Abstract
Comparison of the amino acid sequence of the chicken and human urokinase-type plasminogen activators (uPAs) revealed that the putative PAI-binding site found in the variable region 1 (VR1) loop of mammalian PAs is absent in the homologous region of ch-uPA. ch-uPA, unlike mammalian PAs, also appears to be refractory to inhibition by human PAIs and as a naturally occurring PAI-resistant variant, constitutes a unique model system for assessing the functional relevance of the PAI-binding site. Therefore, we molecularly constructed a ch-uPA, ch-uPARRHR, which contains the putative PAI-binding motif RRHR (residues 192–195) in its VR1 loop. As a result of this substitution, the second-order rate constant of inhibition of PAI-1 increased ≈700-fold from 4.50 × 104 M−1·s−1 for wild-type ch-uPA to 3.02 × 107 M−1·s−1 for ch-uPARRHR, and the ability to form SDS-stable, uPA–PAI-1 complexes increased ≈1000-fold. Furthermore, the interaction of ch-uPARRHR with PAI-2 was also substantially enhanced, while the interaction with other members of the serine proteinase inhibitor superfamily, protein nexin 1, α1-PI, and C1-inhibitor, was unaffected indicating that the RRHR motif is not a general serine proteinase inhibitor binding site. Finally, we show that extracellular matrix degradation by cells expressing ch-uPARRHR is inhibited by PAI-1 in a dose-dependent manner, while matrix breakdown by cells expressing wild-type ch-uPA is unaffected by PAI-1. Thus acquisition of sensitivity to PAI-1 through a structural motif that enhances the specificity of the protease-inhibitor interaction confers to ch-uPA an added level of regulation in the context of the degradative cellular phenotype.
Keywords: proteolytic enzymes/protease nexin 1/serine proteinase inhibitor
The plasminogen activator (PA)/plasmin system is a proteolytic cascade that leads to effective degradation of extracellular protein components (1, 2) and is initiated by the generation of active urokinase-type PA (uPA). Active uPA converts the inactive zymogen substrate plasminogen to plasmin, an enzyme with broad trypsin-like specificity that can degrade several extracellular matrix (ECM) components (3, 4) and in addition can activate prometalloproteinases (5), latent elastase (6), growth factors (7, 8), and cytokines (9).
Specific regulation of PA catalytic activity is achieved by at least four members of the serine protease inhibitor (serpin) superfamily known as plasminogen activator inhibitors or PAIs (PAI-1, -2, and -3) and protease nexin 1 (PN-1) (10). Inactivation of uPA and tissue-type PA (tPA) by PAI-1 and PAI-2 results in the formation of stable equimolar PA–PAI complexes (11–15) that are rapidly removed from the circulation (16) and degraded.
PAI-1 binding to the target PAs uPA and tPA is believed to be mediated predominately by the formation of salt bridges between a series of positively charged residues occupying a surface loop known as variable region 1 (VR1) of the protease, with a complementary series of negatively charged residues in PAI. Deletion of the VR1 residues 296–302 of KHRRSPG (set as 296KHRRSPG302) of tPA produced a mutant that was highly resistant to PAI-1 inhibition (17, 18). Further, mutation of the tPA VR1 sequence 296KHRR299 to either Gly-296, Glu-298, and Glu-299, or to 296AAAA299, conferred resistance to PAI inhibition (18, 19). Deletion of the residues 179RHRGGS184 in the VR1 region of human uPA (h-uPA) is believed to account for the inability of PAI to inhibit proteolytic activity of this mutant (20). Finally, introduction of 296KHRR299 into the VR1 of thrombin, a serine protease inefficiently inhibited by PAI alone, conferred increased sensitivity to PAI inhibition (21), further implicating the basic residue motif in the VR1 region as a PAI binding site.
We have recently reported that, in contrast to several mammalian uPAs, ch-uPA is resistant to human PAI inhibition and interestingly, lacks any positively charged residues in its corresponding VR1 region (22). Thus ch-uPA would appear to be a natural variant within this family of enzymes in which the putative PAI-binding region is missing from the molecule. In the present report we demonstrate that introduction of the putative PAI-binding site, RRHR, into ch-uPA, results in a gain-of-function alteration rendering the mutant highly susceptible to PAI inhibition. Furthermore, we report that in ECM degradation assays, addition of PAI-1 differentially inhibits matrix degradation mediated by cells expressing the RRHR-containing ch-uPA mutant when compared with cells expressing wild-type (wt) ch-uPA.
MATERIALS AND METHODS
Site-Specific Mutagenesis of ch-uPA.
