo-D (29), c-Fos (30), and steroid receptors (31, 32) (for review, see ref. 33). The interactions between the transcriptional activator and coactivator can be phosphorylation-dependent (e.g., CREB) or -independent (e.g., c-Jun, YY1, or c-Myb) (25, 28, 29). CBP/p300 also bind to the basal transcription factor TFIIB, which in turn contacts the TATA box binding protein (TBP) of the TFIID complex in the basal apparatus (22, 25), as well as an associated H3 and H4 histone acetylating enzyme, p300/CBP-associated factor (P/CAF) (34). CBP/p300 coactivators may perform an important role in the integration of diverse signaling pathways that result in changes in gene expression. For example, competition for limiting amounts of CBP/p300 by different transcription factors activated by diverse signaling pathways may allow specific cellular responses to appropriate signals (31).
Previous work had suggested that members of the Rel family interact directly with components of the basal transcriptional apparatus, in that c-Rel was reported to bind directly to TBP (35). In the present study, we identify CBP and p300 as coactivators for the NF-κB component p65. Recruitment of this versatile coactivator may be a key step linking NF-κB activation with the changes in gene expression seen during inflammatory responses.
MATERIALS AND METHODS
Cells and Transfections.
SL-2 Schneider Drosophila cells were obtained from the American Type Culture Collection (ATCC) and cultivated in Schneider 1× Medium (GIBCO/BRL) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, and antibiotics. Cells were cultured at 23°C in room air. COS-7 cells were obtained from the ATCC and grown on 10-cm dishes in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 2 mM l-glutamine. Transfections were performed by the calcium phosphate procedure as described in the manufacturer’s insert for the Mammalian Matchmaker two-hybrid assay kit (CLONTECH). Whole cell extracts were prepared from the transfected cells and chloramphenicol acetyltransferase (CAT) activity determined as described (36). For all of the data presented, at least three transfections and CAT assays were performed. Human umbilical vein endothelial cells (HUVECs) were isolated and cultured as described (37). For experiments evaluating the effects of cytokine induction, confluent cells were incubated with or without recombinant human TNF-α (Genetech) at a final concentration of 100 units/ml.
Plasmids.
Full-length p300 and CBP expression vectors were provided by R. Eckner and D. Livingston (Dana–Farber Cancer Institute, Boston) and R. Goodman (Oregon Health Science University, Portland). E1A expression plasmids, N-(amino acids 1–596), M-(amino acids 744–1571), and C-(amino acids 1572–2414) terminal p300GAL4 fusion constructs, and pGST300 fusion constructs were prepared as described (28, 38). The plasmid construct p-578 ESelCAT contains the region of residues −578 to +35 of the E-selectin promoter subcloned into the SmaI site of the reporter plasmid pCAT3 (39). The F3 VCAM-1 CAT contains the region of residues −98 to +42 of the VCAM-1 promoter cloned into the SmaI site for the reporter plasmid pCAT3 (36).
Immunprecipitation/Western Blot.
Whole-cell extracts were prepared from HUVECs or COS cells in lysis buffer [25 mM Hepes·KOH, pH 7.2/150 mM potassium acetate/2 mM EDTA/0.1% Nonidet P-40/10 mM sodium fluoride/1 mM phenylmethylsulfonyl fluoride/leupeptin (1 μg/ml)/100 × 103 international units of aprotinin/pepstatin (1 μg/ml)/1 mM dithiothreitol]. Extracts were precleared with nonimmune rabbit IgG (Cappel; 1.5 μg/ml) and 50 μl of protein A-agarose. Extracts equivalent to 2 × 107 cells were incubated 16 hr at 4°C with rabbit anti-p65 (Rockland, Gilbertsville, PA; 5 μg/ml), anti-CBP (Santa Cruz Biotechnology; 25 μg/ml), or a 1:200 dilution of mouse hybridoma supernatant (Ac240, provided by R. Eckner, Dana–Farber Cancer Institute) and then incubated 4 hr with 30 μl of protein A-agarose beads (Pierce). The cleared extracts were subjected to centrifugation at 15,000 × g for 10 min. The pellets were washed four times with 1 ml of lysis buffer, then resuspended in SDS sample buffer, boiled for 5 min, and analyzed on SDS/6% polyacrylamide gels. Proteins were transferred to nitrocellulose (Schleicher & Schuell), the membranes were blocked with 5% nonfat dry milk in TBST buffer (20 mM Tris·HCl, pH 7.6/137 mM NaCl/0.5% Tween 20), and incubated with either p65 or Sp1 (Santa Cruz Biotechnology) antisera for 16 hr at 4°C. Blots were washed three times with TBST buffer, then incubated for 1 hr with a secondary antibody conjugated to horseradish peroxidase (Amersham), and then washed three times in TBST. The antigen–antibody interactions were visualized by incubation with ECL chemiluminescence reagent (Amersham). Blots were exposed to x-ray film for 30 sec to 10 min.
Glutathione S-Transferase (GST) Binding Assay.
