Abstract
The Notch family of cell surface receptors and its ligands are highly conserved proteins that regulate cell fate determination, including those involved in mammalian vascular development. We report that Notch induces VEGFR-3 expression in vitro in human endothelial cells and in vivo in mice. In vitro, Notch in complex with the DNA-binding protein CBF-1/suppressor of hairless/Lag1 (CSL) bound the VEGFR-3 promoter and transactivated VEGFR-3 specifically in endothelial cells. Through induction of VEGFR-3, Notch increased endothelial cell responsiveness to VEGF-C, promoting endothelial cell survival and morphological changes. In vivo, VEGFR-3 was upregulated in endothelial cells with active Notch signaling. Mice heterozygous for null alleles of both Notch1 and VEGFR-3 had significantly reduced viability and displayed midgestational vascular patterning defects analogous to Notch1 nullizygous embryos. We found that Notch1 and Notch4 were expressed in normal and tumor lymphatic endothelial cells and that Notch1 was activated in lymphatic endothelium of invasive mammary micropapillary carcinomas. These results demonstrate that Notch1 and VEGFR-3 interact genetically, that Notch directly induces VEGFR-3 in blood endothelial cells to regulate vascular development, and that Notch may function in tumor lymphangiogenesis.
Introduction
The Notch family of receptors and its ligands are cell surface proteins that regulate cell fate determination through direct cell-cell interactions. In vertebrates, there are 4 Notch genes (Notch1–4) and 2 families of ligands, Jagged (Jagged1, 2) and Delta-like (Dll1, 3, 4) (1, 2). Notch signaling modulates the ability of cells to respond to extracellular cues. The finding that genes of the Notch signaling cascade are robustly expressed in the vasculature suggests that Notch signaling guides endothelial cells and associated mural cells through the cell fate decisions needed to form and maintain the vascular system. In fact, embryos nullizygous for components of the Notch signaling cascade (Notch1, Notch1/Notch4, Sel-10, presenilin 1/2, and jagged 1) die midgestation due to defects in vascular remodeling (3–7). Similarly, embryos in which Notch4 signaling has been activated within the endothelium die at E10.5 with severely disorganized vasculature (8).
Notch regulates cell fate decisions by altering patterns of gene expression. Upon ligand activation, the cytoplasmic domain of Notch is proteolytically released and translocates to the nucleus, where it interacts with the CBF1/suppressor of hairless/Lag-2 (CSL) transcriptional repressor and converts it to a transcriptional activator (9, 10). The hairy enhancer of split (HES) and HES-related protein (Hey) families of transcriptional repressors are direct targets of Notch/CSL-dependent signaling (11, 12). Disruption of Notch or the Hey2 homolog, gridlock, results in a loss of ephrin-B2 expression in the endothelial cells of the dorsal aorta of zebrafish (13–15). In primary human endothelial cell cultures, Notch activation upregulates the expression of HES1, Hey1, and Hey2 as well as ephrin-B2 (16). In mice nullizygous for Hey1 and Hey2, ephrin-B2 expression is lost in the aorta, demonstrating a conserved function for Hey proteins in regulating ephrin-B2 (17). Thus in endothelial cells, Notch may regulate ephrin-B2 expression through the induction of Hey1 and Hey2. Notch also signals via a CSL-independent pathway that is poorly understood (18).
The VEGFR family is composed of 3 type I tyrosine kinase receptors that regulate angiogenesis and lymphangiogenesis (19). Among these, VEGFR-3 selectively promotes lymphangiogenesis. Transgenic mice that mis-express the VEGFR-3 ligand, VEGF-C, in skin develop aberrant lymphatic vessels (20). In experimental tumors, VEGF-C overexpression also correlated with the development of lymphatic metastases (21–23). In fact, expression of a VEGFR-3 antagonist can block lymph node metastasis of aggressive metastatic lung cancer cells (24). Thus VEGFR-3 clearly functions to regulate both physiological and pathological lymphangiogenesis.
However, VEGFR-3 function has also been shown to be essential for embryonic angiogenesis. Embryos nullizygous for VEGFR-3 die at E9.5 due to defects in remodeling of the primary vessel networks (25). These VEGFR-3 nullizygous embryos die prior to the initiation of lymphangiogenesis, demonstrating a distinct role for VEGFR-3 signaling in angiogenesis. In breast carcinomas, VEGFR-3 expression was upregulated in the endothelium of tumor blood vessels, while VEGF-C was expressed in the intraductal and invasive cancer cells (26, 27), supporting a role for VEGFR-3 in tumor angiogenesis as well as lymphangiogenesis. The promoter of the VEGFR-3 gene contains elements that drive VEGFR-3 expression specifically to blood endothelial cells (BECs) and lymphatic endothelial cells (LECs) (28). However, little is known of the mechanisms that regulate expression of VEGFR-3.
To investigate the molecular mechanisms by which Notch regulates vascular development, we determined the effects of Notch signal activation on the expression of angiogenic ligands and receptors using 3 human primary endothelial cells lines (human umbilical venous endothelial cells [HUVECs], human arterial endothelial cells [HUAECs], and human microvascular endothelial cells [HMVECs]). Notch induced VEGFR-3 transcripts and protein in all endothelial cell lines. Notch-mediated transactivation of VEGFR-3 promoter-luciferase reporters in endothelial cells required the Notch/CSL binding sites. Using chromatin immunoprecipitation (ChIP) assays, we found that the Notch/CSL complex binds and transactivates the VEGFR-3 promoter, providing evidence that the VEGFR-3 gene is a direct transcriptional target of Notch. Via induction of VEGFR-3, Notch signaling made endothelial cells more responsive to VEGF-C and promoted endothelial cell survival and morphological changes. VEGFR-3 was upregulated by Notch activation within the embryonic endothelium. Mice doubly heterozygous for Notch1 and VEGFR-3 had greater than 50% reduced viability, and E9.5 double mutant embryos displayed severe vascular defects. In adult vasculature, Notch1 and Notch4 were coexpressed with VEGFR-3 in the angiogenic vessels of the ovary and vasculature of the dermis and invasive mammary micropapillary carcinomas (IMCs). Finally, we demonstrated that Notch proteins are expressed in the lymphatic endothelium in vivo and an activated form of Notch1 is present in extratumoral LECs of IMCs.
Results
Notch upregulates VEGFR-3 in primary human endothelial cells.
To identify genes regulated by Notch in endothelial cells, we worked with 3 primary endothelial cell lines, HUVECs, HUAECs, and HMVECs. HUVECs and HUAECs are isolated from specialized large vessels, while HMVECs are derived from dermal capillaries and composed of both BECs and LECs (29).
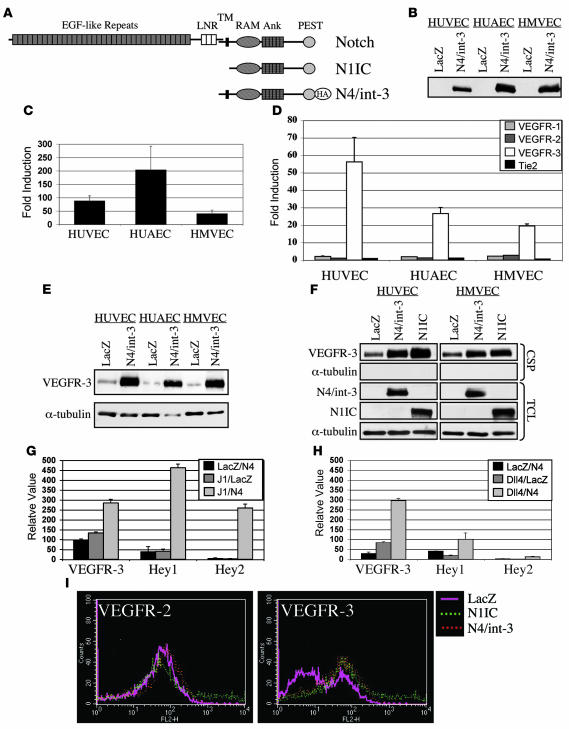
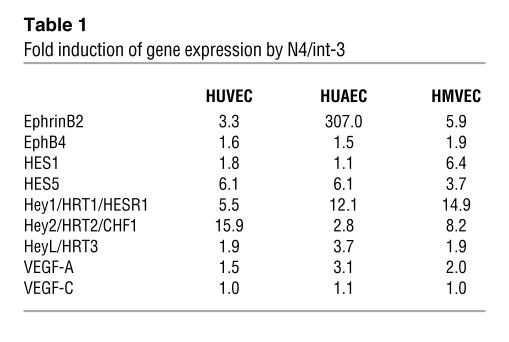
A naturally occurring active allele of Notch4 (N4/int-3) was used to activate Notch signaling within these primary endothelial cell lines. N4/int-3 encodes 30 amino acids upstream of the transmembrane domain and the entire cytoplasmic domain of Notch4 (Figure 1A). We chose to activate Notch4 signaling, as it is highly expressed in the embryonic endothelium (30). HUVECs, HUAECs, and HMVECs were infected with adenoviruses encoding either N4/int-3 (Ad–N4/int-3) or LacZ (Ad-LacZ), cells were plated in complete medium, and total RNA or protein was isolated 24 hours later. N4/int-3 protein was ectopically expressed in Ad–N4/int-3–infected HUVECs, HUAECs, and HMVECs (Figure 1B). N4/int-3 transactivated an engineered CSL reporter encoding 6 CSL binding consensus sequences upstream of the luciferase gene in all 3 endothelial lines, confirming that Notch4 signaling transactivates CSL in endothelial cells (Figure 1C). Quantitative RT-PCR (qRT-PCR) was performed to determine if Notch4 activation affected the expression of the Notch target genes of the HES and Hey families or the angiogenic factors and receptors of the ephrin/Eph, Tie, VEGF families (Figure 1D and Table 1). Although variable in the different endothelial cell lines, Notch4 signal activation induced several established direct Notch transcriptional targets, HES1, HES5, Hey1, Hey2, and HeyL (Table 1). All cell lines expressed VEGFR-1, VEGFR-2, and Tie-2, although Notch activation did not dramatically affect the levels of expression (Figure 1D). Tie-1 was not expressed in any of the endothelial cell lines and was not induced by Notch (data not shown). Strikingly, Notch induced VEGFR-3 transcripts 20- to 55-fold in venous, arterial, and capillary endothelial cells (Figure 1D). Notch did not affect the expression of either VEGF-C or VEGF-D and only moderately upregulated VEGF-A expression in HUAECs (Table 1 and data not shown).