A ch-uPA cDNA subclone was obtained from Jay Degen (Children’s Hospital Research Foundation, Cincinnati, OH) and the coding sequence was amplified with synthetic oligonucleotide primers 5′-GCGGAATTCAAAGGAGAACTTACCAACATG-3′, and 5′-CGCGGATCCTCACTTTGGTTCACGG-3′, in a PCR with VentR DNA Polymerase from New England Biolabs as per the manufacturer’s instructions. The PCR product was then digested with EcoRI and BamHI and inserted into similarly cleaved pBluescript from Stratagene, and transformed into the host strain XL-1 blue (Stratagene). The ch-uPA insert of one clone was sequenced in its entirety and found to match the published sequence (23). A culture of the pBluescript ch-uPAwt clone was infected with helper phage VCS-M13 (Stratagene), and single-stranded circular DNA was prepared. A synthetic oligonucleotide 5′-GGATAGCTGGCATCTTCCGGCGCCACCGTGGTGGCACTGACCAGTTTCTG-3′ was designed to substitute the 192QNIM195 sequence of ch-uPAwt with RRHR, and insert an additional G. The mutagenic oligonucleotide was 5′-phosphorylated and HPLC purified. Mutagenesis was performed using the Amersham Sculptor in vitro mutagenesis system. The ch-uPA insert of the ch-uPARRHR mutant was sequenced completely to verify the presence of the desired mutation.
DNA Sequencing.
DNA sequence was obtained for both strands as described (24).
Expression and Purification of Recombinant ch-uPA in NS0 and Sf9 Cells.
The mouse myeloma cell line NS0 was the host for the cytomegalovirus promoter-driven expression vector pEE12 obtained from C. Bebbington (Celltech, Slough, U.K.). Untransfected NS0 cells were grown in growth medium [DMEM supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum/1 mM sodium pyruvate/4 mM glutamine/100 units/ml penicillin/0.1 mg/ml streptomycin]. ch-uPAwt and ch-uPARRHR inserts were ligated into the EcoRI and BclI restriction sites of the pEE12 expression vector. NS0 cells were transfected by electroporation then grown in the absence of glutamine as described (25), to select for cells expressing glutamine synthetase encoded by the pEE12 vector. Transfected colonies were screened for ch-uPAwt and ch-uPARRHR production. High-level expressors were expanded in culture, transferred to serum-free growth medium, and the conditioned media were collected 24 h later.
Autographica californica nuclear polyhedrosis virus polyhedrin promoter-driven expression vector pVL1392 obtained from PharMingen containing ch-uPA cDNA was used to infect Spodoptera frugiperda (Sf9) cells. High-level expressors were expanded in culture and transferred to serum-free growth medium, and the conditioned media were collected 24 h later.
The recombinant enzymes from both expression systems were purified by benzamidine-Sepharose affinity and gel filtration chromatography (26). Protein concentration was determined by silver staining of electrophoresed samples compared with known quantitities of a protein standard.
Isolation of Chicken Plasminogen.
Chicken plasminogen was purified from chicken serum by lysine-Sepharose affinity chromatography as described (27). Human glu-plasminogen was purchased from Boehringer Mannheim.
Enzyme and Inhibitor Assays.
Quantitation of uPA activities was performed using the indirect chromogenic substrate assay (28) as described (22). h-uPA, ch-uPAwt, or ch-uPARRHR (50 pg) were added to the chromogenic substrate assay at a final concentration of 10 pM, which generates ≈0.3 A405 linearly over 1 h of incubation. For the uPA inhibition studies enzymes were incubated in PBST [PBS/0.1% (vol/vol) Tween 20) for 45 min at 37°C with varying concentrations of benzamidine, recombinant human PAI-1, or recombinant human PAI-2. For the PAI-1 and PAI-2 inhibition studies, one-half of the PBST reaction mixture was assayed for uPA activity in the chromogenic substrate assay, while the other half was analyzed for uPA–PAI complex formation via casein underlay zymography. Recombinant ch-uPA produced by either the baculoviral expression system or the NS0 expression system behaved equivalently in the PAI-1 inhibition assays.
Kinetics of Inhibition.
Second-order rate constants were measured under pseudo-first-order conditions. Briefly, enzyme and inhibitor were preincubated for periods of time varying from 0 to 90 min at 23°C. Following preincubation the mixtures were diluted in TB (0.1 M Tris, pH 8.1/0.1% Triton X-100), and the residual enzymatic activity was measured in an indirect chromogenic substrate assay. The data were fit to a two exponential model as described (29). The results presented are averaged from three separate experiments and are expressed as mean ± SD.
Casein Underlays.
Free uPA and uPA–PAI complexes were visualized using casein-plasminogen underlays as described (22).
Preparation of ECM.
Chicken embryo fibroblasts were used to produce [35S]methionine-labeled ECMs as described (30).
Degradation of 35S-Labeled ECM.