Fragments of CBP and p300 coding sequence were subcloned in-frame into pGEX vectors (Pharmacia). GST fusion proteins were expressed in Escherichia coli DH5α according to the manufacturer’s directions. Approximately 4 μg of fusion proteins was immobilized to glutathione-agarose, washed extensively in binding buffer [150 mM potassium acetate (or 75 mM potassium acetate, low salt formulation)/25 mM Hepes, pH 7.2/2 mM EDTA/5 mM dithiothreitol/1 mM phenylmethylsulfonyl fluoride/10 mM sodium fluoride/leupeptin (10 μg/ml)/aprotinin (10 μg/ml)/pepstatin (1 μg/ml)/0.1% Nonidet P-40], and incubated with 100 μl of COS-7 cell extract at 4°C for 60 min. Bound proteins were washed three times with binding buffer, eluted with 20 μl of SDS sample buffer, boiled 2 min, and subjected to SDS/PAGE. Eluted proteins were detected by Western blot analysis as detailed above.
RESULTS
CBP and p300 Increase p65-Dependent Transactivation.
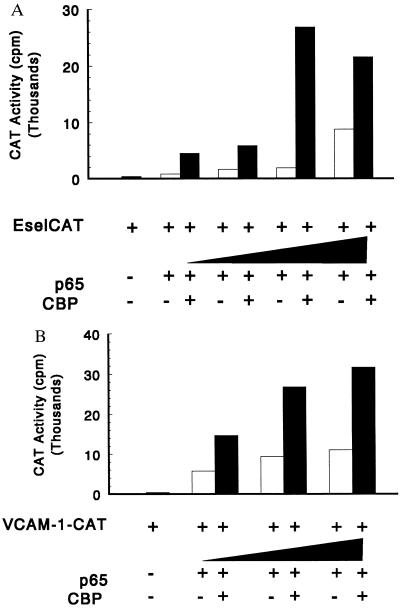
To test the role of CBP/p300 in p65-dependent transcription of E-selectin and VCAM-1, cotransfection experiments were performed. E-selectin (-578 EselCAT) (39) and VCAM-1 (pF3 VCAM-1-CAT) (36) promoter-reporter constructs were activated 5- to 10-fold by cotransfection with a p65 vector in COS cells (Fig. 1). When the reporter constructs were cotransfected with both the CBP and p65 expression plasmids, the activities of the E-selectin and VCAM-1 reporter constructs were raised 3- to 5-fold above those seen with the p65 expression plasmid alone (Fig. 1). In control experiments performed without p65 expression plasmids, the addition of the CBP expression plasmid did not significantly increase the level of CAT activity (data not shown). Identical experiments were performed with p300 expression plasmids and yielded similar results (data not shown). To confirm that overexpressed CBP or p300 did not alter the level of product generated from the p65 expression construct, the level of the transcriptional activator was assessed in transfected cells in the presence or absence of the p300 and CBP expression plasmids. The level of p65 observed in the presence of either CBP or p300 was similar to that observed without the CBP or p300 expression plasmids (data not shown). These studies suggest that CBP/p300 are present in limiting amounts and that overexpression of either coactivator increases p65-dependent reporter gene expression.
Figure 1.
CBP increases transcription of E-selectin (A) and VCAM-1–reporter (B) constructs mediated by p65. COS cells were transfected with 2 μg of reporter gene; 10 μg of the expression plasmid encoding CBP; and 0, 0.1, 0.3, 1, and 3 μg of the expression plasmid encoding p65. Total DNA was kept constant at 12.1 μg by using the empty vector pCR-RSV. The results are representative of three experiments.
E1A Specifically Inhibits p65-Dependent Transactivation.
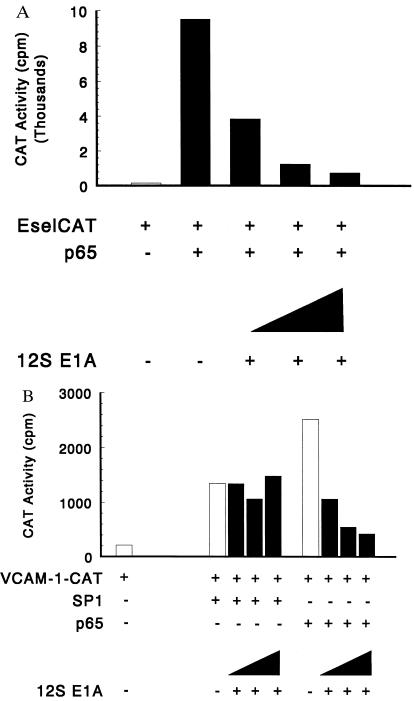
E1A binds to both CBP and p300 and inhibits transcriptional activation mediated by CREB, c-Jun, and Jun B (23–25, 28, 38, 40). The 12S E1A repressed p65-dependent E-selectin and VCAM-1 promoter-reporter activity in a dose-dependent manner in Schneider SL-2 cells (Fig. 2) and in COS cells (data not shown). Optimal VCAM-1 CAT reporter expression requires Sp1, and since SL-2 cells have little or no Sp1, it was possible to demonstrate Sp1-induced VCAM-1 CAT expression. Sp1-dependent VCAM-1 CAT activity was less affected by overexpression of 12S E1A (Fig. 2B). The E1A mutant RG2, which is defective for interacting with p300, did not inhibit p65-mediated transcription. Overexpression of either p300 or CBP relieved the repressive effect of E1A (data not shown). To confirm that overexpressed E1A did not alter the level of product generated from the p65 expression construct, the level of the transcriptional activator was assessed in transfected cells in the presence or absence of the E1A expression plasmid. The level of p65 observed in the presence of E1A was similar to that observed without the E1A expression plasmid (data not shown). The inhibition by 12S E1A suggests that an E1A-sensitive binding protein is required for p65-dependent transactivation. Additionally, these findings suggest that p65 and Sp1 stimulate VCAM-1 expression through different pathways.