Figure 1. Notch upregulates VEGFR-3 in cultured endothelial cells.
(A) Diagram of Notch protein structure. N1IC encodes the cytoplasmic domain of Notch1. N4/int-3 encodes 30 amino acids upstream of the transmembrane domain, the transmembrane domain, and the cytoplasmic domain of Notch4 fused to a HA tag. (B) Western blot analysis of Ad-LacZ or N4/int-3 endothelial cells using anti-HA antibody. (C) Activation of a CSL luciferase reporter in endothelial cells expressing N4/int-3, represented as fold induction in Ad–N4/int-3 cells relative to Ad-LacZ. Data are an average of 3 independent experiments. (D) qRT-PCR of Tie and VEGFRs in Ad-infected endothelial cells. Values were normalized to β-actin qRT-PCR. Data of 3 independent qRT-PCR analyses are represented as fold induction in Ad–N4/int-3 cells relative to Ad-LacZ. (E) Western blot analysis of cell surface proteins isolated from Ad-LacZ and Ad–N4/int-3 endothelial cells immunoblotted with anti–VEGFR-3 antibody and total cell lysates immunoblotted with anti–α-tubulin antibody. (F) Western blot analysis of cell surface proteins (CSPs) isolated from Ad-LacZ–, Ad–N4/int-3–, and Ad-N1IC–infected endothelial cells immunoblotted with antibodies against VEGFR-3 and α-tubulin. Total cell lysates (TCLs) immunoblotted with antibodies against the HA epitope of N4/int-3 (12CA5), N1IC, and α-tubulin. (G and H) VEGFR-3, Hey1, and Hey2 qRT-PCR of Jagged1/Notch4 (J1/N4) or Dll4/Notch4 (Dll4/N4) HUVEC cocultures, respectively. Values were normalized by β-actin qRT-PCR and are presented as relative values. (I) VEGFR-2 and VEGFR-3 FACS of Ad-LacZ, Ad-N1IC, and Ad–N4/int-3 BECs.
Table 1 .
Fold induction of gene expression by N4/int-3
We next determined if ligand-mediated Notch4 signaling also induced VEGFR-3. HUVECs were infected with Ad-Notch4, Ad-Jagged1, Ad-Dll4, or Ad-LacZ and cocultured for 48 hours, and the RNA was isolated. Cocultures containing either Jagged1- or Dll4-expressing HUVECs mixed with Notch4-expressing HUVECs resulted in the induction of VEGFR-3, Hey1, and Hey2, as determined by qRT-PCR (Figure 1, G and H, respectively). Thus ligand-mediated Notch4 signal activation also induced VEGFR-3.
We then determined if the induction of VEGFR-3 mRNA by Notch4 correlated with an increase in VEGFR-3 protein. HUVECs, HUAECs, and HMVECs were infected with either Ad-LacZ or Ad–N4/int-3, and 48 hours after infection, cells were surface biotinylated, biotinylated proteins were purified, and immunoblotting was performed (Figure 1E). In culture, all primary endothelial cell lines expressed a low level of VEGFR-3. N4/int-3 upregulated the expression of VEGFR-3 at the cell surface of all 3 endothelial cell lines.
In the mouse, both Notch1 and Notch4 function in embryonic vascular development (4). Therefore, we asked if Notch1 could also upregulate VEGFR-3. To activate Notch1 signaling, an adenovirus encoding the constitutively active human Notch1 intracellular domain (Ad-N1IC; Figure 1A) was used to infect HUVECs and HMVECs. Twenty-four hours after infection, Ad-N1IC– Ad–N4/int-3–, and Ad-LacZ–infected endothelial cells were surface biotinylated, biotinylated proteins were purified, and immunoblotting was performed (Figure 1F). Both Notch1 and Notch4 signaling upregulated VEGFR-3 to a similar level. Notch1-mediated induction of VEGFR-3 also occurred at the level of transcription as Notch1 signaling upregulated VEGFR-3 transcripts (data not shown).
VEGFR-3 has been shown to function in both angiogenesis and lymphangiogenesis (20, 25). The Notch-mediated induction of VEGFR-3 in HUVECs and HUAECs established that Notch signaling regulates VEGFR-3 expression in BECs. We next sought to determine if Notch induced VEGFR-3 in the BEC and LEC components of HMVECs. BECs purified from HMVECs (29) were infected with Ad-N1IC, Ad–N4/int-3, or Ad-LacZ. The following day, Ad-infected BECs were analyzed by FACS for VEGFR-2 and VEGFR-3 expression. The percentage of cells expressing VEGFR-2 remained unchanged between LacZ, N1IC, and N4/int-3–expressing BECs (77.31%, 78.34%, and 79.76% respectively; Figure 1I). In contrast, Notch1 and Notch4 signaling increased the percentage of VEGFR-3–expressing BECs (LacZ, 40.08%; N1IC, 76.71%; N4/int-3, 70.61%; Figure 1I). FACS analysis was also performed on LECs infected with Ad-N1IC, Ad–N4/int-3, or Ad-LacZ. Notch signal activation did not alter the percentage of LECs positive for VEGFR-3 (data not shown), consistent with the fact that the majority of LECs already express high levels of VEGFR-3 (29).
Notch/CSL complex binds and activates VEGFR-3.
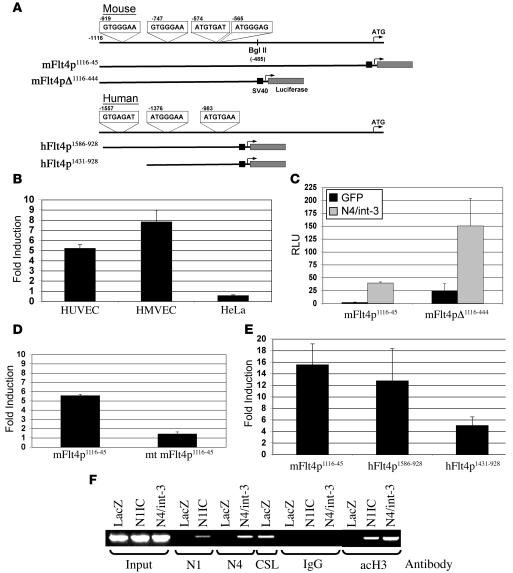
The induction of VEGFR-3 by Notch occurred within 24 hours of the adenoviral infection, suggesting that VEGFR-3 was a direct target of Notch/CSL transactivation. There are 4 putative CSL binding sites upstream of the transcriptional start site of the murine VEGFR-3 gene (Figure 2A). Previous studies have shown that murine VEGFR-3 reporter constructs that encode 3 of the 4 CSL binding sites are strongly expressed in cultured murine brain endothelial cells and the developing lymphatic and blood microvascular endothelium in vivo (28). Thus we designed 2 murine VEGFR-3 luciferase reporter constructs encoding bases –1116 to –45 (mFlt4p1116–45) or bases –1116 to –444 (mFlt4pΔ1116–444) of the VEGFR-3 upstream regulatory region (Figure 2A). The VEGFR-3 reporters were introduced by lipofection into Ad-GFP– and Ad–N4/int-3–infected HUVECs, HMVECs, or HeLa cells and luciferase activity determined 24 hours later. N4/int-3 transactivated the mFlt4p1116–45 reporter 5-fold in HUVECs and nearly 8-fold in HMVECs (Figure 2B). In HeLa cells, N4/int-3, which transactivated the CSL reporter (data not shown), did not induce the mFlt4p1116–45 reporter. In HMVEC, the mFlt4pΔ1116–444 reporter that encodes the 4 CSL binding sites retained Notch responsiveness (Figure 2C). In the control Ad-GFP–infected HMVECs, the mFlt4pΔ1116–444 reporter had higher basal activity than the mFlt4p1116–45 reporter (Figure 2C), suggesting that the sequences between –45 and –444 contain cis repressor elements. Next, we mutated the 4 CSL binding sites in the mFlt4p1116–45 reporter. Wild-type and mutant mFlt4p1116–45 reporters were introduced into Ad-LacZ– and Ad–N4/int-3–infected HMVECs. N4/Int-3 transactivated the wild-type mFlt4p1116–45 reporter 5.5-fold and the mutant mFlt4p1116–45 reporter only 1.4-fold relative to the Ad-LacZ control (Figure 2D). In the Ad-LacZ–infected HMVECs, the mutant mFlt4p1116–45 reporter had higher basal activity than the wild-type reporter, consistent with a loss of CSL transcriptional repression (data not shown).
Figure 2. Notch directly binds and activates the VEGFR-3 promoter in endothelial cells.
(A) Diagrams of murine and human VEGFR-3 5′-UTR, the murine mFlt4p1116–45 and mFlt4pΔ1116–444 luciferase reporters, and the human hFlt4p1586–928 and hFlt4p1431–928. Putative CSL binding sites –919, –747, –574, and –565 in mouse and –1557, –1376, and –983 in human are highlighted. (B) Transactivation of mFlt4p1116–45 luciferase reporter in Ad-GFP– or Ad–N4/int-3–infected HUVECs, HMVECs, and HeLa. Data are presented as fold induction of Ad–N4/int-3 relative to Ad-GFP. (C) mFlt4p1116–45 and mFlt4pΔ1116–444 luciferase reporter assays in Ad-GFP and Ad–N4/int-3 HMVECs. Data are presented as relative luciferase units (RLU). (D) Transactivation of wild-type mFlt4p1116–45 and the mutant (mt) mFlt4p1116–45 luciferase reporters in Ad-LacZ– and Ad–N4/int-3–infected HMVECs. Data are presented as fold induction in Ad-N4/int-3 relative to Ad-LacZ. (E) Transactivation of mFlt4p1116–45, hFlt4p1586–928, and hFlt4p1431–928 luciferase reporters in Ad-GFP– or Ad–N4/int-3–infected HMVECs. Data are presented as fold induction of Ad–N4/int-3 relative to Ad-GFP. (F) ChIP assay of Ad-LacZ–, Ad-N1IC–, and Ad–N4/int-3–infected HMVECs. PCR products amplified from input or N1-, N4-, CSL-, IgG-, or acetyl-H3–immunoprecipitated DNAs.