Culture dishes (35 mm) containing radiolabeled ECM were washed twice with serum free growth medium before seeding cells. Transfected or untransfected NS0 cells (2 × 106 cells) were seeded onto the ECM in 2 ml of serum-free growth medium in the absence or presence of uPA inhibitors. Because native, human PAI-1 is relatively unstable at 37°C (t½ of 2 h), a recombinant mutant PAI-1 (31) that exhibits increased stability (t½ of 145 h) was used in the ECM degradation experiments. The cultures were incubated at 37°C, and at selected time intervals 200 μl of the culture media were removed and centrifuged at 10,000 RPM for 2 min, and the supernatants were counted in a scintillation counter. Cell-mediated release of radioactivity into the culture supernatant has been shown previously to be a manifestation of proteolytic degradation of the ECM (30).
Reagents.
Reagents for enzyme and inhibitor assays and tissue culture were obtained from the previously indicated sources (22). All oligonucleotides were from Operon Technologies (Almaeda, CA). Lysine-Sepharose and benzamidine-Sepharose were from Pharmacia. Active, high-molecular-weight h-uPA, and recombinant human PAI-2 were from American Diagnostica (Greenwich, CT). Recombinant human PAI-1 was a kind gift from Robert Gerard (Katholieke Universiteit Leuven, Leuven, Belgium). PN-1 and C1-inhibitors were kindly provided by William Van Nostrand, and Sanford Simon (State University of New York, Stony Brook) generously provided the α1-PI. Recombinant, stable PAI-1 [mutant 14–1B (31)] was a kind gift from David Ginsburg (University of Michigan, Ann Arbor).
RESULTS
Construction of a ch-uPA Containing the RRHR Motif.
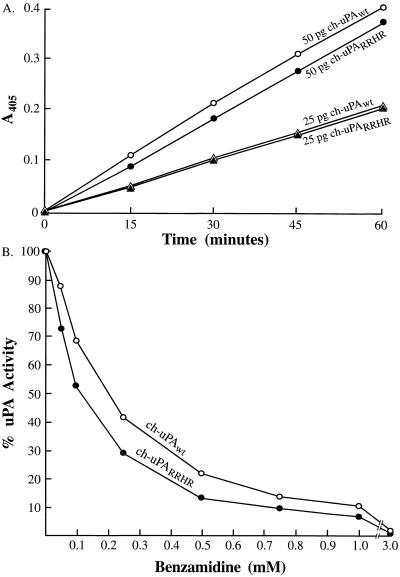
The amino acid sequence RRHR, located in the VR1 region on a surface loop of h-uPA, comprises the putative PAI binding site of uPA. This sequence of basic residues is absent in the corresponding region of ch-uPA and has been linked circumstantially with ch-uPA’s resistance to inhibition by human PAI (22). This four-residue motif therefore was targeted for substitution into ch-uPA as shown in Fig. 1. An additional glycine was inserted along with RRHR to optimize homology of the mutant ch-uPARRHR with h-uPA. The mutant and wt enzymes were expressed in NS0 mouse myeloma cells and purified by benzamidine affinity and gel filtration chromatography. The catalytic activities of equivalent amounts of the purified mutant and wt enzymes, are identical as determined by a time- and dose-dependent activation of chicken plasminogen (Fig. 2A). In addition, sensitivity of ch-uPAwt and ch-uPARRHR to the reversible, active site PA inhibitor, benzamidine, is nearly identical (Fig. 2B).
Figure 1.
Alignment of ch-uPA sequence with the putative PAI binding site of h-uPA. ch-uPA was aligned with Pro-171 in h-uPA (32). The positively charged amino acids of the putative PAI binding site in h-uPA (20) are underlined. The ch-uPA amino acid residues targeted for substitution are in boldface type. In the comparison of chicken and human uPA sequences, identical amino acids are indicated by a vertical line and conservative substitutions are indicated by a diamond.
Figure 2.
Activation of chicken plasminogen by ch-uPAwt and ch-uPARRHR, and sensitivity of ch-uPAwt and ch-uPARRHR to benzamidine. (A) ch-uPAwt (▵, 25 pg; ○, 50 pg) or ch-uPARRHR (▴, 25 pg; •, 50 pg) was assayed for plasminogen activation in a coupled chromogenic substrate assay where the cleaved product is monitored at A405 (22). (B) Inhibition of chicken mutant uPA (ch-uPARRHR) and chicken wt uPA (ch-uPAwt) by benzamidine. ch-uPARRHR (•, 50 pg), or ch-uPAwt (○, 50 pg) was incubated with varying concentrations of benzamidine and assayed for activity in a chromogenic substrate assay. The activity of ch-uPA in the absence of added benzamidine was assigned a value of 100%.
PAI-1 Inhibition of ch-uPA Catalytic Activity and PAI-1–ch-uPA Complex Formation.