Figure 2.
E1A inhibits p65-mediated transcription. (A) 12S E1A inhibits p65-mediated transcription of E-selectin reporter gene expression. E-selCAT reporter plasmid (5 μg) and p65 expression plasmids (25 ng) were cotransfected into Schneider cells along with 1, 3, or 10 ng of 12S E1A expression plasmids. (B) 12S E1A inhibits p65-mediated transcription of VCAM-1–CAT reporter gene expression. VCAM-1–CAT reporter plasmid (5 μg) and p65 or Sp1 expression plasmids (25 ng and 5 μg, respectively) were cotransfected into Schneider cells along with 1, 3, or 10 ng of 12S E1A expression plasmids. The results are representative of three experiments.
CBP/p300 Functionally Interact with p65.
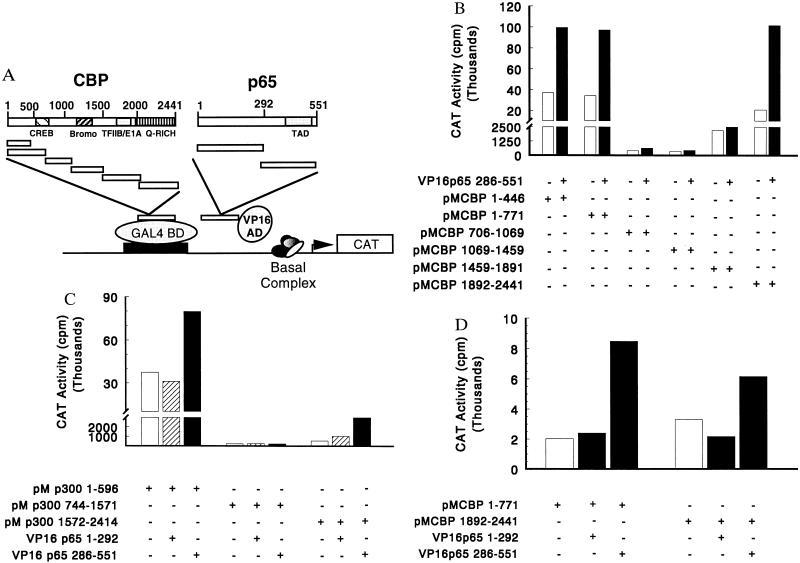
To demonstrate physical interactions between CBP, p300, and p65, three approaches were used: a mammalian two-hybrid system, GST fusion protein interactions, and immunoprecipitation/Western blotting. For the two-hybrid studies, a series of CBP and p300 fragments were generated and fused to the DNA binding protein GAL4. Similarly, two p65 fragments were generated and fused to the transcriptional activator VP16 (see schematic, Fig. 3A). The interacting domains of p65 and CBP were then defined by using the mammalian two-hybrid system in transfected COS cells. The ability of CBP– and p300–GAL4 fusion proteins to activate the CAT reporter gene was examined first. The N-terminal and C-terminal portions of CBP (CBP residues 1–446 and 1–771, as well as CBP residues 1892–2441) and also of p300 were capable of transactivation. The central regions of CBP (amino acids 706-1069, amino acids 1069–1459, and amino acids 1459–1891) and p300 were not capable of activating transcription (Fig. 3 B and C). These findings are in agreement with several reports that CBP or its homolog p300 may function as a transcriptional activator when tethered to DNA through a heterologous DNA binding domain (23–25, 28, 40). To determine which portions of CBP and p300 interact with p65, the ability of the CBP and p300 fusion proteins to activate the CAT reporter gene in the presence of either the N (aa 1–292) or C (amino acids 286–551) terminus of p65 fused to VP16 was examined. As shown in Fig. 3 B and C, the N- and C-terminal portions of both CBP and p300, when cotransfected with the C terminus of p65, significantly increased activated transcription compared with that of VP16 alone or the N- and C-terminal portions of CBP/p300 alone. The activity of the C-terminal region of p300 was weaker than that observed for the N terminus. The C terminus of p65 fused to VP16 did not activate target gene transcription when GAL4 alone or GAL4 fusions with the middle portions of CBP or p300 were used in the assay (Fig. 3 B–D). These results indicate that the C-terminal region of p65, containing the transcriptional activation domains, interacts with both the N and C termini of CBP/p300.
