Abstract
An assay was developed to assess early intermediates arising in λ’s Red recombination pathway. Double-strand breaks were delivered in vivo to nonreplicating λ chromosomes. Analysis by blot hybridization of total DNA extracts revealed the following: (i) long (>1.4 kilobases) single-strand DNA (ssDNA) intermediates; (ii) resection proceeding bidirectionally from the break site; (iii) single-strand overhangs of 3′ polarity; and (iv) in the absence of λ’s ninR functions, a requirement of the redα gene product for the production of ssDNA. Therefore, the physical characteristics exhibited by these ssDNA molecules are consistent with their being an early recombination intermediate in the Red recombination pathway as proposed previously from genetic and in vitro biochemical analyses.
When phage λ infects Escherichia coli, the principal recombination pathway (RecBCD) of the host is inactivated by the action of λ’s gam gene product (Redγ). Phage recombination is then effected by the λ-encoded Red recombination pathway. When DNA replication is blocked, recombination is focused toward the ends of λ’s linkage map (1). The localization of recombination events at the ends of the map reflects the presence of DNA ends arising from cos-cutting by the λ enzyme terminase, which cleaves the chromosome in readiness for packaging (2).
Analysis of recombinants produced under replication-blocked conditions in vivo has provided evidence for heteroduplex DNA splices (3–5). The strand polarity of these splices suggested the existence of an intermediate with a single-strand 3′ overhang, which could result from the action of λ’s redα gene product (λ Red exonuclease). The redα gene product is a 5′-to-3′ double-strand-specific exonuclease; the redβ gene product is a DNA-melting protein (6–8). In cells lacking RecA protein, mutation in either redα or redβ eliminates recombination. In RecA+ cells the recombination defect is less severe, presumably because of the activity of secondary bacterial recombination pathways.
Genetic studies of λ’s Red pathway have demonstrated that recombination is initiated not only at cos but also at other duplex ends (9). When breaks were introduced into λ’s chromosome through cleavage with a type II restriction endonuclease acting in vivo, recombination was stimulated and was focused around the incision. These observations were fortified by physical analyses of recombinant progeny phage particles (2). Similar observations have been made with the budding yeast Saccharomyces cerevisiae, where mating-type switches and meiotic recombination can be initiated by a double-strand break (DSB) in one of two interacting homologous chromosomes or regions (10–14).
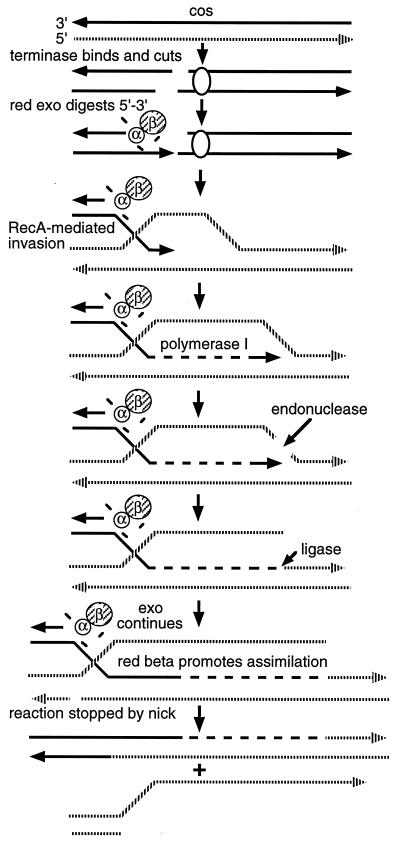
The models proposed for λ’s Red pathway operating under RecA+ conditions (Fig. 1) and for events during meiotic recombination in yeast (15) are remarkably similar. A central feature of both models is that recombination involves invasion of an intact chromosome by a 3′ single-stranded region created at the site of the DSB. In yeast, meiotic recombination intermediates with 3′ single-stranded regions were identified at recombination hotspots (10, 11, 14, 16). Intermediates arose at a sufficiently well-defined time and were sufficiently stable (hours) (10, 17) to be amenable to characterization (14).
Figure 1.
The Red model for lytic cycle λ crosses conducted with blocked DNA replication in rec+ cells (1). (i) λ’s chromosome is cleaved at cos by the λ-encoded enzyme terminase; (ii) the cleaved chromosome is now a substrate for λ’s redα/β gene products, resulting in single-strand overhangs of 3′ polarity; (iii) the single-strand overhang now invades a homologous uncut chromosome; (iv) DNA polymerase I extends the invasive strand; (v) a nick is introduced, possibly through endonucleolytic activity residing in DNA polymerase I; DNA ligase then ligates the invading strand with the nicked strand; and (vi) the Redβ protein completes assimilation of the invading strand until assimilation is stopped by encounter with an incidental nick.