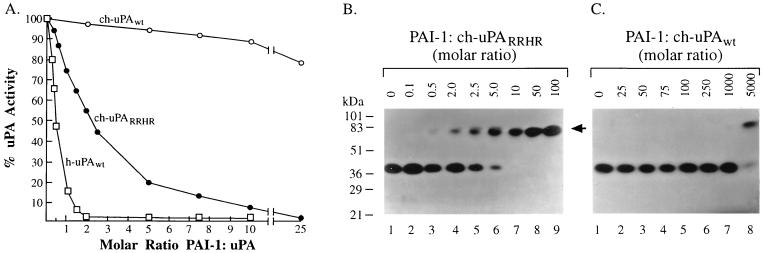
The mutant ch-uPARRHR was examined for its sensitivity to the serpin PAI-1 and compared with wt ch-uPA and h-uPA. Equal amounts of catalytically active h-uPA, ch-uPAwt, and ch-uPARRHR were incubated with increasing amounts of human recombinant PAI-1 (rPAI-1) and assayed for plasminogen activation in a coupled chromogenic substrate assay, and for SDS-stable complex formation. At a PAI/PA molar ratio of 0.5:1, 50% of the h-uPA activity was inhibited (Fig. 3A). The ch-uPAwt was relatively resistant to PAI-1 because a 25-fold molar excess had little effect on catalytic activity. However the ch-uPARRHR mutant required only a 2-fold molar excess of rPAI-1 to yield 50% inhibition.
Figure 3.
The effect of PAI-1 on h-uPA, ch-uPAwt, and ch-uPARRHR catalytic activity and electrophoretic mobility. ch-uPARRHR (A and B, 50 pg), or ch-uPAwt (A and C, 50 pg) was incubated either without (B and C, lane 1), or with human rPAI-1 in the various molar amounts indicated and analyzed for residual uPA activity (A) and electrophoretic mobility by casein-plasminogen underlay (B and C). The underlay was incubated at 37°C for 3 days. The positions of prestained molecular weight markers are given in kDa on the left. The arrowhead indicates the position of the uPA–PAI-1 complexes.
The interaction of PAI-1 with h-uPA results in the formation of equimolar, SDS-stable complexes, a characteristic of serpin interaction with target proteases (33). However, ch-uPAwt does not appear to form stable complexes with human PAI-1 (22). To test for complex formation of ch-uPARRHR with PAI-1, increasing amounts of rPAI-1 were incubated with both wt and mutant ch-uPA and the mixtures were analyzed by casein underlay zymography (Fig. 3 B and C). In the absence of rPAI-1, the lytic zone due to ch-uPA activity appears at ≈40 kDa (Fig. 3 B and C, lane 1). With increasing concentrations of rPAI-1, the ch-uPARRHR activity at 40 kDa is diminished and a corresponding activity at 80–85 kDa, the electrophoretic position of the uPA–PAI-1 complex, progressively increases (Fig. 3B, lanes 2–6). Even though uPA and PAI-1 form inactive complexes, uPA activity is seen in the underlays at 80–85 kDa because prolonged incubation of the uPA–PAI complex eventually leads to hydrolysis of the uPA acyl-intermediate of PAI, yielding active enzyme and an inactive PAI (34, 35). When 1 fmol of ch-uPARRHR is incubated with 10 fmol of rPAI-1, all of the ch-uPARRHR activity shifts up to the uPA–PAI-1 complex (Fig. 3B, lane 7). In contrast, ch-uPAwt under identical incubation conditions did not exhibit significant formation of uPA-PAI-1 complexes, even when 1000 fmol rPAI-1 was added (Fig. 3C, lane 7). Only when 5000 fmol rPAI-1 were added to ch-uPAwt were complexes formed (Fig. 3C, lane 8). Thus, the addition of the RRHR motif confers to uPA an ≈1000-fold enhanced ability to form SDS-stable complexes with PAI-1. The extent of uPA–PAI-1 complex formation detected on casein underlays was confirmed by Western blot analysis, with anti-ch-uPA polyclonal antiserum. A lack of activity at 40–45 kDa on casein underlay zymography corresponded with a lack of immunoreactivity at 40–45 kDa on Western blots, indicating that all of the ch-uPAwt or ch-uPARRHR had shifted into complex formation and that little or no catalytically inactive (non-PAI-1-reactive) recombinant ch-uPA was present in the preparations (data not shown).
The increase in susceptibility to PAI inhibition and SDS-stable complex formation of ch-uPARRHR versus ch-uPAwt was analyzed further by measuring the second-order rate constant of PAI-1 inhibition. Table 1 shows that ch-uPARRHR was inhibited by PAI-1 at a 671-fold higher association rate (k1) than ch-uPAwt. The introduction of the RRHR motif into ch-uPA thus confers a substantially increased rate of inhibition by PAI-1, 3.02 × 107 M−1·s−1, which is in the range of reported values for the rapid rate of PAI-1 inhibition of h-uPA (Table 1).
Table 1.