Figure 3.
CBP/p300 and p65 functionally interact in a mammalian two-hybrid system. (A) Structures of the CBP-GAL4 and p65 VP16 constructs. The indicated regions of CBP were cloned into a vector (pM) containing the GAL4 DNA binding domain (BD). Similarly, overlapping regions of p65 were inserted into a vector containing the VP16 activation domain (AD). (B) The N and C termini of CBP interact with the C terminus of p65. COS cells were cotransfected with 2 μg of the pG5CAT reporter gene, 10 μg of the indicated GAL4(pM), and 10 μg of VP16 expression plasmids. Total DNA was kept constant at 22 μg. Data are representative of five experiments. When pooled data were analyzed by the nonparametric Wilcoxon signed rank test, the CBP 1–771 versus CBP 1–771 and p65 286–551 were significantly different (P = 0.015), as were the CBP 1892–2441 versus CBP 1892–2441 and p65 286–551 (P = 0.031). (C) The N and C termini of p300 interact with the C terminus of p65. Cells were cotransfected as described above (B). Data are representative of five experiments. (D) The C terminus of p65 containing the transcriptional activation domain is required for interaction with either the N or C terminus of CBP. Cells were cotransfected as above (B). Data are representative of three experiments.
Interactions between CBP/p300 and p65.
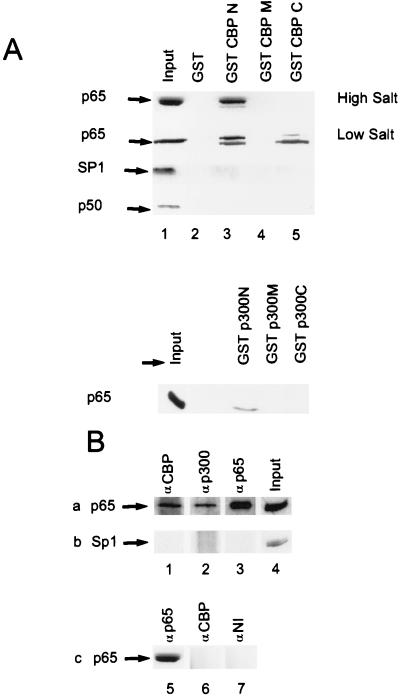
The use of GST fusion proteins prepared from both coactivators provided support for an interaction between CBP/p300 and p65. GST fusion proteins containing the N-terminal (amino acids 1–771), C-terminal (amino acids 1892–2441), and middle (amino acids 1069–1459) portions of CBP and the N-terminal (amino acids 1–596), C-terminal (amino acids 1582–2414), and middle (amino acids 744–1571) portions of p300 were prepared and expressed as described (28). These proteins were linked to glutathione-agarose beads and incubated with lysates prepared from p65-transfected COS cells. The beads were washed and bound material was analyzed by SDS/PAGE followed by Western blotting. p65-immunoreactive material from COS-cell lysates comigrated with the p65 bound to the N-terminal fragment of both CBP and p300 (Fig. 4A). Under low salt conditions, an interaction between the C-terminal fragment of CBP and p65 could be detected (Fig. 4A, lane 5). Binding between CBP/p300 and p65 was specific because no interaction of p65 was observed with either GST alone or with middle fragments of either CBP or p300 (Fig. 4A). Further evidence for the specificity of these interactions was obtained by analysis of control and p50-transfected COS cells. Sp1, present constitutively in COS cells, and p50, present in lysates of p50-transfected COS cells, did not interact with any of the GST fusion proteins or GST alone (Fig. 4A). Similar findings with recombinant bacterially expressed p50/p65 (39) demonstrate a direct interaction between NF-κB and the N terminus of CBP (data not shown). Collectively, these studies demonstrate physical interactions between N- and C-terminal regions of CBP/p300 and p65.
Figure 4.
Physical interaction of CBP/p300 with p65 in vitro and in vivo. (A) Association of CBP/p300 and p65. GST–CBP or GST–p300 fusion constructs were used as ligands and tested for interaction with p65, p50, and Sp1 from programmed COS-cell lysates. Agarose-resin containing GST–CBP [N (amino acids 1–771), M (amino acids 1069–1459), C (amino acids 1892–2441)], or GST–p300 [N (amino acids 1–596), M (amino acids 744-1571), C (amino acids 1572–2370)] were mixed with COS cell lysates. After washing at either high or low salt, bound proteins were released and analyzed by SDS/PAGE on 10% gels followed by Western blot analysis for p65. (B) Physical interaction of CBP/p300 with p65 in endothelial cells. Rows a and b are from TNF-α-treated (100 units/ml, 30 min) HUVECs; row c is from unstimulated HUVECs. Immunoblot with anti-p65 (row a and c) or anti-Sp1 antibody (row b) after immunoprecipitation of whole cell extracts with CBP (lanes 1 and 6), p300 (lane 2), p65 (lanes 3 and 5) antiserum or nonimmune (NI) serum (lane 7). Input, whole cell lysate without immunoprecipitation (lanes 4 and 8).