In this study, we demonstrate an early recombination intermediate that arises in vivo following the delivery of a defined DSB to a λ chromosome in the presence of the Red recombination pathway.
MATERIALS AND METHODS
Bacterial Strains and λ Genetic Elements.
Crosses were performed in Escherichia coli strains FA77 (dnaBts Su−) (18), FWS2520 (dnaBts recJ::Tn10 Su−) (our collection), and FWS3282 (dnaBts D(srlR-recA) 306::Tn10 Su−) each containing pPAORM3.8 (19). All phage (Tables 1 and 2) were constructed by using standard in vivo techniques (27).
Table 1.
Phage genotypes
| Phage | Relevant genotype | Ref. |
|---|---|---|
| STU4 | cI857 P80 S7 | This study |
| MMS2560 | gam210 cI857 P80 S7 | This study |
| MMS2597 | gam210 cI857 P80 nin5 S7 | This study |
| MMS2408 | b1451 cI857 P80 S7 | This study |
| MMS2453 | red329 cI857 P80 S7 | This study |
| MMS2451 | b1451 cI857 P80 nin5 S7 | This study |
| MMS2649 | red113 cI857 P80 S7 | This study |
| MMS2484 | red113 cI857 P80 nin5 S7 | This study |
| MMS2650 | red15 cI857 P80 S7 | This study |
| MMS2485 | cI857 P80 nin5 S7 | This study |
| MMS2531 | XhoI::EcoRI cI857 P80 S7 | This study |
| MMS2444 | SR1::XhoI XhoI::EcoRI cI857 P80 S7 | This study |
Table 2.
λ genetic elements
| Element | Description and ref. |
|---|---|
| cI857 | Temperature-sensitive allele of cI for heat induction of lysogens (20) |
| P80 | Suppressor-sensitive (sus) allele of P that provides a partial replication block (21) |
| S7 | sus allele of S suppressible only by SuIII (22) |
| b1451 | Δredα (23) |
| red329 | sus mutation in redα (24) |
| red113 | sus mutation in redβ (24) |
| gam210 | sus mutation in redγ (25) |
| red15 | Δredα/β (23) |
| nin5 | ΔninR region (26) |
| SR1::XhoI | Insertion of an XhoI linker at position 21226 (∗) |
| XhoI::EcoRI | Mutant XhoI site (∗) |
F.W.S. and M.M.S., unpublished work.
Preparation of Phage Stocks.
The temperature-sensitive cI857 allele, in conjunction with the S7 mutation, allowed high-titer phage stocks to be prepared by heat induction. XhoI-modified phage stocks were prepared by inducing lysogens carrying the plasmid pPAORM3.8, which encodes the PaeR7 restriction–modification system (PaeR7 is an isoschizomer of XhoI) (19). Lysogenic cultures (50 ml) were grown at 34°C in LBK broth (1% tryptone/0.5% yeast extract/1% NaCl/75 mM CaCl2/4 mM FeCl2/2 mM MgSO4/0.1 mg/ml vitamin B1) to a density of approximately 1 × 108 cells per ml. The culture was then incubated at 43°C for 15 min, followed by incubation with vigorous shaking at 37°C for 3 hr. Cells were harvested by centrifugation, and lysed in 2 ml of 10 mM Tris·HCl, pH 8.0/300 mM NaCl/5 mM sodium citrate by the combined action of lysozyme and chloroform. After lysis, 10 units of DNase I (molecular biology grade, United States Biochemical) were added, followed by incubation at 37°C for 30 min. Cell debris was removed by centrifugation, and the phage suspension was stabilized by the addition of MgSO4 to a final concentration of 20 mM. Phage were further purified by banding in a CsCl equilibrium gradient, followed by dialysis against TM buffer (10 mM Tris·HCl, pH 8.0/10 mM MgSO4). Phage were titered by plaque assay following serial dilution in TM buffer.
Cross Conditions.
Host cells carrying the plasmid pPAORM3.8 were grown from an overnight culture at 26°C in cross broth (1% tryptone/0.5% NaCl supplemented with 0.2% maltose and 0.1 mg/ml vitamin B1 plus 100 μg/ml carbenicillin) to a density of ≈3 × 108 per ml. Ten-millliter aliquots were centrifuged at 4°C and cells were resuspended in 10 ml of prewarmed (43°C) TM buffer. Prewarmed XhoI-modified phage were added at a multiplicity of infection (moi) of 8, and the culture was supplemented with 10 ml of prewarmed 2× cross broth. After a 15-min incubation of the infected culture at 43°C (to express the red gene products), prewarmed unmodified phage were added at a moi of 8, and the infection was allowed to proceed for various lengths of time. Crosses were terminated by removing aliquots of cells and immediately placing them in an ethanol/water ice bath; cells were then concentrated by centrifugation at 4°C and stored at −20°C prior to DNA extraction. Infected cells blocked for λ DNA replication do not lyse spontaneously.