Second-order rate constant of PAI-1 inhibition of uPA
| Enzyme | k1, M−1·s−1 |
|---|---|
| ch-uPAwt | 4.50 ± 1.81 × 104 |
| (n = 3) | |
| ch-uPARRHR | 3.02 ± 1.76 × 107 |
| (n = 3) | |
| h-uPA* | 1.8 × 107 to 1.6 × 108 |
h-uPA rate constant values were taken from references 36 and 37.
Reactivity of ch-uPARRHR and ch-uPAwt with Other Serpins.
It was of interest to determine if the substitution of the RRHR motif into the VR1 region of ch-uPA also rendered the avian enzyme more sensitive to other serpins. PAI-2, a serpin that like PAI-1 reacts specifically with mammalian uPAs, was incubated with ch-uPARRHR and ch-uPAwt. Formation of SDS-stable complexes between the uPAs and human rPAI-2 was monitored as described for uPA–PAI-1 complexes. At a 15- to 25-fold molar excess of rPAI-2 over ch-uPARRHR, a 40–60% diminishment in the 40-kDa uPA activity was observed with a corresponding appearance of a 75- to 80-kDa complex. In contrast, ch-uPAwt was completely resistant to rPAI-2 up to a 250-fold molar excess (data not shown).
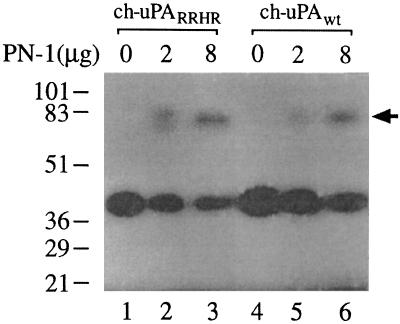
The mutant and wt enzymes were also incubated with PN-1, another serpin that inhibits uPA. As shown in Fig. 4, complex formation occurs to the same extent with both enzymes. Likewise, complex formation between ch-uPAwt and the serpins α1-PI, or C1-inhibitor required large excesses of the serpins but nevertheless were unaffected by introduction of the RRHR motif (data not shown). Thus the differential sensitivities observed when ch-uPAwt and ch-uPARRHR are incubated with PAIs are not manifested when the enzymes are incubated with other serpins indicating that the RRHR motif is not a general serpin binding site.
Figure 4.
The effect of PN-1 on the electrophoretic mobility of ch-uPARRHR, and ch-uPAwt. Fifty picograms of ch-uPARRHR or ch-uPAwt was incubated for 45 min at 37°C without (lanes 1 and 4, respectively) or with 2 or 8 μg PN-1 (lanes 2 and 5 and lanes 3 and 6, respectively). After incubation, the samples were analyzed by zymography as in Fig. 3. The positions of prestained molecular weight markers are given in kDa on the left. The arrowhead indicates the position of uPA–PN-1 complexes.
ECM Degradation by Cells Expressing Mutant or wt ch-uPA.
The participation of the RRHR motif in controlling a cellular phenotype was assessed by monitoring the effect of PAI-1 on ECM degradation mediated by cells producing mutant or wt ch-uPA. Extensive matrix degradation was observed over a 48-h time course when transfected NS0 cells expressing recombinant wt ch-uPA (NS0–ch-uPAwt) or mutant ch-uPA (NS0–ch-uPARRHR) were cultured on radiolabeled matrices (Fig. 5). Matrix breakdown by untransfected NS0 cells is 30–35% of that observed with NS0–ch-uPAwt or NS0–ch-uPARRHR, indicating that the catalytic activity of ch-uPA is responsible for much of the degradation. In addition, NS0 cells producing an active site mutant of ch-uPA, which is catalytically inactive, degrade the matrix at a rate similar to that of untransfected NS0 cells (data not shown). These results are consistent with previous work which has shown that uPA is a major catalytic mediator of degradation of these matrices (30).
Figure 5.