Interactions between CBP/p300 and p65 in intact cells were demonstrated by using immunoprecipitation/Western blot analysis. To examine these interactions in a physiologically relevant setting, lysates from TNF-α-stimulated HUVECs were immunoprecipitated with anti-CBP or p65 and then immunoblotted with anti-p65 antisera. Additionally, because the previous functional studies and two-hybrid analyses were done in COS cells, CBP and p65 were overexpressed in COS cells and programmed lysates were analyzed in parallel with the endothelial extracts. Both the TNF-α-stimulated HUVEC and the COS cell lysates immunoprecipitated with anti-CBP or anti-p300 antiserum demonstrated an immunoreactive band at ≈65 kDa that was recognized with anti-p65 antisera (Fig. 4B, lanes 1 and 2, and data not shown). This band was identical to that observed in the input lane (Fig. 4B, lane 4) and in cell lysates that were immunoprecipitated and immunoblotted with anti-p65 antisera (Fig. 4B, lane 3). To demonstrate the specificity of these interactions, the Western blots were stripped and reprobed with an antisera to Sp1. Sp1 was observed in the input lanes (Fig. 4B, lane 4) but not in the lysates immunoprecipitated with antisera directed against p65, CBP, or p300 (lanes 1–3). Control unstimulated HUVEC lysate immunoprecipitated with anti-CBP antiserum did not demonstrate an immunoreactive band at 65 kDa that was recognized by the anti-p65 antiserum (Fig. 4B, compare lanes 5 and 6). These results demonstrate an association between CBP and p65 only in TNF-α-treated endothelial cells, when p65 is localized in the nuclear compartment (41). Collectively, both types of approaches suggest that an interaction between p65 and CBP/p300 occurs in intact cells.
DISCUSSION
In this study, we demonstrate that CBP and p300 are transcriptional coactivators for p65. The results may provide important insights into the assembly of cytokine-induced transcriptional enhancers, as will be illustrated below for the E-selectin gene. E-selectin is a 115-kDa member of the selectin family whose expression is endothelial-cell-specific and only observed after induction with lipopolysaccharide or the inflammatory cytokines (TNF-α and IL-1) (42). E-selectin plays an important role in “leukocyte rolling,” a critical and initial interaction of leukocytes with vascular endothelial cells (for review, see ref. 43). The structure of the E-selectin gene and the 5′ flanking region have been defined (for review, see ref. 44). Mutational analysis of the E-selectin promoter identified four positive regulatory domains (PDs) necessary for cytokine-induced gene expression. Three of these domains (PDI, -III, and -IV) contain NF-κB recognition sequences and bind a p50/p65 heterodimeric form of NF-κB (39, 41). The fourth element, PDII, is recognized by several members of the CREB/ATF family of transcription factors (45), although c-Jun/activating transcription factor 2 (ATF-2) heterodimers are the preferred complex bound to the site (46). Both c-Jun and ATF-2 are phosphorylated in response to inflammatory cytokines by components of the stress-activated family of mitogen-activated protein kinases (46, 47). In addition to the transcriptional activators, the chromatin architectural protein high mobility group protein I(Y) [HMG I(Y)] binds to several sites of the E-selectin promoter, potentiating the binding of NF-κB to PDIII and PDIV and the binding of ATF-2 to PDII (39, 48).
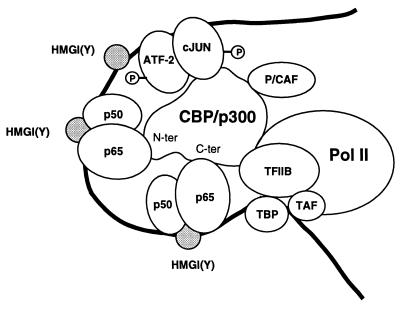
A proposed model representing the cytokine-induced enhancer on the E-selectin promoter is provided in Fig. 5. This model of E-selectin transcriptional regulation assumes that coactivators and the basal transcriptional machinery are present in limiting amounts and that differences in promoter activity result from differences in the affinity of the collection of transcriptional activators for a fixed concentration of basal components. After induction by cytokine, heterodimers of NF-κB bind to PDI, -III, and -IV in the promoter. The binding of HMG I(Y) to multiple sites increases the binding affinity of NF-κB and bends DNA in a way that facilitates the formation of a higher-order complex necessary for transcriptional activation (49, 50). Results from the current study suggest that in addition to the transcription factors NF-κB, ATF-2, and c-Jun and the architectural protein HMG I(Y), this higher-order complex also binds coactivators (i.e., CBP and/or p300). In the model of the E-selectin-induced enhancer (Fig. 5), the alignment between transcriptional activators and CBP/300 has been assembled to maximize potential associations. The N- and C-terminal portions of CBP/p300 interact with p65. The presence of multiple functional κB elements in the E-selectin promoter (PDI, -III, and -IV) suggests that both sites on the coactivator may be bound by p65 (Fig. 5). p65 binding at one site on the coactivator may facilitate occupancy of the other p65 site and binding of additional partners that interact with CBP/p300. The region of CBP between residues 1621 and 1891 contains binding sites for TFIIB, as well as E1A, c-Fos, and pp90rsk (for review, see ref. 33). Occupancy of the N- and C-terminal p65 binding sites on CBP/p300 may increase the interactions of CBP with the TFIIB component of the basal transcriptional machinery. Previously described interactions between members of the Rel family and the C-terminal basic domain of TBP in the TFIIB complex (35) may also facilitate κB-mediated transcription.