DNA Extraction.
DNA extraction was performed as described by Ausubel et al. (28). Infected cells were resuspended in 2.5 ml of TE buffer (10 mM Tris·HCl, pH 8.0/1 mM EDTA) to which 25 μl of lysozyme at 5 mg/ml was added, followed by 5 min incubation at 37°C; 100 ml of 10% sodium dodecyl sulfate (SDS) and 5 μl of proteinase K (20 mg/ml) were then added, and incubation was continued at 37°C for 2 hr. A 300-μl aliquot of 5 M NaCl was added followed by gentle mixing and addition of 300 μl of 10% cetyltrimethylammonium bromide (CTAB; United States Biochemical) in 0.7 M NaCl. The lysate was shaken gently and incubated at 60°C for 10 min. The mixture was extracted with 2.5 ml of chloroform/isoamyl alcohol (20:1, vol/vol) and then centrifuged. The aqueous phase was further extracted with 2.5 ml of saturated phenol/chloroform. The DNA was precipitated with an equal volume of isopropyl alcohol, washed one time with 70% ethanol, and air dried. The DNA was then resuspended in 100 μl of TE buffer, pH 8.0.
DNA Analysis.
Equivalent aliquots of total DNA were digested with restriction endonucleases, fractionated by agarose gel electrophoresis, and transferred through capillary action to nitrocellulose membranes either in the absence of prior denaturation [native conditions (29)] or after denaturation and neutralization (30). When DNA samples were treated with S1 nuclease, they were first digested with restriction endonuclease, precipitated with ethanol, and resuspended in nuclease buffer (10 mM Tris·HCl, pH 7.9/10 mM MgCl2/200 mM NaCl/1 mM ZnSO4/1 mM dithiothreitol). One unit of S1 nuclease (Boehringer Mannheim) was added followed by incubation at 30°C for 30 min. Blots were probed either with defined oligonucleotides that had been end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase (New England Biolabs) or with various defined random primed DNA (either to the left or the right of the XhoI cut site) fragments labeled with [α-32P]ATP by using a random prime labeling kit as specified by the manufacturer (Promega). The sequences of the oligonucleotides used were as follows:
probe 54 5′-TTGCAGGGTGGCCTGTTGCTGGCTG-3′;
probe 53 5′-CAGCCAGCAACAGGCCACCCTGCAA-3′;
probe 249 5′-GCGCCAGCATGATTAATACAGC-3′; and
probe 250 5′-GCTGTATTAATCATGCTGGCGC-3′.
The blots were visualized by autoradiography at −70°C using Kodak X-Omat film. Densitometric scanning of autoradiographs utilized an LKB 2222–020 UltroScan XL (Pharmacia Biotech, Piscataway, NJ).
RESULTS
Experimental Design.
The following considerations influenced the experimental design: (i) in λ phage crosses blocked for DNA replication a DSB delivered by a restriction enzyme acting in vivo on an unmodified parent promoted recombination focused at the break site (2); and (ii) the identification of yeast meiotic recombination intermediates was possible using yeast cultures synchronized for meiosis (10, 11, 14, 16). Highly synchronous λ phage infections can be achieved by adsorbing phage to cells resuspended in TM buffer at 4°C, followed by a rapid elevation of temperature to 37°C (31). However, crosses conducted under these conditions do not allow for the maintenance of a tight replication block (achieved with a P80 mutation in the phage chromosome and a dnaBts host mutation). Consequently, we sacrificed tight synchrony for maintenance of the double replication block by adding the unmodified parent phage in TM buffer plus growth medium at 43°C. Measurements of unabsorbed phage showed that about 70% of the unmodified phage preparation infected the host cells within the first 15 min; the remainder continued to infect throughout the course of the experiment. Generally <1% remained in the culture supernatant at the end of the experiment (data not shown).
Identification of Single-Strand DNA (ssDNA) Recombination Intermediates.