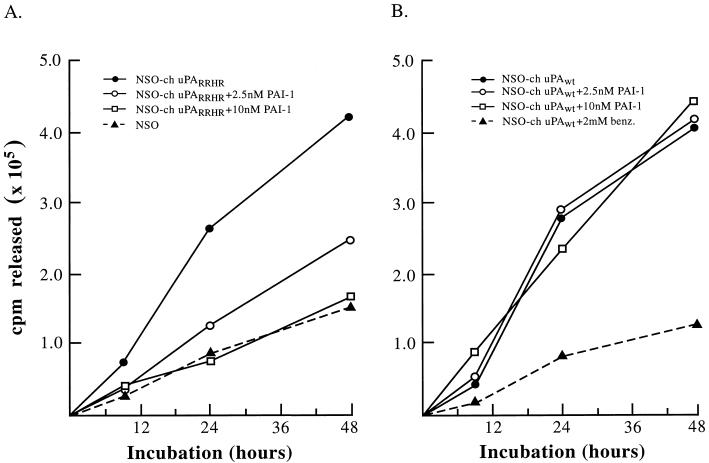
Effect of stable PAI-1 on ECM degradation by transfected NS0 cells expressing ch-uPARRHR and ch-uPAwt. Transfected or untransfected NS0 cells were seeded (2 × 106 cells/2 ml) onto 35S-labeled ECM (9 × 105 cpm/35-mm dish) in the absence or presence of recombinant, stable PAI-1 (31), or benzamidine (benz.). The cultures were incubated at 37°C in serum-free growth medium and at the indicated times, 200 μl of the medium was removed and centrifuged to remove cell debris, and the supernatant was counted. The total radioactivity released into the culture supernatant was calculated for each time point after subtracting the cpm released from the control dishes incubated in the absence of cells. The radioactivity released from the ECM in the absence of cells totaled 5–10% that in the presence of untransfected NS0 cells (0.04–0.20 × 105 cpm/35-mm dish). (A) ECM degradation mediated by untransfected NS0 cells or NS0 cells transfected with and expressing the ch-uPA mutant (NS0-ch-uPARRHR). Recombinant, stable PAI-1 was added to the cultures at the indicated concentrations. (B) Degradation of radiolabeled ECM by NS0 cells transfected with and expressing wt ch-uPA (NS0–ch-uPAwt). Recombinant stable PAI-1 or benzamidine was added to the cultures as indicated.
When NS0–ch-uPARRHR cells are added to the labeled matrices in the presence of nanomolar levels (2–10 nM) of a recombinant stable PAI-1 (31), the extensive and progressive degradation of the ECM is inhibited in a dose-dependent manner to the levels seen with untransfected NS0 cells (Fig. 5A). In contrast, stable PAI-1 had no effect on the ability of NS0–ch-uPAwt cells to degrade ECM (Fig. 5B). Matrix degradation by these cells was extensive and progressive in either the absence or presence of up to 10 nM stable PAI-1, but was inhibited by the reversible uPA inhibitor, benzamidine. Thus, the differential sensitivity to PAI-1 observed in vitro with purified mutant and wt molecules can be recapitulated in a physiological, cellular setting, indicating that the presence of the RRHR motif on a uPA molecule can allow for PAI-1-mediated control of a cellular phenotype, namely ECM degradation.
DISCUSSION
Several studies have implicated a sequence of basic residues in the VR1 surface loop of both tPA (17–19) and uPA (20) as the major, if not only, PAI-1 binding site on these two serine proteases. ch-uPA would appear to be an appropriate test molecule for demonstrating a requirement of the basic residue motif for PAI-1 reactivity and also for examining its possible physiological significance. ch-uPA is a potent activator of plasminogen (38), possesses the identical domain structure of all mammalian uPAs, (23, 39), and has been directly linked to the invasive phenotype (30, 38), but surprisingly its VR1 region, although positionally homologous to h-uPA, is completely devoid of basic residues (Fig. 1). Furthermore, ch-uPA appears to be refractory to mammalian PAI-1 and PAI-2 (22). Introduction into ch-uPA of the precise RRHR motif that is found in human, bovine, porcine, and ovine uPAs renders the resulting ch-uPA ≈700-fold more rapidly inhibitable by PAI-1 (Table 1) and ≈1000-fold more readily able to form SDS-stable uPA–PAI-1 complexes (Fig. 3B).
The chicken mutant uPA, ch-uPARRHR, was similarly more sensitive to another PA-specific serpin, PAI-2, but exhibited little or no difference from wt ch-uPA in its reactivity to the serpins PN-1, α1-PI, and C1-inhibitor. The very low efficiency of ch-uPA inhibition and complex formation with PN-1 and the absence of any differential effect of PN-1 on ch-uPARRHR and ch-uPAwt (Fig. 4) indicate that the RRHR motif may mediate a specific electrostatic interaction only with PAI-1 and possibly PAI-2. It may be that the relative electrostatic and hydrophobic composition of the VR1 region of specific serine proteases is one of the evolutionary modes of selecting out those serpins that will function physiologically as highly specific and highly efficient inhibitors of a given serine protease.
Possibly the more physiologically significant gain of function that results from the mutational introduction of the RRHR motif into the VR1 region of ch-uPA is the ability of PAI-1 to modulate ECM degradation mediated by cells expressing and secreting catalytically active ch-uPARRHR. It has been shown previously that when highly transformed chicken cells (RSVCEF) are cultured on connective tissue matrix, they produce elevated levels of active, two-chain ch-uPA (30, 40). The uPA was shown to mediate almost complete degradation of the matrix either indirectly when plasminogen was present and plasmin was generated, or when plasminogen was not available, the active uPA directly cleaved specific ECM components such as fibronectin (38, 41). Even in the presence of serum, a rich source of serpins, the transformed RSVCEF cultures continued to degrade the matrix via a uPA-dependent mechanism (30). When the RRHR motif is introduced into ch-uPA, matrix degradation by cells expressing this mutant can now be controlled by the presence of nanomolar levels of PAI-1 in the system (Fig. 5). The reduction in ECM breakdown brought about by 10 nM PAI-1, compared with the lack of a PAI-1 effect on ch-uPAwt-expressing cells, indicates that the RRHR motif under these defined conditions can control cellular phenotype.