Figure 5.
Model of the TNF-α-induced E-selectin enhancer. After induction by cytokine, heterodimers of NF-κB bind to the promoter, which is constitutively occupied by an ATF-2/c-Jun heterodimer. In parallel with nuclear accumulation of NF-κB, ATF-2 and c-Jun are phosphorylated by p38 kinase and c-Jun N-terminal kinase, respectively (46). The binding of HMG I(Y) at multiple sites increases the binding of NF-κB (39) and bends DNA (49) in a way that facilitates the formation of a higher-order complex. The transcriptional activators make extensive protein–protein contacts with the coactivator and the basal complex. Indicated are interactions between the transactivation region of p65 and the N- and C-terminal regions of CBP, an association between c-Jun and the coactivator, and interactions between TFIIB and CBP. Collectively, these events place multiple transcriptional activators in a favorable architecture to complete for the coactivator that is present in limiting amounts. Because RNA polymerase II is constitutively associated with CBP/p300 (51), binding of the coactivator to the E-selectin enhancer may also efficiently recruit the polymerase.
In parallel with the nuclear accumulation of NF-κB, activation of the c-Jun N-terminal kinase and p38 kinases by inflammatory cytokines results in the rapid phosphorylation of the constitutively bound c-Jun and ATF-2 heterodimer in the E-selectin promoter (46). c-Jun is phosphorylated by c-Jun N-terminal kinase and ATF-2 is phosphorylated by the p38 kinases (46). CBP/p300 are coactivators for c-Jun, while ATF-2 appears to function independently of this class of proteins (25, 28). The binding site for c-Jun on CBP/p300 is located near those of CREB, c-Myb, Sap-1a/Elk-1, and Tax and is positioned between residues 461 and 661 (33). The location of a p65 binding site in the N-terminal activation region (residues 1–446) of the coactivator (Fig. 3 A and B) might permit simultaneous occupancy of both the c-Jun site and the N-terminal p65 site (Fig. 5). Collectively, these initial events place multiple transcriptional activators in a favorable architecture to compete for CBP/p300. The constitutive association of CBP with RNA polymerase II (51) may in turn facilitate the recruitment of the polymerase as part of a holoenzyme complex (52) to the E-selectin cytokine-induced enhancer.
This model has general implications for the induced expression of other κB-dependent genes. (i) Like the E-selectin cytokine-response region, a variety of other inducible genes have multiple elements for transcription factors that could bind CBP/p300. These promoters could include those with multiple NF-κB elements or a series of different transcriptional activators capable of interacting with CBP/p300. Inducible enhancers with multiple NF-κB elements include cell surface molecules such as VCAM-1 (53) and class I major histocompatibility complex (54), regulatory proteins such as IκB-α (55), chemokines such as the mouse KC (MIP-1) (56) and RANTES (57), and viral enhancers such as the HIV long terminal repeat (58). Alternatively, multiple different transcriptional activators may effectively recruit the coactivator. Typical of this class of promoter would be that of the β-interferon gene that, like E-selectin (39), also contains NF-κB and ATF2/c-Jun elements (59). The presence of multiple transcriptional activators capable of binding CBP may lead to synergistic recruitment of a coactivator present at limiting concentrations. (ii) CBP/p300 have been identified as multifaceted coactivators, serving as integrators for information from different signal transduction pathways and physically bridging a variety of DNA binding factors to the basal transcription machinery (31, 60). Signaling pathways may alter the phosphorylation state of transcriptional activators capable of interacting with CBP/p300 or may result in changes in the phosphorylation of the coactivator. These events may alter the ability of the transcriptional activators to compete for limiting amounts of CBP. Thus these coactivators may play an important role in NF-κB-dependent gene expression and in coordinating the signals required to generate an inflammatory response.
Acknowledgments
We thank Dr. Richard Goodman, as well as Drs. Richard Eckner and David Livingston, for generously providing CBP and p300 expression vectors, respectively, and Dr. Dimitris Thanos for p50 and p65 expression constructs. We are grateful to Kay Case in the Vascular Research Division for excellent technical assistance with human endothelial cells. This work was supported by research grants from the National Institutes of Health to Y.S. (GM 73848) and to T.C. (HL 35716, HL45462, and PO1 HL36028).