The 5′UTR of the human VEGFR-3 gene encodes 9 putative CSL binding sites within 4 kb of the initiating ATG. To evaluate the activity of a subset of the human CSL binding sites, we constructed 2 human VEGFR-3 luciferase reporters, one encoding 3 CSL binding sites hFlt4p1586–928 and the other encoding 2 CSL binding sites hFlt4p1431–928 (Figure 2A). Reporter plasmids hFlt4p1586–928, hFlt4p1431–928, and mFlt4p1116–45 were introduced into HMVECs that were infected with either Ad-LacZ or Ad–N4/int-3 and luciferase reporter assays performed the following day. The human hFlt4p1586–928 reporter was induced 13-fold by Notch4 activation, similar to the mouse mFlt4p1116–45 reporter (15-fold; Figure 2E). The hFlt4p1431–928 reporter was induced nearly 3-fold less than the hFlt4p1586–928 reporter, consistent with this reporter containing 1 less CSL binding site. Therefore, the upstream promoters of both human and mouse VEGFR-3 genes contain Notch-responsive CSL elements.
To demonstrate that the N4/int-3 transactivation occurred via a direct interaction between the Notch/CSL complex and VEGFR-3 promoter, ChIP assays were performed. After cross-linking of protein and DNA, cell lysates from Ad-LacZ–, Ad-N1IC–, or Ad–N4/int-3–infected HMVECs were subjected to immunoprecipitation with antibodies against N1IC, N4/int-3, or CSL. The 5′UTR of the human VEGFR-3 gene contains multiple repetitive elements, making it difficult to amplify by PCR (ref. 28 and data not shown). Thus we chose primers that flank 3 CSL binding sites (–3818, –3687, and –3456) outside of the repetitive elements to amplify the precipitated DNA. PCR products were specifically amplified from N1IC and N4/int-3 lysates precipitated with antibodies against N1IC or N4/int-3 (Figure 2F). A PCR product was detected from the LacZ lysate immunoprecipitated with an antibody against CSL, consistent with CSL being bound to the VEGFR-3 promoter in the absence of Notch (Figure 2F). PCR products were not amplified from lysates precipitated with nonspecific IgG. The Notch/CSL transcriptional activation complex includes histone acetylases, such as p300 and CBP (31). PCR products were detected from N1IC and N4/int-3 lysates precipitated with antibodies against acetylated histone 3 (H3; Figure 2F), suggesting that the Notch/CSL complex is functional on the VEGFR-3 promoter.
Notch promotes VEGF-C–mediated HUVEC survival.
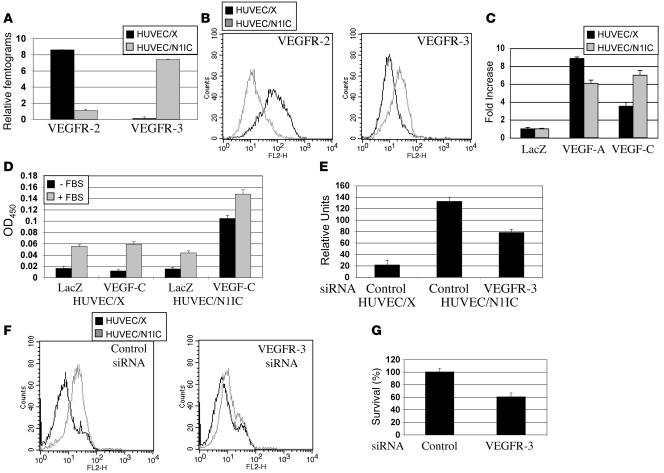
Previous studies have separately implicated Notch and VEGFR-3 signaling in the promotion of endothelial cell survival (32–35). Because Notch signaling induced VEGFR-3, we hypothesized that Notch signaling may promote cell survival in response to the VEGFR-3 ligand, VEGF-C. For these studies, populations of stable HUVEC lines were generated by retroviral transduction with either an empty retroviral vector (HUVEC/X) or a retrovirus encoding N1IC (HUVEC/N1IC). HUVEC lines were plated at confluency on a type I collagen in complete medium. After 7 days, total RNA was isolated and qRT-PCR performed to determine the expression of the VEGFRs, VEGFR-2 and VEGFR-3 (Figure 3A). Consistent with previous studies (32, 36), VEGFR-2 was downregulated by Notch, while VEGFR-3 was strongly induced in the HUVEC/N1IC relative to HUVEC/X. FACS analysis of the HUVEC lines using antibodies against VEGFR-2 and VEGFR-3 confirmed that the protein levels were also altered in response to Notch activation (Figure 3B). Thus Notch signal activation dramatically changed the expression pattern of the VEGFRs in HUVECs.
Figure 3. Notch promotes HUVEC survival in response to VEGF-C.
(A) qRT-PCR of VEGFR-2 and VEGFR-3 in HUVEC/X and HUVEC/N1IC cultured on type I collagen for 7 days. Values were normalized by β-actin expression determined by qRT-PCR, and data are represented as relative femtograms. (B) VEGFR-2 and VEGFR-3 FACS of HUVEC/X and HUVEC/N1IC. (C) WST-8 assay of HUVEC/X or HUVEC/N1IC infected with Ad-LacZ, Ad–VEGF-A165, or Ad–VEGF-C and cultured 6 days in complete medium. Data represented as percentage of viable cells relative to Ad-LacZ controls. (D) WST-8 assay of HUVEC/X or HUVEC/N1IC Ad-infected with LacZ or VEGF-C and cultured 6 days in endothelial serum-free medium with or without FBS. Data are presented as OD450, a measure of cell viability. (E) VEGFR-3 qRT-PCR and (F) FACS of HUVEC/X and HUVEC/N1IC lipofected with a control or VEGFR-3 siRNA. qRT-PCR values were normalized by β-actin expression determined by qRT-PCR, and data are represented as relative units. (G) WST-8 assay of HUVEC/N1IC lipofected with a control or VEGFR-3 siRNA. Data are presented as percentage of viable cells relative to HUVEC/N1IC containing control siRNA.
To determine whether Notch-mediated VEGFR-3 signaling promoted endothelial cell survival, HUVEC/X and HUVEC/N1IC were infected with Ad-LacZ, Ad–VEGF-A165, or Ad–VEGF-C and plated at subconfluency in complete medium containing serum and growth factors. After 6 days, HUVEC viability was ascertained by a WST-8 assay that measured dehydrogenase activity. Relative to the LacZ-expressing HUVEC/X, exogenous VEGF-A or VEGF-C promoted endothelial cell viability of HUVEC/X by 8.8- and 3.5-fold, respectively (Figure 3C). HUVEC/N1IC numbers were reduced 1.5-fold in response to VEGF-A and increased 2-fold in response to VEGF-C relative to HUVEC/X controls (Figure 3C). These experiments indicate that Notch modulates the response of HUVEC to angiogenic factors by making them more responsive to VEGF-C and less responsive to VEGF-A through its induction of VEGFR-3.
We next asked if the Notch-mediated induction of VEGFR-3 promoted serum-free survival in response to VEGF-C. Ad-LacZ– and Ad–VEGF-C–infected HUVEC/X and HUVEC/N1IC were plated at subconfluency in basal medium with or without serum and cell viability determined 3 days later. In the absence of serum, VEGF-C did not promote survival of the HUVEC/X controls (Figure 3D). In contrast, VEGF-C increased HUVEC/N1IC cell survival more than 5-fold (Figure 3D). The addition of serum increased endothelial cell survival of VEGF-C–expressing HUVEC/N1IC similar to the increase observed for LacZ- and VEGF-C–expressing HUVEC/X and LacZ-HUVEC/N1IC. Taken together, these data suggest that induction of HUVEC survival by VEGF-C is dependent on the induction of VEGFR-3 by Notch.
VEGF-C can bind and activate both VEGFR-2 and VEGFR-3, but Notch-mediated HUVEC survival is most likely to occur via VEGFR-3, as VEGFR-2 expression is downregulated by Notch (Figure 3, A and B). To confirm this hypothesis, we introduced a siRNA against VEGFR-3 into the HUVEC/N1IC line. The VEGFR-3 siRNA suppressed the Notch-mediated induction of VEGFR-3 by nearly 39% relative to a control siRNA (Figure 3, E and F). When the VEGFR-3 siRNA was introduced into the Ad–VEGF-C–infected HUVEC/NI1C, endothelial cell survival was reduced 40%, consistent with the inhibition of VEGFR-3 expression by the VEGFR-3 siRNA (Figure 3G). Thus we conclude that Notch promotes VEGF-C–mediated endothelial survival via the upregulation of VEGFR-3.
Notch promotes VEGF-C–mediated morphological changes in HUVECs.
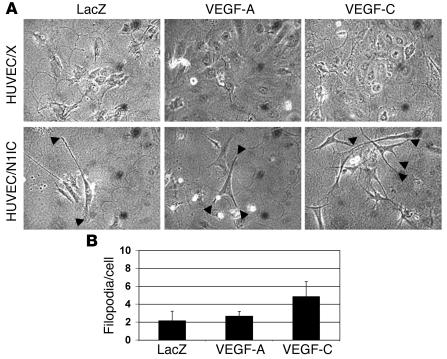
We have found that Notch signaling promotes HUVEC morphological changes as seen by the extension of processes into the underlying extracellular matrix (Figure 4; ref. 37). We asked whether Notch signaling would affect HUVEC morphological changes in response to VEGF-C. HUVEC/X and HUVEC/N1IC infected with Ad-LacZ, Ad–VEGF-A165, or Ad–VEGF-C were plated at subconfluency on a fibrin matrix in complete medium. After 6 days, none of the Ad-infected HUVEC/X extended processes into the underlying matrix (Figure 4A). VEGF-A– and VEGF-C–expressing HUVEC/X proliferated relative to the Ad-LacZ endothelial control consistent with the known stimulatory function of these angiogenic factors (data not shown). All adenovirally infected HUVEC/N1IC lines underwent morphological changes, including elongation of the cells and extension of processes into the underlying fibrin matrix (Figure 4B). However, the number of extensions differed between cell types. Ad-LacZ–infected HUVEC/N1IC developed an average of 2.14 ± 1.07 processes per cell, as compared with Ad–VEGF-A (2.67 ± 0.52 per cell) and Ad–VEGF-C (4.83 ± 1.69 per cell) (Figure 4B). Thus VEGF-C was a more potent inducer of endothelial cell morphological changes than VEGF-A when Notch signaling was activated in HUVECs.