The absence of a PAI binding site in native ch-uPA raises the question of how the plasminogen activating and degradative potential of ch-uPA is regulated in the different in vivo tissue remodeling and pathological situations where active uPA might be generated. Avian PAI homologues have not yet been described, and thus the precise negative regulatory mechanisms for ch-uPA are unknown. Perhaps ch-uPA, despite its many structural and catalytic similarities with uPAs from other species, is inhibited through a unique mechanism. It may be that specific cofactors are utilized that enhance the binding of avian serpins to uPA, substituting for the missing RRHR motif. It is interesting that thrombin, which also does not contain a prototypic basic residue motif in its VR1 region and does not react efficiently with PAI-1, is enhanced 100- to 200-fold in its reactivity to PAI-1 by the cofactors vitronectin and heparin (42, 43). It is also possible that avian serpins exist that interact with avian uPA via a different surface loop and through a different array of amino acid residues, but still culminating in the reactive center of the serpin inserting into the active site of the enzyme yielding complete inhibition. The exact mechanism of how ch-uPA is regulated awaits the isolation and identification of specific avian uPA inhibitors.
Acknowledgments
We thank Dr. J. Jesty (Department of Medicine) for use of his Molecular Devices microplate reader, software, and help with the kinetic analysis, and also thank L. Abrescia for excellent technical assistance.
ABBREVIATIONS
- PA
plasminogen activator
- uPA
urokinase-type PA
- ch-uPA
chicken uPA
- h-uPA
human urokinase-type PA
- tPA
tissue-type PA
- ECM
extracellular matrix
- PAI
plasminogen activator inhibitor
- rPAI
recombinant PAI
- serpin
serine proteinase inhibitor
- VR1
variable region 1
- PN-1
protease nexin 1
- wt
wild type
References
- 1.Mignatti P, Rifkin D B. Physiol Rev. 1993;73:161–195. doi: 10.1152/physrev.1993.73.1.161. [DOI] [PubMed] [Google Scholar]
- 2.Testa J E, Quigley J P. Cancer Metastasis Rev. 1990;9:353–367. doi: 10.1007/BF00049524. [DOI] [PubMed] [Google Scholar]
- 3.Reich R, Thompson E W, Iwamoto Y, Martin G R, Deason J R, Fuller G C, Miskin R. Cancer Res. 1988;48:3307–3312. [PubMed] [Google Scholar]
- 4.Mignatti P, Robbins E, Rifkin D B. Cell. 1986;47:487–498. doi: 10.1016/0092-8674(86)90613-6. [DOI] [PubMed] [Google Scholar]
- 5.Werb Z, Mainardi C, Vater A, Harris E D. N Engl J Med. 1977;296:1017–1023. doi: 10.1056/NEJM197705052961801. [DOI] [PubMed] [Google Scholar]
- 6.Chapman H A, Stone O L. Biochem J. 1984;222:721–728. doi: 10.1042/bj2220721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lyons R M, Keski-Oja J, Moses H L. J Cell Biol. 1988;106:1659–1665. doi: 10.1083/jcb.106.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato Y, Rifkin D B. J Cell Biol. 1989;109:309–315. doi: 10.1083/jcb.109.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laiho M, Keski-Oja J. Cancer Res. 1989;49:2533–2553. [PubMed] [Google Scholar]
- 10.Kruithof E K O. Enzyme. 1988;40:113–121. doi: 10.1159/000469153. [DOI] [PubMed] [Google Scholar]
- 11.Coleman P L, Patel P D, Cwikel B J, Rafferty U M, Sznycer-Laszuk R, Gelehrter T D. J Biol Chem. 1986;261:4352–4357. [PubMed] [Google Scholar]
- 12.Sprengers E D, Kluft C. Blood. 1987;69:381–387. [PubMed] [Google Scholar]
- 13.Åstedt B, Lecander I, Ny T. Fibrinolysis. 1987;1:203–208. [Google Scholar]
- 14.van Meijer M, Pannekoek H. Fibrinolysis. 1995;9:263–276. [Google Scholar]
- 15.Kruithof E K O, Baker M S, Bunn C L. Blood. 1995;86:4007–4024. [PubMed] [Google Scholar]
- 16.Colucci M, Paramo J A, Collen D. J Clin Invest. 1985;75:818–824. doi: 10.1172/JCI111777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madison E L, Goldsmith E J, Gerard R D, Gething M J H, Sambrook J F. Nature (London) 1989;339:721–724. doi: 10.1038/339721a0. [DOI] [PubMed] [Google Scholar]