ABBREVIATIONS
- ATF-2
activating transcription factor 2
- CBP
CREB- binding protein
- CREB
cAMP response element binding protein
- GST
glutathione S-transferase
- CAT
chloramphenicol acetyltransferase
- VCAM-1
vascular cell adhesion molecule 1
- TNF-α
tumor necrosis factor α
- HUVEC
human umbilical vein endothelial cell
- TBP
TATA box binding protein
- IL
interleukin
- PD
positive regulatory domain
- HMG Y(I)
high mobility group protein I(Y)
References
- 1.Tjian R, Maniatis T. Cell. 1995;70:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 2.Verma I, Stevenson J, Schwarz E, Van Antwerp D, Miyamoto S. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 3.Thanos D, Maniatis T. Cell. 1995;80:529–532. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 4.Siebenlist U, Franzuso G, Brown R. Annu Rev Cell Biol. 1994;10:405–455. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 5.Baeuerle P, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 6.Grilli M, Chiu J-S, Lenardo M. Int Rev Cytol. 1993;143:1–62. doi: 10.1016/s0074-7696(08)61873-2. [DOI] [PubMed] [Google Scholar]
- 7.Finco T S, Baldwin A S. Immunity. 1995;3:263–373. doi: 10.1016/1074-7613(95)90112-4. [DOI] [PubMed] [Google Scholar]
- 8.Schmitz M, dos Santos Silva M, Baeuerle P. J Biol Chem. 1995;270:15576–15584. doi: 10.1074/jbc.270.26.15576. [DOI] [PubMed] [Google Scholar]
- 9.Palombella V, Rando O, Goldberg A, Maniatis T. Cell. 1994;78:773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 10.Rice N, Ernst M. EMBO J. 1993;12:4685–4695. doi: 10.1002/j.1460-2075.1993.tb06157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Z, Hagler J, Palombella V, Melandri F, Scherer D, Ballard D, Maniatis T. Genes Dev. 1995;9:1586–1597. doi: 10.1101/gad.9.13.1586. [DOI] [PubMed] [Google Scholar]
- 12.Chen Z, Parent L, Maniatis T. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- 13.Beg A, Finco T, Nantermet P, Baldwin A. Mol Cell Biol. 1993;13:3301–3310. doi: 10.1128/mcb.13.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henkel T, Machleidt T, Alkalay I, Kronke M, Ben-Neriah Y, Baeuerle P. Nature (London) 1993;365:182–185. doi: 10.1038/365182a0. [DOI] [PubMed] [Google Scholar]
- 15.Sha W, Liou H, Tuomanen E, Baltimore D. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 16.Beg A, Sha W, Bronson R, Ghosh S, Baltimore D. Nature (London) 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 17.Weih F, Carrasco D, Durham S, Barton D, Rizzo C, Ryseck R, Lira S, Bravo R. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 18.Kontgen F, Grumont R, Strasser A, Metcalf D, Li R, Tarlington D, Gerondakis S. Genes Dev. 1995;9:1965–1977. doi: 10.1101/gad.9.16.1965. [DOI] [PubMed] [Google Scholar]
- 19.Beg A, Sha W, Bronson R, Baltimore D. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 20.Klement J, Rice N, Car B, Abbondanzo S, Powers G, Bhatt H, Chen C-H, Rosen C, Stewart C. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chrivia J, Kwok R, Lanb N, Hagiwara M, Montmony M, Goodman R. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 22.Kwok R, Lundblad J, Chrivia J, Richards J, Bachinger H, Brennan R, Roberts S, Green M, Goodman R. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 23.Eckner R, Ewen M, Newsome D, Gerdes M, DeCaprio J, Lawrence J, Livingston D. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 24.Lundblad J, Kwok R, Laurance M, Harter M, Goodman R. Nature (London) 1995;374:85–88. doi: 10.1038/374085a0. [DOI] [PubMed] [Google Scholar]
- 25.Lee J-S, See R H, Deng T, Shi Y. Mol Cell Biol. 1996;16:4312–4326. doi: 10.1128/mcb.16.8.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arias J, Alberts A, Brindle P, Claret F, Smeal T, Karin M, Feramisco J, Montminy M. Nature (London) 1994;370:226–229. doi: 10.1038/370226a0. [DOI] [PubMed] [Google Scholar]
- 27.Dai P, Akimaru H, Tanaka Y, Hou D-X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. Genes Dev. 1996;10:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 28.Lee J-S, Zhang X, Shi Y. J Biol Chem. 1996;271:17666–17674. [PubMed] [Google Scholar]
- 29.Yuan W, Condorelli G, Caruso M, Felsani A, Giordano A. J Biol Chem. 1995;271:9009–9013. doi: 10.1074/jbc.271.15.9009. [DOI] [PubMed] [Google Scholar]