Figure 4. Notch promotes HUVEC morphological changes in response to VEGF-C.
(A) 3D assay of HUVEC/X or HUVEC/N1IC Ad-infected with LacZ, VEGF-A165, or VEGF-C and cultured in complete medium on a fibrin gel. The development of processes extending into the underlying fibrin matrix of Ad-infected HUVEC/X and HUVEC/N1IC after 6 days is shown. Original magnification, ×10. (B) Average number of processes per cell for Ad-infected HUVEC/N1IC.
Notch upregulates VEGFR-3 in murine embryonic vasculature.
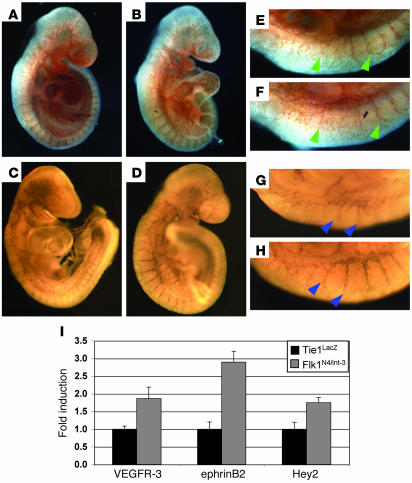
Having established that Notch induced VEGFR-3 in vitro, we examined whether Notch also upregulated VEGFR-3 in the vasculature of murine embryos. We used transgenic embryos in which Notch4 had been constitutively activated specifically within the vascular endothelium under control of the VEGFR-2 promoter (Tie1LacZ;Flk1N4/int-3). These transgenic embryos die at E10.5 due to hemorrhaging and vascular defects (8). Day 9.5 Tie1LacZ;Flk1N4/int-3 and phenotypically normal Tie1LacZ embryos were isolated and whole-mount immunostaining performed with antibodies against VEGFR-3 or the endothelial cell marker PECAM. We focused our analysis on less severely affected double transgenic embryos (Tie1LacZ;Flk1N4/int-3) so as to minimize the difference of endothelial cell numbers between the 2 groups. As compared with Tie1LacZ controls, Tie1LacZ;Flk1N4/int-3 embryos have less PECAM, as well as less VEGFR-3, staining in the head and brachial arches, commensurating with the loss of capillary vessels that results from defects in vascular remodeling (Figure 5, A and B). Consistent with this observation, qRT-PCR of RNA isolated from pooled embryos revealed a 2-fold decrease in VE-cadherin expression in the Notch4 endothelial gain-of-function embryos (data not shown).
Figure 5. VEGFR-3 is upregulated in E9.
embryos in which Notch signaling has been activated within the endothelium. (A–H) Control Tie1LacZ (A, C, E, and G) and gain-of-function Tie1LacZ;Flk1N4/int-3 (B, D, F, and H) embryos were immunostained with antibodies against PECAM (A, B, E, and F) or VEGFR-3 (C, D, G, and H). (E–H) The dorsal region consisting of the intersomitic vessels was enlarged to demonstrate the similar morphology of the intersomitic vessels (PECAM; green arrowheads) and the induction of VEGFR-3 (blue arrowheads) in the Tie1LacZ;Flk1N4/int-3 embryos relative to the controls. (I) VEGFR-3, ephrinB2, and Hey2 qRT-PCR of pooled Tie1LacZ or Tie1LacZ;Flk1N4/int-3 E9.5 embryos. Values were normalized by β-actin and VE-cadherin expression and are presented as fold expression Tie1LacZ;Flk1N4/int-3 relative to Tie1LacZ. Original magnification, ×5 (A–D); ×7 (E–H).
In contrast to the caudal portions of the embryos, the PECAM staining of intersomitic vessels appeared to be similar between the 2 embryonic phenotypes (Figure 5, C and D), whereas VEGFR-3 was more strongly expressed in the intersomitic vessels of the Tie1LacZ;Flk1N4/int-3 embryos as compared with the Tie1LacZ controls (Figure 5, G and H). In zebrafish, the sprouting intersomitic vessels are one of the sites of VEGFR-3 expression outside of the venous endothelium (38). qRT-PCR analysis of RNA isolated from pooled embryos confirmed a nearly 2-fold increase in VEGFR-3 transcripts in the Tie1LacZ;Flk1N4/int-3embryos relative to the Tie1LacZ embryos (Figure 5I). This was similar to the induction of the known endothelial Notch target genes, EphrinB2 and Hey2 (Figure 5I).
Notch1 and VEGFR-3 genetically interact to regulate embryonic vascular development.
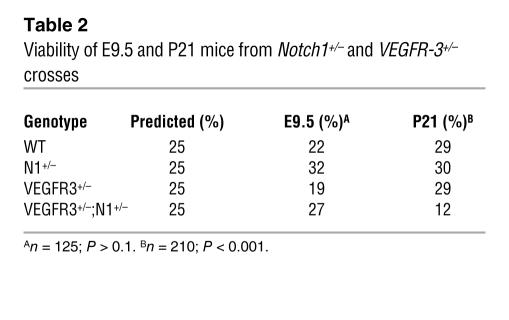
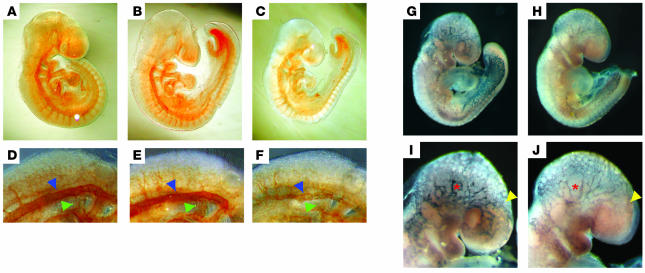
To determine if Notch and VEGFR-3 genes interact to regulate vascular development, we mated Notch1+/– (4) and VEGFR-3+/– (25) mice and analyzed the genotypes of the offspring. As expected, Notch1+/– or VEGFR3+/– mice were viable. We observed a greater than 50% reduction in the viability of Notch1+/–;VEGFR-3+/– doubly heterozygous mice (Table 2). As embryos nullizygous for either Notch1 or VEGFR-3 display severe vascular abnormalities at E9.5 (4, 25), embryos from Notch1+/– and VEGFR-3+/– crosses were isolated at this stage and whole-mount immunohistochemistry for PECAM performed (Figure 6, A–F). Vascular development of Notch1+/– and VEGFR3+/– embryos was similar to wild-type littermates. The majority Notch1+/–;VEGFR-3+/– embryos were smaller in size than their littermates but were not developmentally delayed, as they contain the same number of somites (Figure 6, A–C, and data not shown). In more than half of these double mutant embryos, vascular development was severely disrupted. In the affected embryos, the overall PECAM staining was reduced compared with Notch1+/– and VEGFR-3+/– embryos (Figure 6, A–C). These Notch1+/–;VEGFR-3+/– embryos had coarctation and disorganization of the dorsal aorta (Figure 6F). The intersomitic vessels of the double heterozygous embryos appeared structural, similar to their heterozygous littermates, but stained weaker for PECAM (Figure 6, D–F). We also observed a reduction in the PECAM staining in the left and right atria of the primary heart tube of the double mutant embryos, indicative of reduced endothelium (Figure 6F), though their hearts were beating.
Table 2 .
Viability of E9.5 and P21 mice from Notch1+/– and VEGFR-3+/– crosses
Figure 6. Notch1 and VEGFR-3 genetically interact to regulate vascular development.
Notch1+/– (A and D), VEGFR-3+/– (B and E), and Notch1+/–;VEGFR-3+/– (C and F) E9.5 embryos were immunostained for PECAM. (D–F) The dorsal region of embryos was enlarged to visualize the dorsal aorta (blue arrowheads), atria of the primary heart tube (green arrowheads), and intersomitic vessels. E9.5 VEGFR-3+/– (G and I) and Notch1+/–;VEGFR-3+/– (H and J) embryos were β-gal stained to visualize the VEGFR-3–positive vasculature. (I and J) The cranial region of the embryos was enlarged to demonstrate the decrease in VEGFR-3 expression (red asterisks) and reduction of VEGFR-3–positive vessel density (yellow arrowheads). Original magnification, ×5 (A–C, G, and H); ×15 (D–F); ×10 (I and J).
As the VEGFR-3+/– gene is disrupted by the introduction of LacZ (25), we also performed whole-mount β-galactosidase staining on E9.5 embryos (Figure 6, G–J). Strong vascular β-galactosidase staining was observed in the cranial and posterior blood vasculature, and weaker staining was observed in the area of the dorsal aorta and intersomitic vessels of VEGFR-3+/– embryos (Figure 6, G and I). In the Notch1+/–;VEGFR-3+/– embryos, there were fewer vessels that stained for β-galactosidase (Figure 6H). Moreover, vessels that stained positive for β-galactosidase also showed a reduction in the β-galactosidase expression consistent with Notch1 promoting vascular expression of VEGFR-3. These effects were most evident in the cranial vasculature (Figure 6, I and J). There was reduced density of β-galactosidase–positive vessels in the anterior portion and almost complete loss of these vessels in the ventral region of the head (Figure 6J). Taken together, these results indicate that the reduced VEGFR-3 expression of the wild-type allele in Notch1+/–;VEGFR-3+/– embryos compromises vascular development, resulting in embryonic lethality.
Notch and VEGFR-3 are coexpressed in the vasculature of the murine ovary.