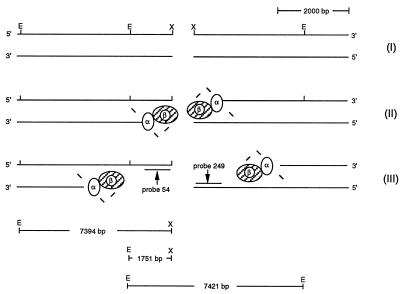
A defined break was introduced in vivo at λ’s unique XhoI site by superinfecting strain FA77 (rec+) carrying the PaeR7 restriction system on plasmid pPAORM3.8 (henceforth referred to as the XhoI system) with STU4 phage (wild type for λ’s Red functions; from now on referred to as wild-type λ). Prior infection with modified phage allowed for the expression of λ’s Red functions (9) and also provided intact homology. If early recombination events proceed as outlined in Fig. 2, then the following intermediates should be demonstrable: intermediate I, the XhoI double strand cut fragment (expected size following in vitro EcoRI digestion of total DNA extracts, 1751 bp); intermediate II, a ssDNA fragment where resection has not proceeded past the proximal EcoRI site (expected mobility approximately equivalent to 1751 bp); and intermediate III, an ssDNA fragment in which resection has proceeded past the proximal EcoRI site (expected mobility approximately equivalent to 7394 bp).
Figure 2.
Formation of ssDNAs after delivery of a double chain break at λ’s XhoI site. A schematic representation of events following the delivery of a double chain break at λ’s XhoI (X) site (intermediate I). After cleavage, the 5′ DNA end is resected using λ’s Redα/β complex (intermediate II) exposing 3′ single-strand overhangs. Once the α/β complex passes the proximal EcoRI (E) site, this site becomes unavailable for restriction in vitro, leading to the formation of intermediate III. The sizes of the anticipated DNA fragments after in vitro digestion with EcoRI are presented below the figure.
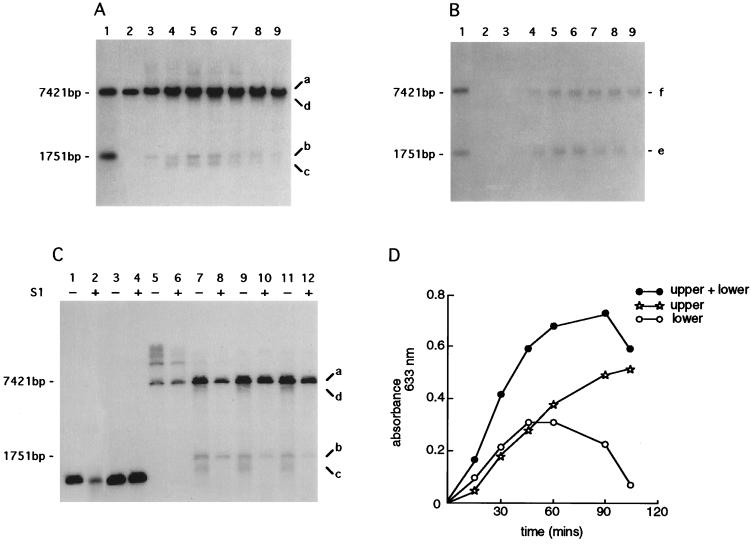
Samples were collected at various times after the addition of the unmodified (cuttable) phage. Total DNA extracts were prepared from each collected sample and digested in vitro with EcoRI (Fig. 3). Fragments were separated by gel electrophoresis and blotted under nondenaturing (29) or denaturing (30) conditions. After hybridization with appropriate probes (see Materials and Methods and Fig. 2), denatured blots (Fig. 3A) showed the following: (i) a signal that corresponded in mobility to the modified uncuttable helper phage (band a, 7421 bp; Fig. 3A); (ii) a signal that corresponded in mobility to the double-stranded DNA XhoI-cut fragment (band b, 1751 bp; Fig. 3A); and (iii) two other less intense signals (bands c and d; Fig. 3A) that migrated slightly faster than the signals designated a and b (Fig. 3A). The double-stranded DNA XhoI-cut fragment (band b; Fig. 3A) could be detected immediately after the addition of the unmodified phage stock (<30 sec) (data not shown).
Figure 3.