- 30.Bannister A, Kouzarides T. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S-C, Heyman R, Rose D, Glass C, Rosenfeld M. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 32.Chakravarti D, Lamorte V, Nelson M, Nakasjima T, Schulman I, Juguilon H, Montminy M, Evans R. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 33.Janknecht R, Hunter T. Curr Biol. 1996;6:951–954. doi: 10.1016/s0960-9822(02)00636-x. [DOI] [PubMed] [Google Scholar]
- 34.Yang X-J, Ogryzkov V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 35.Kerr L, Ransone L, Wamsley P, Schmitt M, Boyer T, Zhou Q, Berk A, Verma I. Nature (London) 1993;365:412–419. doi: 10.1038/365412a0. [DOI] [PubMed] [Google Scholar]
- 36.Neish A, Williams A, Palmer H, Whitley M, Collins T. J Exp Med. 1992;176:1583–1593. doi: 10.1084/jem.176.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimbrone M J. Prog Hemost Thromb. 1976;3:1–28. [PubMed] [Google Scholar]
- 38.Lee J-S, Galvin K M, See R H, Eckner R, Livingston D, Moran E, Shi Y. Genes Dev. 1995;9:1188–1198. doi: 10.1101/gad.9.10.1188. [DOI] [PubMed] [Google Scholar]
- 39.Whitley M, Thanos M, Read M, Maniatis T, Collins T. Mol Cell Biol. 1994;14:6464–6475. doi: 10.1128/mcb.14.10.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arany Z, Newsome D, Oldread E, Livingston D, Eckner R. Nature (London) 1995;374:81–84. doi: 10.1038/374081a0. [DOI] [PubMed] [Google Scholar]
- 41.Read M, Whitley M, Williams A, Collins T. J Exp Med. 1994;179:503–512. doi: 10.1084/jem.179.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bevilaqua M P, Stengelin S, Gimbrone M A, Seed B. Science. 1989;243:1160–1165. doi: 10.1126/science.2466335. [DOI] [PubMed] [Google Scholar]
- 43.Springer T. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 44.Collins T, Read M, Neish A, Whitley M, Thanos D, Maniatis T. FASEB J. 1995;9:899–909. [PubMed] [Google Scholar]
- 45.Kaszubska W, Hooft van Huijsduijnen R, Ghersa P, DeRaemy-Schenk A, Chen B, Hai T, DeLamarter J, Whelan J. Mol Cell Biol. 1993;13:7180–7190. doi: 10.1128/mcb.13.11.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Read M A, Whitley M Z, Gupta S, Pierce J W, Best J, Davis R D, Collins T. J Biol Chem. 1997;272:2753–2761. doi: 10.1074/jbc.272.5.2753. [DOI] [PubMed] [Google Scholar]
- 47.Gupta S, Campbell D, Derijard B, Davis R D. Science. 1995;267:389–393. doi: 10.1126/science.7824938. [DOI] [PubMed] [Google Scholar]
- 48.Lewis J, Kaszubska W, DeLamarter J, Whelan J. Mol Cell Biol. 1994;14:5701–5709. doi: 10.1128/mcb.14.9.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Falvo J, Thanos D, Maniatis T. Cell. 1995;83:1101–1111. doi: 10.1016/0092-8674(95)90137-x. [DOI] [PubMed] [Google Scholar]
- 50.Meacock S, Pescini-Gobert R, DeLamarter J, van Huijsduijnen R. J Biol Chem. 1994;269:31756–31762. [PubMed] [Google Scholar]
- 51.Kee B, Arias J, Montmony M. J Biol Chem. 1996;271:2373–2375. doi: 10.1074/jbc.271.5.2373. [DOI] [PubMed] [Google Scholar]
- 52.Ossipow V, Tassan J-P, Nigg E A, Schibler U. Cell. 1995;83:137–146. doi: 10.1016/0092-8674(95)90242-2. [DOI] [PubMed] [Google Scholar]
- 53.Neish A, Read M, Thanos D, Pine R, Maniatis T, Collins T. Mol Cell Biol. 1995;15:2558–2569. doi: 10.1128/mcb.15.5.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mansky P, Brown W, Park J, Choi J, Uang S. J Immunol. 1994;153:5082–5090. [PubMed] [Google Scholar]
- 55.Cheng Q, Cant C, Mott T, Hofer-Warbinek R, Wagner E, Birnstiel M, Bach F, de Martin R. J Biol Chem. 1994;269:13551–13557. [PubMed] [Google Scholar]
- 56.Ohmori Y, Fukomoto S, Hamilton T. J Immunol. 1995;155:3593–3600. [PubMed] [Google Scholar]
- 57.Danoff T, Lalley P, Change Y, Heeger P, Neilson E. J Immunol. 1994;152:1182–1189. [PubMed] [Google Scholar]
- 58.Perkins N, Edwards N, Duckett C, Agranoff A, Schmid R, Nabel G. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thanos D, Maniatis T. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 60.Nakajima T, Fukamizu A, Takahashi J, Gage F, Fischer T, Blenis J, Montminy M. Cell. 1996;86:465–474. doi: 10.1016/s0092-8674(00)80119-1. [DOI] [PubMed] [Google Scholar]