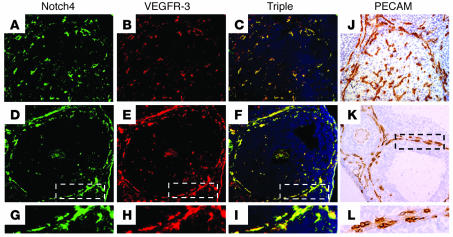
In the adult, the ovary is an organ of robust physiological angiogenesis, required for development of follicles and corpus luteum formation. During folliculogenesis, a dramatic increase in vessel density occurs in the theca layer surrounding the granulosa cells and oocyte that later regresses after ovulation (Figure 7, K and L; refs. 39–41). After oocyte release, the follicle granulosa cells proliferate rapidly to form the corpus luteum. As the corpus luteum matures, a vascular network of blood vessels derived from the thecal layer grow into the corpus luteum via the process of angiogenesis (Figure 7J; refs. 42, 43). Since the ovary is an active site of adult physiological angiogenesis, we determined the expression patterns for Notch4 and VEGFR-3 in the vasculature of the theca layer and corpus luteum. Notch4 and VEGFR-3 had an overlapping expression pattern in the angiogenic vessels of the corpus luteum (Figure 7, A–C) and in mature and angiogenic vessels of the theca layer (Figure 7, D–I). Notch1 was also expressed in the BECs of the ovary (data not shown; ref. 44). Thus Notch1 and Notch4 and VEGFR-3 are expressed in the vasculature of the ovary, suggesting a role for Notch/VEGFR-3 signaling in follicular angiogenesis.
Figure 7. Notch4 and VEGFR-3 are coexpressed in the vasculature of ovarian follicles.
Murine ovary sections immunostained with antibodies against PECAM (J–L), Notch4 (A, D, and G), and VEGFR-3 (B, E, and H). (C, F, and I) Triple overlay of Notch4 and VEGFR-3 and DAPI. (A–C and J) Angiogenic blood vessels of the corpus luteum. (D–F and K) Endothelial cells in the theca layer of the ovarian follicles. (A–F, J, and K) Original magnification, ×25. (G–I and L) Magnified (×50) view of theca layer vasculature.
Notch, VEGFR-3, and LYVE-1 are coexpressed in the vasculature of the murine skin.
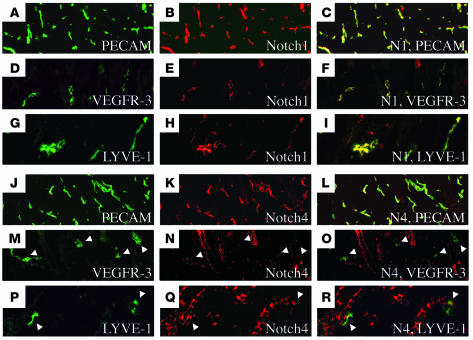
After the formation of the embryonic blood vasculature, VEGFR-3 expression becomes mostly restricted to the lymphatic endothelium and regulates lymphangiogenesis (20, 29, 45, 46). Thus we explored whether Notch1 or Notch4 were coexpressed with VEGFR-3 in the lymphatic vasculature. We evaluated the expression of Notch1 and Notch4 in the vasculature of the dorsal skin isolated from P4 mice, as the dermal vasculature consists of both blood and lymphatic vessels (29). Five-micron cross-sections of skin were costained with antibodies against Notch1 or Notch4, and VEGFR-3 or PECAM (Figure 8). Notch1 and Notch4 share an overlapping pattern of expression with the BEC and LEC marker PECAM (Figure 8, A–C and J–L). We also observed expression of Notch4 with 2 different antibodies in the hair follicles (Figure 8, K, N, and Q, and data not shown). As we observed in the ovary, Notch1 and Notch4 were coexpressed with VEGFR-3 in the dermal vasculature (Figure 8, D–F and M–O). As VEGFR-3 has been reported to be restricted to the lymphatic vessels of the skin, we also evaluated the expression of Notch1 and Notch4 and the LEC specific marker, LYVE-1. Both Notch1 and Notch4 were coexpressed with LYVE-1 in the dermal lymphatic vessels (Figure 8, G–I and P–R). Thus we report for the first time that Notch1 and Notch4 are expressed in the lymphatic vessels of the neonatal dermis.
Figure 8. Notch1 and Notch4 are coexpressed with VEGFR-3 and LYVE-1 in the dermal vasculature.
Murine P4 dermal sections immunostained with PECAM (A and J), VEGFR-3 (D and M), LYVE-1 (G and P), Notch1 (B, E, and H), or Notch4 (K, N, and Q) antibodies. Notch1 is coexpressed with PECAM (C), VEGFR-3 (F), and LYVE-1 (I). Notch4 is coexpressed with PECAM (L), VEGFR-3 (O), and LYVE-1 (R). (M–R) White arrowheads highlight Notch4 staining in the dermal lymphatic endothelium. Original magnification, ×12.5 (A–C and J–L); ×40 (D–I and M–R).
Notch, VEGFR-3, and LYVE-1 are coexpressed in human invasive micropapillary breast carcinomas.
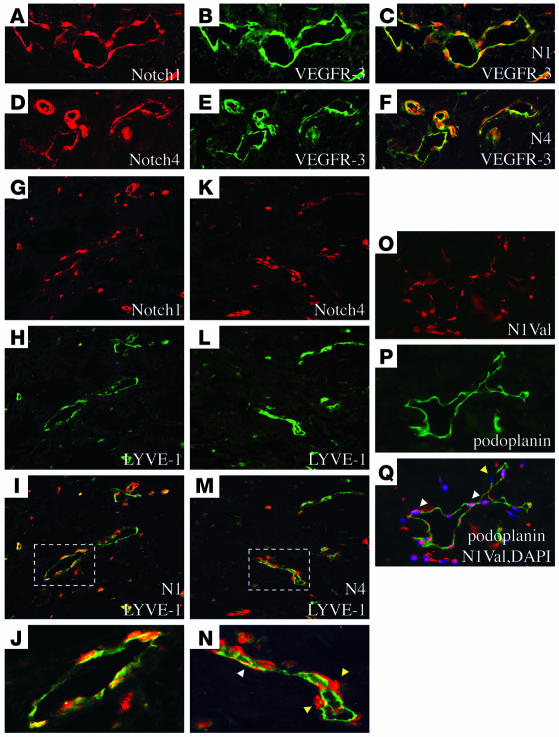
Previous studies have shown that both the Notch ligand, Dll4, and VEGFR-3 are strongly expressed in the vasculature of breast ductal carcinomas, suggesting a role for Notch and VEGFR-3 in tumor angiogenesis and lymphangiogenesis (26, 47). Thus we next analyzed the expression of Notch1 and Notch4 in human invasive micropapillary breast carcinomas (IMCs). IMCs are aggressive tumors that have a high propensity for high lymphatic vessel density, lymphovascular invasion, and lymph node metastasis (48). Five-micron paraffin sections were costained with antibodies against Notch1 or Notch4, and VEGFR-3 or LYVE-1 (Figure 9). Notch1 and Notch4 shared an overlapping expression pattern with VEGFR-3 in both the blood and lymphatic vasculature of IMC (Figure 9, A–F). Costaining with antibodies against Notch1 or Notch4 and LYVE-1 revealed that both Notch proteins are expressed in the extratumoral LECs (Figure 9, G–N). Notch4 expression was also observed in cells adjacent to the lymphatic vessels (Figure 9N). As expression of the Notch receptors does not necessarily mean that the Notch is actively signaling, we also costained with the lymphatic endothelial marker podoplanin and an antibody that recognizes the active Notch1 peptide that has been processed by γ-secretase/presenilin (N1Val; Figure 9, O–Q). Expression of the activated Notch1 peptide was observed in the majority of the lymphatic endothelial nuclei, indicating that Notch1 is expressed and actively signaling in the lymphatic vessels of IMCs (Figure 9Q).
Figure 9. Notch1 and Notch4 are coexpressed with VEGFR-3 and LYVE-1 in IMC.
Human IMC immunostained with VEGFR-3 (B and E), LYVE-1 (H and L), podoplanin (P), Notch1 (A and G), Notch4 (D and K), or activated Notch1 (N1Val) (O) antibodies. Notch1 (C) and Notch4 (F) are coexpressed with VEGFR-3 in the extratumoral IMC vessels. Notch1 (I) and Notch4 (M) are coexpressed with LYVE-1 in the extratumoral lymphatics. J and N represent magnified view of boxed areas in I and M, showing lymphatic vessels. (N) White arrowhead indicates Notch4-positive LEC. Yellow arrowheads highlight Notch4-positive non-endothelial cells adjacent to the lymphatic endothelium. (Q) The activated Notch1 peptide is localized to the LEC nuclei. White arrowheads highlight positive nuclei, and yellow arrowhead indicates a negative nucleus. Original magnification, ×50 (A–F and O–Q); ×40 (G–I and K–M); ×120 (J and N).
Discussion
Mouse genetic studies have demonstrated that Notch signaling is required for proper patterning of the embryonic vasculature (49). Notch in complex with the CSL transcription factor is thought to regulate cell fate decisions by inducing tissue-specific genes that modulate the ability of cells to respond to extracellular cues. Consistent with this hypothesis, Notch has been proposed to regulate angiogenesis via the induction of ephrinB2 (15, 16) and the suppression of VEGFR-2 (32, 36, 50). Both of these Notch target genes likely lie downstream of the HES/Hey proteins. Here we describe VEGFR-3 as a direct target of Notch/CSL transactivation in endothelial cells, and what we believe to be a novel mechanism of angiogenic regulation by Notch. Like Notch, VEGFR-3 is essential for embryonic blood vessel remodeling (3–7, 25). We found that Notch1 and VEGFR-3 genetically interact to regulate embryonic vascular development. In addition, Notch signaling through the direct upregulation of VEGFR-3 promotes BEC survival and morphological changes in response to VEGF-C. Our studies further suggest that both Notch1 and Notch4 may function in the lymphatic vessels as well as blood vessels.
Notch in complex with CSL binds to and transactivates VEGFR-3.