Time course of the formation of ssDNA intermediates. An infection was established by wild-type λ (STU4) in strain FA77 (rec+) carrying pPAORM3.8. Cells were harvested at 15-min intervals; total DNA extracts were prepared and digested in vitro with EcoRI. Gels were blotted onto nitrocellulose membranes under either Southern conditions (i.e., with denaturation) or native conditions (i.e., without denaturation). (A) Southern blot analysis of total DNA extracts digested in vitro. (B) Native blot of the same DNA samples as used in A. Lane 1, size standards for EcoRI digestion (7421 bp) and EcoRI/XhoI double digestion (1751 bp) of total λ DNA (see restriction map, Fig. 2); lanes 2–9, samples harvested at 15-min intervals; lane 2, time 0; lane 3, 15 min; lane 4, 30 min; lane 5, 45 min; lane 6, 60 min; lane 7, 75 min; lane 8, 90 min; and lane 9, 105 min. Each blot was probed with oligonucleotide 54 (position indicated in Fig. 2) designed to hybridize with single-strand overhangs of 3′ polarity. Signals designated a, b, c, d, e, and f indicate DNA intermediates referred to in the text. (C) Southern analysis of DNAs treated with S1 nuclease. DNA samples were either untreated (−) or treated (+) with 1 unit of S1 nuclease, except for lane 2, which was treated with 10 units. Lanes 1, 2, 3, and 4, represent a gel-purified XhoI/PstI λ fragment and serve as the control for nuclease digestion; lanes 5–12, samples harvested at 15-min intervals and digested in vitro with EcoRI; lanes 5 and 6, time 0; lanes 7 and 8, 15 min; lanes 9 and 10, 30 min; and lanes 11 and 12, 60 min. The blot was probed with oligonucleotide 54. Signals designated a, b, c, and d correspond to those in A. (D) Densitometric scanning of the native blot presented in B. “Lower” refers to the signal at 1751 bp; “upper” refers to the signal at 7421 bp; “upper + lower” indicates the sum of the two signals.
In contrast, the corresponding native blot showed only two signals (bands e and f), with mobilities approximately corresponding to molecular sizes 1751 bp and 7421 bp, respectively (Fig. 3B).
To determine the nature of the DNA species designated c and d, the DNA samples restricted by EcoRI in vitro were further digested with S1 nuclease and examined by Southern blotting (Fig. 3C). DNA species designated c and d were both susceptible to S1 nuclease digestion, indicating that these DNAs contained single-stranded regions, and probably correspond to the DNA species designated e and f on the native blots.
The kinetics of ssDNA formation was assessed by densitometry of the autoradiogram from the native blot (i.e., DNA species e and f; Fig. 3B). The signal designated e increased and then decreased in intensity, in contrast to the signal designated f, which increased over the 105-min time course of the experiment (Fig. 3D).
These data indicated that resection from the XhoI cut site produced a ssDNA intermediate, and that resection proceeded past the proximal EcoRI site over time (manifest by the shift upward in molecular size of band e to band f; Fig. 3B).
Strand polarity was assigned to the ssDNA intermediates by hybridization of native blots with strand-specific oligonucleotide probes (Fig. 4). The progress of in vivo cutting by XhoI was monitored by probing a denatured blot containing PstI-cut DNA isolated at 0, 15, and 60 min. In the latter two samples, the DNA bound probes specific for each strand and for each side of the XhoI cut (Fig. 4 A, C, E, and G). Probing the same DNAs under native conditions revealed binding of only those probes that were specific for chains ending 3′ at the XhoI cut site (Fig. 4 B, D, F, and H). These data demonstrate that the DNA restricted in vivo is subject, on both sides of the cut, to resection of the 5′-ended chains.
Figure 4.
Strand specificity of ssDNA intermediates. Wild-type λ (STU4)-infected strain FWS2520 (recJ) carrying pPAORM 3.8. Cells were harvested at time 0, 15 min, and 60 min into the infection. The series of blots presented in A–H are of total DNA extracts digested in vitro with PstI (expected DNA fragment sizes following in vitro PstI digestion: uncut modified phage DNA, 4749 bp; XhoI–PstI fragment to the right of the XhoI cut site, 3507 bp; and XhoI–PstI fragment to the left of the XhoI cut site, 1249 bp). Gels A–D examine events to the right of the XhoI cut site; gels E–H examine events to the left. A, C, E, and G are denatured blots; B, D, F, and H are native blots. A and B are hybridized with probe 250 (5′ specificity); C and D, with probe 249 (3′ specificity); E and F, with probe 54 (3′ specificity); and G and H, with probe 53 (5′ specificity). Lanes 1 and 5 in each gel are the size standards PstI/XhoI (3507 bp and 1249 bp) and PstI (4749 bp) (denatured native blot size standards were obtained by boiling the restricted DNAs immediately prior to electrophoresis); experimental samples are in lane 2 (time 0), lane 3 (15 min), and lane 4 (60 min).
The above results were generalized by mutating λ’s single XhoI cut site (through the insertion of an EcoRI linker at position 33498) and introducing a novel XhoI cut site into λ’s leftmost EcoRI site (through insertion of an XhoI linker at position 21226; Tables 1 and 2; data not shown).
Involvement of λ’s Red Functions in the Formation of ssDNA Intermediates.