- 18.Madison E L, Goldsmith E J, Gerard R D, Gething M J H, Sambrook J F, Bassel-Duby R S. Proc Natl Acad Sci USA. 1990;87:3530–3533. doi: 10.1073/pnas.87.9.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bennet W F, Paoni N F, Keyt B A, Botstein D, Jones A J S, Presta L, Wurm F M, Zoller M J. J Biol Chem. 1991;266:5191–5201. [PubMed] [Google Scholar]
- 20.Adams D S, Griffin L A, Nachajko W R, Reddy V B, Wei C. J Biol Chem. 1991;266:8476–8482. [PubMed] [Google Scholar]
- 21.Horrevoets A J G, Tans G, Smilde A E, van Zonneveld A-J, Pannekoek H. J Biol Chem. 1993;268:779–782. [PubMed] [Google Scholar]
- 22.Testa J E, Stefansson S, Sioussat T, Quigley J P. Fibrinolysis. 1995;9:93–99. [Google Scholar]
- 23.Leslie N D, Kessler C A, Bell S M, Degen J L. J Biol Chem. 1990;265:1339–1344. [PubMed] [Google Scholar]
- 24.Sipley J D, Menninger J C, Hartley K O, Ward D C, Jackson S P, Anderson C W. Proc Natl Acad Sci USA. 1995;92:7515–7519. doi: 10.1073/pnas.92.16.7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bebbington C R, Renner G, Thomson S, King D, Abrams D, Yarranton G T. Bio/Technology. 1992;10:169–175. doi: 10.1038/nbt0292-169. [DOI] [PubMed] [Google Scholar]
- 26.Goldfarb R H, Quigley J P. Biochem J. 1980;19:5463–5471. doi: 10.1021/bi00565a001. [DOI] [PubMed] [Google Scholar]
- 27.Quigley J P. J Cell Biol. 1976;71:472–486. doi: 10.1083/jcb.71.2.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friberger P, Knös M, Gustavsson S, Aurell L, Claeson G. Haemostasis. 1978;7:138–145. doi: 10.1159/000214252. [DOI] [PubMed] [Google Scholar]
- 29.Jesty J, Chen T-Z, Lorenz A. Biochemistry. 1994;33:12686–12694. doi: 10.1021/bi00208a020. [DOI] [PubMed] [Google Scholar]
- 30.Fairbairn S, Gilbert R, Ojakian G, Schwimmer R, Quigley J P. J Cell Biol. 1985;101:1790–1798. doi: 10.1083/jcb.101.5.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkenpas M B, Lawrence D A, Ginsburg D. EMBO J. 1995;14:2969–2977. doi: 10.1002/j.1460-2075.1995.tb07299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verde P, Stoppelli M P, Galeffi P, Di Nocera P, Blasi F. Proc Natl Acad Sci USA. 1984;81:4727–4731. doi: 10.1073/pnas.81.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potempa J, Korzus E, Travis J. J Biol Chem. 1994;269:15957–15960. [PubMed] [Google Scholar]
- 34.Philips M, Juul A-G, Thorsen S. Biochim Biophys Acta. 1984;802:99–110. doi: 10.1016/0304-4165(84)90039-4. [DOI] [PubMed] [Google Scholar]
- 35.Kruithof E K O, Tran-Thang C, Ransijn A, Bachmann F. Blood. 1984;64:907–913. [PubMed] [Google Scholar]
- 36.Thorsen S, Philips M, Selmer J, Lecander I, Åstedt B. Eur J Biochem. 1988;175:33–39. doi: 10.1111/j.1432-1033.1988.tb14162.x. [DOI] [PubMed] [Google Scholar]
- 37.Hekman C M, Loskutoff D J. Arch Biochem Biophys. 1988;262:199–210. doi: 10.1016/0003-9861(88)90182-8. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan L M, Quigley J P. Cell. 1986;45:905–915. doi: 10.1016/0092-8674(86)90565-9. [DOI] [PubMed] [Google Scholar]
- 39.Alexander D S, Aimes R T, Quigley J P. Enzyme Protein. 1996;49:38–58. doi: 10.1159/000468615. [DOI] [PubMed] [Google Scholar]
- 40.Berkenpas M B, Quigley J P. Proc Natl Acad Sci USA. 1991;88:7768–7772. doi: 10.1073/pnas.88.17.7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quigley J P, Gold L I, Schwimmer R, Sullivan L M. Proc Natl Acad Sci USA. 1987;84:2776–2780. doi: 10.1073/pnas.84.9.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keijer J, Linders M, Wegman J J, Ehrlich H J, Mertens K, Pannekoek H. Blood. 1991;78:1254–1261. [PubMed] [Google Scholar]
- 43.Naski M, Lawrence D A, Mosher D F, Podor T J, Ginsburg D. J Biol Chem. 1993;268:12367–12372. [PubMed] [Google Scholar]