It has been suggested that the endothelial expression of VEGFR-3 is regulated at both the transcript and protein levels (28, 29), yet little is known about these regulatory mechanisms. The portion of VEGFR-3 promoter that specifically drives expression of VEGFR-3 in BECs and LECs has been previously described (28). We identified 3 CSL consensus sequences (–747, –574, and –565) in this same VEGFR-3 regulatory region (Figure 2A). The mFlt4p1116–45 reporter encoding these same 3 CSL consensus sites was transactivated by Notch in endothelial cells but not in non-endothelial cells. Notch and additional endothelial cell–specific transcription factors may interact to regulate expression of VEGFR-3 in endothelial cells. Further confirming that the Notch/CSL complex directly activates VEGFR-3, Histone3 was acetylated upstream of VEGFR-3 upon Notch activation. In transgenic embryos, Notch4 signaling induced VEGFR-3 within the intersomitic endothelium but not within other endothelial cell types, suggesting that other angiogenic factors cooperate with Notch to transactivate VEGFR-3. Thus we present a mechanism for driving expression of VEGFR-3 in BECs. These studies further indicate that VEGFR-3 signaling may be elemental to the function of Notch in the development of vascular systems.
Notch1 and VEGFR-3 cooperate to regulate vascular development.
Disruption of either Notch1 or VEGFR-3 signaling in mice results in embryonic lethality prior to E10.5 (4, 25). Both knockout embryos display collapsed dorsal aorta and cardinal veins and disrupted cranial and yolk sac vascular remodeling. We observed that disruption of one allele of Notch1 and VEGFR-3 phenocopied the dorsal aorta and cranial vascular defects present in either single nullizygous embryo. At this embryonic time point, Notch1 and VEGFR-3 have been described to have an overlapping expression pattern in the aorta and cranial vasculature (25, 45, 51, 52). In addition, these double heterozygous mutant embryos were growth retarded, similar to what has been described for VEGFR-3–/– embryos (25). As Notch directly transactivates VEGFR-3, we thus propose that Notch1 and VEGFR-3 signaling cooperate to regulate embryonic aortic and cranial vascular development. In the adult vasculature, we show that Notch1 and Notch4 are coexpressed with VEGFR-3 in the angiogenic vessels of the murine corpus luteum and theca layer of ovary. Therefore, we speculate that the Notch-mediated induction of VEGFR-3 has a role in regulating the vascular development of a subset of blood vessels in the embryo and may also have a function in adult physiological angiogenesis. Alternatively, Notch1 and VEGFR-3 may function in parallel pathways to effect vascular development.
Notch via its induction of VEGFR-3 mediates VEGF-C induced endothelial cell survival and morphological changes.
The concept that Notch and VEGFR-3 signaling may cooperate to regulate endothelial cell fate determination is supported by our observations that mice doubly heterozygous for null alleles of VEGFR-3 and Notch1 display embryonic vascular remodeling defects and that Notch activation within the endothelium results in VEGFR-3 misexpression. Consistent with this hypothesis, we find that Notch, via induction of VEGFR-3, promoted endothelial cell survival and morphological changes in response to VEGF-C. In endothelial cells, activation of Notch has been shown to inhibit apoptosis and promote survival of endothelial cells (53, 54). We propose that Notch can regulate VEGF-C–mediated endothelial cell survival by inducing VEGFR-3, as blocking the expression of VEGFR-3 after Notch activation suppressed endothelial cell survival in response to VEGF-C. VEGFR-3 signaling has also been shown to promote differentiation of BEC in culture (55). We found that VEGF-C was a potent inducer of endothelial cell morphological changes when Notch signaling was activated. Therefore, Notch may also function to promote VEGF-C–induced BEC morphological changes via its induction of VEGFR-3. However, it remains to be determined if Notch alone or through its regulation of VEGFR-3 expression also promotes LEC survival.
Notch modulates the ability of endothelial cells to respond to their extracellular environment.
Notch signaling is thought to regulate multipotent cell fate decisions by ensuring that the proper number of cells responds to the milieu of extracellular factors. Angiogenesis requires endothelial cells to respond to a variety of external stimuli such as those provided by bFGF, VEGF, ephrins, and angiopoietins (56, 57). Thus we propose that Notch achieves this by altering the expression of receptors at the cell surface of endothelial cells. We show that Notch signaling suppresses VEGFR-2 expression, while dramatically increasing VEGFR-3 expression in primary endothelial cells. Consistent with this reciprocal change in expression, the survival of HUVECs in which Notch signaling was activated was reduced in response to VEGF-A and increased in response to VEGF-C. Thus Notch signaling may regulate endothelial cell fate decisions by decreasing VEGFR-2– and increasing VEGFR-3–based signaling. A similar role for Notch signaling in regulating expression of VEGFRs has been proposed during postnatal retinal angiogenesis. In the retina of Dll4+/– mice, the disruption of Notch signaling correlated with an increase in vascular density, an increase in VEGFR-2, and a reduction in VEGFR-1 endothelial expression (50).
VEGFR-2 and VEGFR-3 are able to form a heterodimeric complex (58), and this VEGFR complex has been suggested to maintain vascular integrity and promote angiogenesis (59). In fact, VEGFR-3 cooperates with VEGFR-2 during morphogenesis of the intersomitic arteries in zebrafish (38), indicating a wider role for VEGFR-3 in the vasculature than previously appreciated. Loss of the zebrafish Hey2 homolog, gridlock, inhibits the sprouting of the intersomitic arteries, similar to the effect observed when VEGFR-2 and VEGFR-3 signaling is disrupted (13), suggesting a role for Notch signaling in regulating VEGFR-2 and VEGFR-3 function. We demonstrate that Notch activation within the murine embryonic endothelium promoted VEGFR-3 expression within the developing intersomitic vessels. Thus we propose that Notch may differentially regulate VEGFR-2 and VEGFR-3 expression to guide the formation of specific vessels such as the intersomitic vasculature.
Notch in physiological and pathological lymphangiogenesis.
VEGFR-3 expression has been reported to be restricted to the lymphatic endothelium in the dermis (29). Within the dermis, we found that Notch1 and Notch4 are coexpressed with VEGFR-3 and the LEC marker LYVE-1, demonstrating that Notch1 and Notch4 are expressed in the lymphatic endothelium. The coexpression of Notch1 and Notch4 within the lymphatic endothelium may indicate that these receptors compensate for each other and may explain why a lymphatic phenotype has not yet been observed in the viable Notch4 knockout mice (4).
Similar to findings in murine skin, Notch1 and Notch4 were coexpressed with VEGFR-3 in the extratumoral blood and lymphatic vessels of IMCs. The cleaved and activated Notch1 peptide was observed in a subset of the lymphatic endothelial nuclei, indicating that Notch1 is not only expressed but is activated in tumor lymphatic vessels. In IMCs of breast, VEGF-C, via VEGFR-3 signal activation, has been proposed to promote LEC proliferation, tumor lymphatic invasion, and tumor metastasis (60). Thus our findings implicate Notch1 signaling in tumor lymphangiogenesis. Could Notch signaling participate in pathological tumor lymphangiogenesis and contribute to metastatic disease in the breast? If so, therapeutic Notch antagonists may block breast tumor metastasis by disrupting VEGFR-3 signaling, in turn perturbing tumor lymphangiogenesis and angiogenesis.
Methods
Constructs and cell culture.
N4/int-3 (Genbank accession no. M80456; ref. 30), GFP, and LacZ cDNAs were engineered in the vector pAd-Lox. The N4/int-3 construct encodes 1 HA epitope at the 3′ end. The Ad-N1IC encodes the cytoplasmic domain of human Notch1, generously provided by Paulo Dotto (University of Lausanne, Lausanne, Switzerland) (61). Human VEGF-A165 and VEGF-C cDNAs were generated by RT-PCR and engineered in pAdLox. Adenovirus was generated and tittered as previously described (62). For retroviral infection, LacZ or the entire murine cytoplasmic domain of Notch1 was engineered into pHyTc.
HUVECs, HUAECs, and neonatal dermal HMVECs were purchased from Cambrex. GP2-293 cells (BD Biosciences) and HeLa were maintained in DMEM, 10% FBS. BECs were isolated from HMVECs (29).
Adenoviral infections.
Early passage HUVECs, HUAECs, and HMVECs were trypsinized. 1 × 106 to 3 × 106 cells in 2 ml EGM-2 Bulletkit medium (Cambrex) were transferred to a 10-cm2 bacterial plate coated with 5% BSA and infected with adenoviruses at 25 pfu/cell for 1 hour at 37°C. Then cells were plated in growth medium for RNA or protein isolation or for reporter assays. For coculture assays, HUVECs were Ad-infected in suspension and mixed in a 1:3 ligand/receptor ratio of infected cells. Ad-LacZ– or Ad-GFP–infected HUVECs were used as negative controls.
Retroviral infections.
GP2-293 cells were CaPO4 transfected with pVSV-G vector, and either pHyTC LacZ or pHyTC N1IC and supernatants were collected and used to infect early passage HUVECs, as previously described (37).
Luciferase reporter assays.
The pGA981-6 (63) luciferase reporter was used to measure CSL activity. For the mFlt4p1116–45 luciferase reporter, a DNA fragment encoding –1116 to –45 upstream of the initiating ATG was PCR amplified and subcloned into pGL3-Promoter vector (Promega), and mFlt4pΔ1116–444 was generated by removing a BglII fragment. Mutant mFlt4p1116–45 was generated by mutating the CSL sites, RTGDGAD to RTGCTGC (64), using QuickChange Site-Directed Mutagenesis Kit (Stratagene). The hFlt4p1586–928 and hFlt4p1431–928 DNA fragments were generated by PCR, and amplicons were cloned into the pGL3-Promoter vector.
Ad-infected cells were seeded at 1.5 × 105 cells per well in 12-well plates. The following day 750–775 ng of reporter plasmid and either 125 ng pHyTC LacZ or 25 ng phRL-SV40 renilla (Promega) were introduced using Lipofectamine2000 (Invitrogen) in triplicate. Lysates were harvested 24 hours later. For CSL reporter assays, luciferase activity was determined (Enhanced Luciferase Assay Kit; BD Biosciences — Pharmingen) and normalized by β-galactosidase activity (Galacto-light Plus; Tropix). For VEGFR-3 reporter assays, luciferase and renilla activity was determined with Dual-Luciferase Reporter Assay System (Promega). All values were normalized by renilla values.
ChIP assays.