Table 3 summarizes ssDNA formation after infections with various Red mutants. In the absence of the Redα polypeptide (b1451 and red329 crosses), in both rec+ and recJ hosts, ssDNA species of 3′ polarity were detected. ssDNA signals were absent when crosses were performed with phage that were doubly mutant for redα and λ’s ninR region (b1451 nin5 cross). To establish whether the Red gene products were sufficient to produce ssDNA recombination intermediates of 3′ polarity, crosses were performed with phage that carried simply a nin5 deletion; in these crosses ssDNAs of 3′ polarity were observed. As identical observations were obtained in either E. coli wild-type or recJ backgrounds, the results demonstrate that the 3′ overhangs were not created by a helicase-assisted RecJ nuclease (32).
Table 3.
Effect of Red mutations on ssDNA formation
| λ genotype | Red mutation | ssDNA rec+*† | ssDNA recA* |
|---|---|---|---|
| Wild type | + | + | |
| gam210 | γ | − | − |
| gam210 nin5 | γ | − | − |
| b1451 | α | + | − |
| red329 | α | + | ND |
| b1451 nin5 | α | − | − |
| red113 | β | + | + |
| red113 nin5 | β | + | + |
| red15 | α/β | − | ND |
| nin5 | + | + |
ND, not determined.
Presence or absence of signal on native blots. For all genotypes, the corresponding denatured blot showed a strong signal corresponding to the XhoI double strand cut fragment.
Identical observations in E. coli rec+ and recJ hosts.
ssDNA intermediates could not be demonstrated in the following infections of a rec+ host: (i) in the absence of the Redγ polypeptide (gam210; gam210 nin5 crosses; Table 3); and (ii) in the simultaneous absence of both the Redα and Redβ polypeptides (red15 cross; Table 3). However, somewhat surprisingly, ssDNA intermediates were observed under rec+ and recA conditions with phage carrying a sus mutation in the redβ gene (red113 mutation).
Kinetic Analysis of ssDNA Formation.
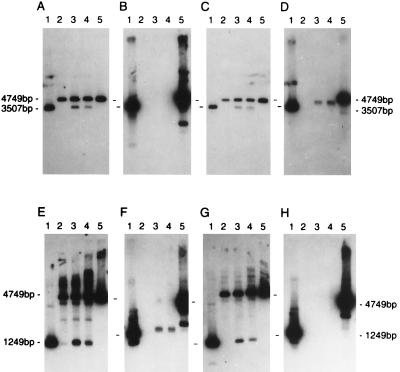
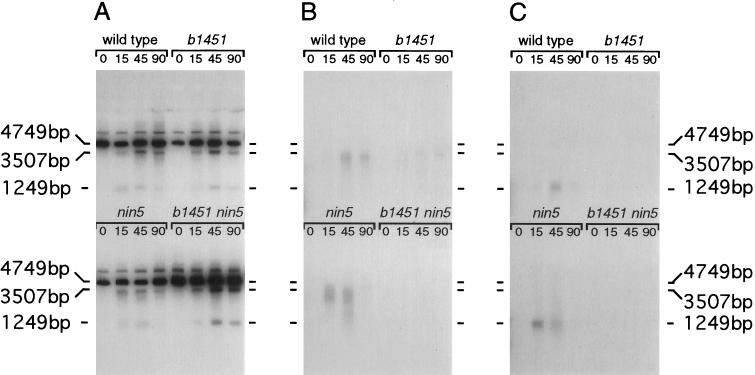
The preceding data indicate that both the Redα exonuclease and a ninR-encoded function(s) are likely contributors to the ssDNA intermediates demonstrated in Fig. 3B. To elucidate the separate contributions of Red and NinR on the formation of ssDNA intermediates, a kinetic analysis was undertaken on infections (wild-type, b1451, nin5, and b1451 nin5 phage) performed simultaneously on the same batch of cells (Fig. 5). A comparison of the native hybridization patterns (Fig. 5 B and C) allows the following conclusions: (i) ssDNA intermediates that are detectable at early time points (15 min) arise primarily through the action of Redα exonuclease (nin5 cross); (ii) ssDNA that is detected at later time points (90 min) results primarily from the action of the ninR function (b1451 cross; (iii) Redα exonuclease contributes more to resection from DSBs than its ninR counterpart; and (iv) the pattern of ssDNA formation generated with wild-type phage is a composite consisting of ssDNA generated by the Redα exonuclease and the ninR function.
Figure 5.
Kinetic analysis of ssDNA formation. Concurrent rec+ infections were established with λ: wild type, b1451, nin5, and b1451 nin5. Total DNA extracts were prepared from cells recovered at times 0, 15 min, 45 min, and 90 min. Each sample was digested in vitro with PstI prior to electrophoresis and blotting. (A) Denatured blot probed with two random-primed DNA fragments that recognize λ DNA to the right and to the left of the XhoI cut site. (B) Native blot probed with a random-primed DNA fragment that recognizes λ DNA to the right of the XhoI cut site. (C) Native blot probed with a random-primed DNA fragment that recognizes λ DNA to the left of the XhoI cut site. Size markers align with λ DNA cleaved with PstI (4749 bp) and PstI/XhoI (3507 bp and 1249 bp).
DISCUSSION
Genetic and biochemical analyses of λ’s Red recombination pathway had predicted ssDNA intermediates of 3′ polarity (1, 3, 5, 6). The physical analyses performed in this in vivo study accord with these predictions. Furthermore, the ssDNA intermediates are long-lived (possibly >105 min), may exceed 1.4 kb in length, and are produced bidirectionally from a DSB, in the two intervals examined.
The expected dependence of ssDNA formation on Redα was demonstrable only in a nin5 background. The ninR region may contribute an exonuclease with properties similar to those of Redα. Alternatively, the ninR region may contribute an inhibitor of an unspecified nuclease that is active on 3′-ended ssDNA. Phage carrying the nin5 deletion enjoy wild-type levels of recombination, implying that NinR-dependent ssDNAs do not participate in any major way in the formation of recombinants in Red+ RecA+ infections when DNA replication is blocked (T. Tarkowski, personal communication). The data in Fig. 5 show that the redα- and ninR exonucleolytic effects are temporally separated. Early (Redα-generated) ssDNA intermediates rise and fall in intensity as the infection proceeds; late (NinR-generated) ssDNA intermediates accumulate over time (cf. b1451 and nin5 crosses, Fig. 5 B and C). These kinetic distinctions suggest that the loss of signal from the Redα-generated ssDNA species was due to the incorporation of these early ssDNA intermediates into intermediates further advanced along the Red pathway, whereas the NinR-generated ssDNAs are unsuitable for recombination. The appearance and disappearance of ssDNAs are also seen in S. cerevisiae after induction of the mating-type switch (33). In λ crosses, using Red functions supplied by a plasmid (34), recombinant molecules are observed as early as 20 min after infection and accumulate for up to 2 hr subsequent to synchronous in vivo restriction cutting (A. Kuzminov, personal communication). These kinetics suggest that the ssDNAs shown in this study do represent the predicted first intermediate in the Red pathway.
The longevity of the ssDNA intermediates invites comment. To survive for so long (possibly >105 min), these ssDNA intermediates must be protected from nucleases. Cassuto and Radding (35) proposed that the formation of single-strand intermediates involving the Redα/β polypeptides is a coupled reaction (35). In their view, Redα exonuclease operates in concert with the Redβ polypeptide, with β protein either coating and protecting the nascent strand as it is being formed or facilitating incorporation of the nascent strand into the homologous duplex. Our data only partially support the Cassuto and Radding model. We found reduced amounts of ssDNA in the absence of Redβ (either with or without the ninR-encoded functions). However, ssDNA intermediates were observed in the absence of Redβ (in both rec+ and recA backgrounds), indicating that the presence of neither Redβ nor RecA is essential for Redα exonuclease to operate.
Several mechanistic parallels have been established between the λ Red pathway and the alternative RecE recombination pathway in E. coli (reviewed in ref. 36). RecE recombination is observed in E. coli recB recC sbcA mutants, where suppressor-mediated derepression of the Rac prophage activates a 5′-3′-exonuclease (the recE gene product) and an annealing protein (the recT gene product) with biochemical properties similar to those of the Redα/β proteins (36). sbcA mutations are able to complement both redα and redβ mutations (37, 38), and genetic analysis has shown that the RecE and RecT polypeptides can operate on DSBs when delivered to defined plasmid DNAs (39), yielding heteroduplexes with predicted 3′ strand polarities (40). However, the RecE/RecT polypeptides appear less efficient at operating at DSBs than the Redα/β proteins (39), which may indicate a difference in how the two sets of proteins recognize a DSB. The in vivo assays employed in this study should be able to establish whether long stable ssDNAs are also found in the RecE pathway.
The longevity of the Red intermediates and the tractability of the λ/E. coli system encourages further investigation of the “in vivo biochemistry” of Red-mediated DSB-induced recombination.
Acknowledgments
We thank members of the Stahl laboratory for helpful comments on this manuscript. Trudee Tarkowski’s suggestions played a major role in the shaping of this work. S.A.H. thanks Jim Sawitzke and Kit Tilly for help with λ biology. Susan Smaus, Gary Hettrick, and Bob Evans helped in the preparation of this manuscript for publication. This work was supported by National Institutes of Health Grant GM-33677 and National Science Foundation Grant MCB-9402695. F.W.S. is American Cancer Society Research Professor of Molecular Biology.