Ad-infected HMVECs (3 × 106) were seeded in 10-cm2 plates. The next day, ChIP assays were performed using the ChIP assay kit (Upstate). Lysates were immunoprecipitated with bTan20 (N1; Developmental Studies Hybridoma Bank), 12CA5, RBPJκ (D-20; Santa Cruz Biotechnology Inc.) or acetyl-H3 (Upstate) antibodies. Precipitated DNA was amplified by PCR using human VEGFR-3–specific primers (GCTCCATCGCAAACAAAAAATAATAG and GGTGCTGGCTGTTGTCCAGAAG).
3D in vitro bioassays.
Retrovirally infected HUVECs (1 × 104) were seeded in 24-well plates. For qRT-PCR, cells were plated on porcine type I collagen (Nitta gelatin). After 7 days in complete media, cells were collagenased and trypsinized and RNA isolated (65). For proliferation/survival and differentiation assay, Ad-infected HUVECs were seeded on fibrin (Sigma-Aldrich) in complete growth medium. After 5–6 days, cell numbers/viability were determined relative to a calibration curve using Cell Counting Kit-8 (CCK-8; Dojindo). HUVEC morphological change was scored as the number of cellular extensions per cell. For assays utilizing siRNAs, Ad-infected HUVECs grown on collagen were lipofected with either negative control siRNA (Ambion) or VEGFR-3 siRNA AM461 (Ambion) with Effectene (Qiagen), and the following day 2 × 104 HUVECs were seeded on fibrin. After 3 days, cell viability was determined.
qRT-PCR.
RNA was isolated 24 hours after adenoviral infections (65). qRT-PCR was performed as described in ref. 16. PCR primers were designed to amplify transcripts for β-actin, Tie1, Tie2, VEGFR-1, VEGFR-2, VEGFR-3, VEGF-A, VEGF-C, VEGF-D, Prox1, and LYVE-1. Human PCR amplicons for each gene in pDrive (Qiagen) were used as reference standards. A standard curve for each gene was generated from serially diluted standards, and values for unknown samples were extrapolated. β-Actin qRT-PCR was used to normalize samples.
Biotinylation and western blot analysis.
Twenty-four hours after adenovirus infection, HUVECs, HUAECs, and HMVECs were incubated with 0.5 mg/ml EZ Link Sulfo-NHS-Biotin (Pierce) at 4°C for 40 minutes, then incubated in DMEM at 4κC for 10 minutes. Cells were lysed in TENT (50 mM Tris pH 8.0, 2 mM EDTA, 150 mM NaCl, 1% Triton X-100). Equal amounts of protein as determined by Bradford were incubated with ImmunoPure Immobilized Streptavidin beads (Pierce Biotechnology), and bound proteins were eluted in protein sample buffer. Streptavidin-purified and total cell lysates were separated on SDS-PAGE and transferred to nitrocellulose (MSI). Membranes were probed with antibodies against HA tag (12CA5), Notch1 (C-20; Santa Cruz Biotechnology Inc.), α-tubulin (Sigma-Aldrich), or VEGFR-3 (C-20; Santa Cruz Biotechnology Inc.). Proteins were visualized by ECL (Amersham) in conjunction with complimentary HRP-conjugated antibodies (Bio-Rad).
FACS.
Ad-infected BECs were plated on type I collagen in triplicate. Twenty-four hours later, cells were detached by scraping, blocked in PBS/5% FBS, and incubated with antibodies against VEGFR-2 (1121; ImClone) or VEGFR-3 (3C5; ImClone). Primary antibodies were detected with RPE anti-human fluorescent antibody (Jackson ImmunoResearch Laboratories) and separated by FACS (FACSCaliber; Becton Dickinson). Total RNA and TENT lysates were also extracted from replica plates for RT-PCR and western blot analyses. For retroviral HUVEC lines, HUVECs were grown on type I collagen and VEGFR-2 and VEGFR-3 detected with the Monoclonal Anti-Human VEGF Receptor Phycoerythrin Sampler (R&D Systems) following the manufacturer’s protocol.
Immunohistochemistry.
All studies using human breast tissue were approved by Columbia University Institutional Review Board. All studies using mice were approved by Columbia University Institutional Animal Care and Use Committee (IACUC). Human IMCs and murine ovary and dorsal skin sections (5 μm) immunostained with antibodies against PECAM (Pharmingen), Notch4 (J. Kitajewski laboratory or Santa Cruz Biotechnology Inc.), VEGFR-3 (eBioscience or R&D Systems), LYVE-1 (R&D Systems), Podoplanin (Angiobio), or Cleaved Notch1 (Val1744) Antibody (Cell Signaling Tech). Primary antibodies were detected with biotinylated antibodies (Vector Laboratories) and visualized with either Vectastain Standard ABC Elite kit (Vector Laboratories) or streptavidin-conjugated Alexa Fluor 488 or Alexa Fluor 594 (Invitrogen). Images were photographed with Nikon ECLIPSE E800 microscope and Nikon DXM 1200 digital camera. Double- and triple-overlay images were made using Photoshop.
Tie1LacZ and Tie1LacZ;Flk1N4/int-3 ES cell embryos were generated (8). Notch1+/– (4) and VEGFR-3+/– (25) mice were mated to generate double mutant E9.5 embryos as well as control littermates. Embryos were fixed in 4% paraformaldehyde, bleached, and stored in 100% methanol. Embryos were rehydrated and immunostained with antibodies against PECAM and VEGFR-3 (eBioscience). Primary antibodies were detected with anti-rat HRP antibody (Sigma-Aldrich) and Vectastain Standard ABC Elite kit. Staining for β-galactosidase activity was done using standard methods (66). Images were photographed with Nikon SMZ1000 microscope and Spot Insight QE digital camera.
Statistics.
CSL-reporter, Flt4-reporter, survival, and qRT-PCR data are expressed as mean standard deviation. P values were determined by 1-tailed Student’s t test. Statistical significance was defined as P value less than 0.05.
Acknowledgments
We thank Anshula Sharma, Carl Nelson, Gina Finan, Sam Pauli, and Ralf Zimmerman for technical assistance. We thank Kari Alitalo for providing the VEGFR-3+/– mice and Darrell Yamashiro, Jessica Kandel, and Brian Zeiler for critical reading of the manuscript. The bTan20 antibody developed by Spyros Artavanis-Tsakonas was obtained from the Developmental Hybridoma Bank developed under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa. This work was supported by grants from the NIH (RO1 HL62454), the American Heart Association (AHA-0150435N), and the Department of Defense (W81XWH-04-1-0540, to J. Kitajewski). C. Shawber was supported by NIH training grant 2T32CA-09503-16 and Department of Defense fellowship DAMD17-031-0218. S.A. Stowell and M. Vorontchikhina were supported by NIH training grant 2T32CA-09503-16. S. Podgrabinska and M. Skobe were supported by Department of Defense grant BC044819.
Footnotes
Nonstandard abbreviations used: Ad, adenovirus; BEC, blood endothelial cell; ChIP, chromatin immunoprecipitation; CSL, CBF1/suppressor of hairless/Lag-2; Dll, Delta-like; HES, hairy enhancer of split; Hey, HES-related protein; HMVEC, human microvascular endothelial cell; HUAEC, human arterial endothelial cell; IMC, invasive mammary micropapillary carcinoma; LEC, lymphatic endothelial cell; N1IC, Notch1 intracellular domain; qRT-PCR, quantitative RT-PCR.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3369–3382 (2007). doi:10.1172/JCI24311
References
- 1.Weinmaster G. The ins and outs of notch signaling. Mol. Cell. Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Herreman A., et al. Presenilin 2 deficiency causes a mild pulmonary phenotype and no changes in amyloid precursor protein processing but enhances the embryonic lethal phenotype of presenilin 1 deficiency. Proc. Natl. Acad. Sci. U. S. A. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krebs L.T., et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 5.Tetzlaff M.T., et al. Defective cardiovascular development and elevated cyclin E and Notch proteins in mice lacking the Fbw-7 F-box protein. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3338–3345. doi: 10.1073/pnas.0307875101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsunematsu R., et al. Mouse Fbw7/Sel-10/Cdc4 is required for notch degradation during vascular development. J. Biol. Chem. 2003;279:9417–9423. doi: 10.1074/jbc.M312337200. [DOI] [PubMed] [Google Scholar]
- 7.Xue Y., et al. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 8.Uyttendaele H., Ho J., Rossant J., Kitajewski J. Vascular patterning defects associated with expression of activated Notch4 in embryonic endothelium. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5643–5648. doi: 10.1073/pnas.091584598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Strooper B., et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 10.Weinmaster G. Notch signaling: direct or what? Curr. Opin. Genet. Dev. 1998;8:436–442. doi: 10.1016/s0959-437x(98)80115-9. [DOI] [PubMed] [Google Scholar]
- 11.Davis R.L., Turner D.L. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa O., et al. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc. Natl. Acad. Sci. U. S. A. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong T.P., Rosenberg M., Mohideen M.A., Weinstein B., Fishman M.C. gridlock, an HLH gene required for assembly of the aorta in zebrafish. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]
- 14.Zhong T.P., Childs S., Leu J.P., Fishman M.C. Gridlock signalling pathway fashions the first embryonic artery. Nature. 2001;414:216–220. doi: 10.1038/35102599. [DOI] [PubMed] [Google Scholar]
- 15.Lawson N.D., et al. Notch signaling is required for arterial-venous differentiation during embryonic vascular development. Development. 2001;128:3675–3683. doi: 10.1242/dev.128.19.3675. [DOI] [PubMed] [Google Scholar]
- 16.Shawber C.J., Das I., Francisco E., Kitajewski J. Notch signaling in primary endothelial cells. Ann. N. Y. Acad. Sci. 2003;995:162–170. doi: 10.1111/j.1749-6632.2003.tb03219.x. [DOI] [PubMed] [Google Scholar]
- 17.Fischer A., Schumacher N., Maier M., Sendtner M., Gessler M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004;18:901–911. doi: 10.1101/gad.291004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shawber C., Boulter J., Lindsell C.E., Weinmaster G. Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev. Biol. 1996;180:370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- 19.Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat. Med. 2000;6:389–395. doi: 10.1038/74651. [DOI] [PubMed] [Google Scholar]
- 20.Jeltsch M., et al. Hyperplasia of lymphatic vessels in VEGF-C transgenic mice. Science. 1997;276:1423–1425. doi: 10.1126/science.276.5317.1423. [DOI] [PubMed] [Google Scholar]
- 21.Mandriota S.J., et al. Vascular endothelial growth factor-C-mediated lymphangiogenesis promotes tumour metastasis. EMBO J. 2001;20:672–682. doi: 10.1093/emboj/20.4.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattila M.M., et al. VEGF-C induced lymphangiogenesis is associated with lymph node metastasis in orthotopic MCF-7 tumors. Int. J. Cancer. 2002;98:946–951. doi: 10.1002/ijc.10283. [DOI] [PubMed] [Google Scholar]
- 23.Skobe M., et al. Concurrent induction of lymphangiogenesis, angiogenesis, and macrophage recruitment by vascular endothelial growth factor-C in melanoma. Am. J. Pathol. 2001;159:893–903. doi: 10.1016/S0002-9440(10)61765-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stalmans I., et al. Arteriolar and venular patterning in retinas of mice selectively expressing VEGF isoforms. J. Clin. Invest. 2002;109:327–336. doi: 10.1172/JCI200214362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dumont D.J., et al. Cardiovascular failure in mouse embryos deficient for VEGF Receptor-3. Science. 1998;282:946–949. doi: 10.1126/science.282.5390.946. [DOI] [PubMed] [Google Scholar]
- 26.Valtola R., et al. VEGFR-3 and its ligand VEGF-C are associated with angiogenesis in breast cancer. Am. J. Pathol. 1999;154:1381–1390. doi: 10.1016/S0002-9440(10)65392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witmer A.N., et al. VEGFR-3 in adult angiogenesis. J. Pathol. 2001;195:490–497. doi: 10.1002/path.969. [DOI] [PubMed] [Google Scholar]
- 28.Iljin K., et al. VEGFR3 gene structure, regulatory region, and sequence polymorphisms. FASEB J. 2001;15:1028–1036. doi: 10.1096/fj.00-0383com. [DOI] [PubMed] [Google Scholar]
- 29.Podgrabinska S., et al. Molecular characterization of lymphatic endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16069–16074. doi: 10.1073/pnas.242401399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uyttendaele H., et al. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 31.Lai E.C. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3:840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henderson A.M., Wang S.J., Taylor A.C., Aitkenhead M., Hughes C.C.W. The basic helix-loop-helix transcription factor HESR1 regulates endothelial cell tube formation. J. Biol. Chem. 2001;276:6169–6176. doi: 10.1074/jbc.M008506200. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z.J., et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol. Cell. Biol. 2003;23:14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Favier B., et al. Neuropilin-2 interacts with VEGFR-2 and VEGFR-3 and promotes human endothelial cells survival and migration. Blood. 2006;108:1243–1250. doi: 10.1182/blood-2005-11-4447. [DOI] [PubMed] [Google Scholar]
- 35.Salameh A., Galvagni F., Bardelli M., Bussolino F., Oliviero S. Direct recruitment of CRK and GRB2 to VEGFR-3 induces proliferation, migration, and survival of endothelial cells through the activation of ERK, AKT, and JNK pathways. Blood. 2005;106:3423–3431. doi: 10.1182/blood-2005-04-1388. [DOI] [PubMed] [Google Scholar]
- 36.Taylor K.L., Henderson A.M., Hughes C.C. Notch activation during endothelial network formation in vitro targets the basic HLH transcription factor HESR-1 and downregulates VEGFR-2/KDR expression. Microvasc. Res. 2002;64:372–383. doi: 10.1006/mvre.2002.2443. [DOI] [PubMed] [Google Scholar]
- 37.Das I., et al. Notch oncoproteins depend on gamma-secretase/presenilin activity for processing and function. J. Biol. Chem. 2004;279:30771–30780. doi: 10.1074/jbc.M309252200. [DOI] [PubMed] [Google Scholar]
- 38.Covassin L.D., Villefranc J.A., Kacergis M.C., Weinstein B.M., Lawson N.D. Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6554–6559. doi: 10.1073/pnas.0506886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goede V., Schmidt T., Kimmina S., Kozian D., Augustin H.G. Analysis of blood vessel maturation processes during cyclic ovarian angiogenesis. Lab. Invest. 1998;78:1385–1394. [PubMed] [Google Scholar]
- 40.Redmer D.A., Reynolds L.P. Angiogenesis in the ovary. Rev. Reprod. 1996;1:182–192. doi: 10.1530/ror.0.0010182. [DOI] [PubMed] [Google Scholar]
- 41.Shweiki D., Itin A., Neufeld G., Gitay-Goren H., Keshet E. Patterns of expression of vascular endothelial growth factor (VEGF) and VEGF receptors in mice suggest a role in hormonally regulated angiogenesis. J. Clin. Invest. 1993;91:2235–2243. doi: 10.1172/JCI116450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann R.C., Hartman T., Bohlen P., Sauer M.V., Kitajewski J. Preovulatory treatment of mice with anti-VEGF receptor 2 antibody inhibits angiogenesis in corpora lutea. Microvasc. Res. 2001;62:15–25. doi: 10.1006/mvre.2001.2312. [DOI] [PubMed] [Google Scholar]
- 43.Fraser H.M., Wulff C. Angiogenesis in the corpus luteum. Reprod. Biol. Endocrinol. 2003;1:88. doi: 10.1186/1477-7827-1-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vorontchikhina M.A., Zimmermann R.C., Shawber C., Tang H., Kitajewski J. Unique patterns of Notch1, Notch4 and Jagged1 expression in ovarian vessels during folliculogenesis and corpus luteum formation. Gene Expr. Patterns. 2005;5:701–709. doi: 10.1016/j.modgep.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 45.Kaipainen A., et al. Expression of the fms-like tyrosine kinase 4 gene becomes restricted to lymphatic endothelium during development. Proc. Natl. Acad. Sci. U. S. A. 1995;92:3566–3570. doi: 10.1073/pnas.92.8.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kukk E., et al. VEGF-C receptor binding and pattern of expression with VEGFR-3 suggests a role in lymphatic vascular development. Development. 1996;122:3829–3837. doi: 10.1242/dev.122.12.3829. [DOI] [PubMed] [Google Scholar]
- 47.Mailhos C., et al. Delta4, an endothelial specific Notch ligand expressed at sites of physiological and tumor angiogenesis. Differentiation. 2001;69:135–144. doi: 10.1046/j.1432-0436.2001.690207.x. [DOI] [PubMed] [Google Scholar]
- 48.Kim M.J., Gong G., Joo H.J., Ahn S.H., Ro J.Y. Immunohistochemical and clinicopathologic characteristics of invasive ductal carcinoma of breast with micropapillary carcinoma component. Arch. Pathol. Lab. Med. 2005;129:1277–1282. doi: 10.5858/2005-129-1277-IACCOI. [DOI] [PubMed] [Google Scholar]
- 49.Shawber C.S., Kandel J.J., Kitajewski J. Notch: cell fate determination from vascular development to human vasculopathy. Drug Discovery Today: Disease Models. 2004;1:351–358. [Google Scholar]
- 50.Suchting S., et al. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dumont D.J., Yamaguchi T.P., Conlon R.A., Rossant J., Breitman M.L. tek, a novel tyrosine kinase gene located on mouse chromosome 4, is expressed in endothelial cells and their presumptive precursors. Oncogene. 1992;7:1471–1480. [PubMed] [Google Scholar]
- 52.Uyttendaele H., et al. Notch4/int-3, a mammary proto-oncogene, is and endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 53.MacKenzie F., Duriez P., Wong F., Noseda M., Karsan A. Notch4 inhibits endothelial apoptosis via RBP-Jkappa-dependent and -independent pathways. J. Biol. Chem. 2004;279:11657–11663. doi: 10.1074/jbc.M312102200. [DOI] [PubMed] [Google Scholar]
- 54.Liu Z.-J., et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: Implication for modulating arteriogenesis and angiogenesis. Mol. Cell. Biol. 2003;23:14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Persaud K., et al. Involvement of the VEGF receptor 3 in tubular morphogenesis demonstrated with a human anti-human VEGFR-3 monoclonal antibody that antagonizes receptor activation by VEGF-C. J. Cell Sci. 2004;117:2745–2756. doi: 10.1242/jcs.01138. [DOI] [PubMed] [Google Scholar]
- 56.Saaristo A., Karpanen T., Alitalo K. Mechanisms of angiogenesis and their use in the inhibition of tumor growth and metastasis. Oncogene. 2000;19:6122–6129. doi: 10.1038/sj.onc.1203969. [DOI] [PubMed] [Google Scholar]
- 57.Yancopoulos G.D., et al. Vascular-specific growth factors and blood vessel formation. Nature. 2000;407:242–248. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 58.Dixelius J., et al. Ligand-induced vascular endothelial growth factor receptor-3 (VEGFR-3) heterodimerization with VEGFR-2 in primary lymphatic endothelial cells regulates tyrosine phosphorylation sites. J. Biol. Chem. 2003;278:40973–40979. doi: 10.1074/jbc.M304499200. [DOI] [PubMed] [Google Scholar]
- 59.Matsumura K., et al. Modulation of VEGFR-2-mediated endothelial-cell activity by VEGF-C/VEGFR-3. Blood. 2003;101:1367–1374. doi: 10.1182/blood-2002-05-1329. [DOI] [PubMed] [Google Scholar]
- 60.Li Y.S., et al. Expression of vascular endothelial growth factor-C and its receptor in invasive micropapillary carcinoma of the breast. Pathol. Int. 2006;56:256–261. doi: 10.1111/j.1440-1827.2006.01961.x. [DOI] [PubMed] [Google Scholar]
- 61.Rangarajan A., et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hardy S., Kitamura M., Harris-Stansil T., Dai Y., Phipps M.L. Construction of adenovirus vector through Cre-lox recombination. J. Virol. 1997;71:1842–1849. doi: 10.1128/jvi.71.3.1842-1849.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kato H., et al. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 64.Jarriault S., et al. Signaling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 65.Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 66.Nagy, A., Gertsenstein, M., Vintersten, K., and Behringer, R. 2003. Staining for beta-galactosidase (lacZ) activity. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 687–691 [Google Scholar]