ABBREVIATIONS
- DSB
double-strand break
- ssDNA
single-strand DNA
References
- 1.Stahl F W, Kobayashi I, Stahl M M. J Mol Biol. 1985;181:199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 2.Stahl F W, Fox M S, Faulds D, Stahl M M. Genetics. 1990;125:463–474. doi: 10.1093/genetics/125.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White R L, Fox M S. Proc Natl Acad Sci USA. 1974;71:1544–1548. doi: 10.1073/pnas.71.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahl F W, McMilin K D, Stahl M M, Crasemann J M, Lam S. Genetics. 1974;77:395–408. doi: 10.1093/genetics/77.3.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiqi I, Stahl M M, Stahl F W. Genetics. 1991;128:7–22. doi: 10.1093/genetics/128.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Little J W. J Biol Chem. 1967;242:679–686. [PubMed] [Google Scholar]
- 7.Carter D M, Radding C M. J Biol Chem. 1971;246:2502–2512. [PubMed] [Google Scholar]
- 8.Sriprakash K S, Lund N, Huh M M, Radding C M. J Biol Chem. 1975;250:5438–5445. [PubMed] [Google Scholar]
- 9.Thaler D S, Stahl M M, Stahl F W. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 10.Connolly B, White C I, Haber J E. Mol Cell Biol. 1988;8:2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White C I, Haber J E. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolodkin A L, Klar A J S, Stahl F W. Cell. 1986;46:733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- 13.Nicolas A, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:35–38. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 14.Sun H, Treco D, Schultes N P, Szostak J W. Nature (London) 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 15.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 16.Sun H, Treco D, Szostak J W. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 17.Rudin N, Sugarman E, Haber J E. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McMilin K D, Russo V E A. J Mol Biol. 1972;68:49–55. doi: 10.1016/0022-2836(72)90261-6. [DOI] [PubMed] [Google Scholar]
- 19.Gingeras T R, Brooks J E. Proc Natl Acad Sci USA. 1983;80:402–406. doi: 10.1073/pnas.80.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sussman R, Jacob F. C R Acad Sci. 1962;254:1517–1519. [PubMed] [Google Scholar]
- 21.Campbell A. Virology. 1961;14:22–32. doi: 10.1016/0042-6822(61)90128-3. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg A R, Howe M. Virology. 1969;38:200–202. doi: 10.1016/0042-6822(69)90148-2. [DOI] [PubMed] [Google Scholar]
- 23.Daniels D L, Schroeder J L, Szybalski W, Sanger F, Blattner F R. In: Lambda II. Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. pp. 469–676. [Google Scholar]
- 24.Shulman M J, Hallick L M, Echols H, Signer E R. J Mol Biol. 1970;52:501–520. doi: 10.1016/0022-2836(70)90416-x. [DOI] [PubMed] [Google Scholar]
- 25.Zissler J, Signer E, Schaefer F. In: The Bacteriophage Lambda. Hersey A D, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1971. pp. 455–469. [Google Scholar]
- 26.Court D, Sato K. Virology. 1969;39:348–352. doi: 10.1016/0042-6822(69)90060-9. [DOI] [PubMed] [Google Scholar]
- 27.Arber W, Enquist L, Hohn B, Murray N E, Murray K. In: Lambda II. Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. pp. 433–466. [Google Scholar]
- 28.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology 1. New York: Greene & Wiley; 1989. [Google Scholar]
- 29.Lichten M J, Fox M S. Nucleic Acids Res. 1983;11:3959–3971. doi: 10.1093/nar/11.12.3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Southern E. J Mol Biol. 1975;98:503–513. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 31.Better M, Freifelder D. Virology. 1983;126:168–182. doi: 10.1016/0042-6822(83)90469-5. [DOI] [PubMed] [Google Scholar]
- 32.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugawara N, Haber J E. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poteete A R, Fenton A C. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cassuto E, Radding C M. Nature (London) New Biol. 1971;229:13–16. doi: 10.1038/newbio229013a0. [DOI] [PubMed] [Google Scholar]
- 36.Kolodner R, Hall S D, Luisi-Luca C. Mol Microbiol. 1994;11:23–30. doi: 10.1111/j.1365-2958.1994.tb00286.x. [DOI] [PubMed] [Google Scholar]
- 37.Gillen J R, Clark A J. In: Mechanism of Recombination. Grell R F, editor. New York: Plenum; 1974. pp. 123–136. [Google Scholar]
- 38.Gillen J R, Willis D K, Clark A J. J Bacteriol. 1981;145:521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokochi T, Kusano K, Kobayashi I. Genetics. 1995;139:5–17. doi: 10.1093/genetics/139.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silberstein Z, Tzfati Y, Cohen A. J Bacteriol. 1995;177:1692–1698. doi: 10.1128/jb.177.7.1692-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]