Abstract
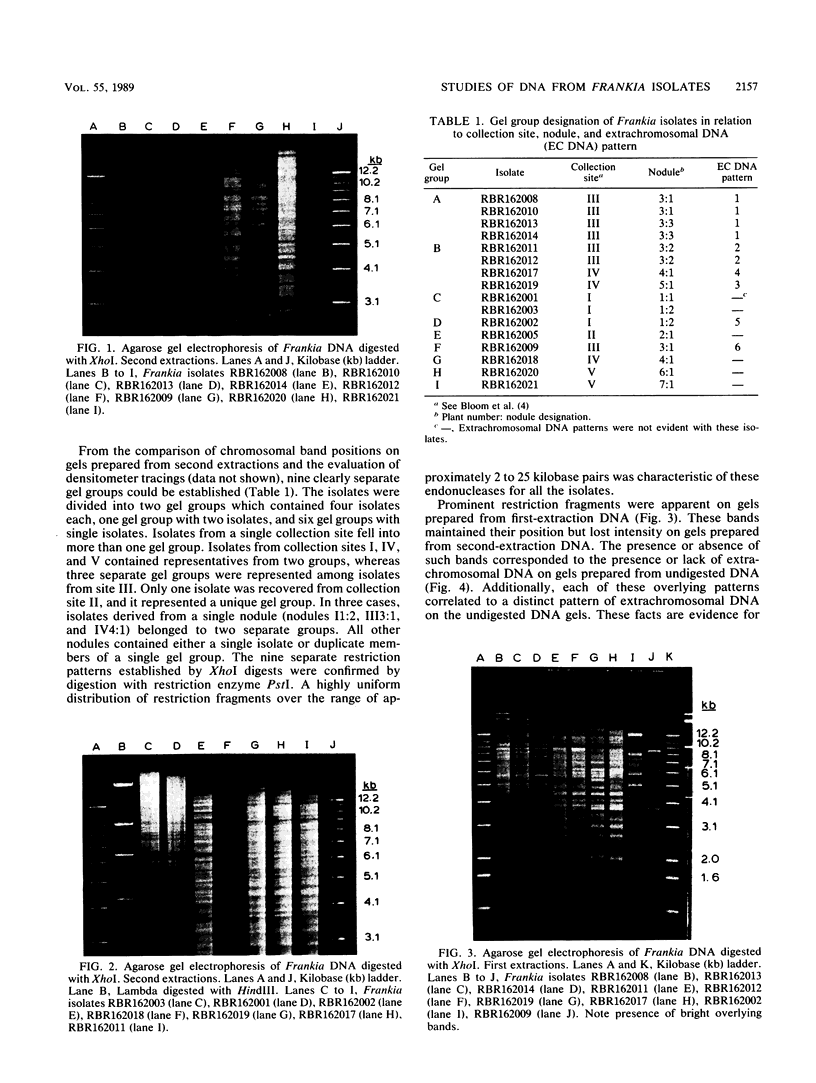
Sixteen Frankia strains were isolated from Myrica pennsylvanica (bayberry) root nodules collected at diverse sites in New Jersey. Restriction pattern analysis of total genomic DNA was used to group the isolates into gel groups, and the genetic relatedness among the isolates was evaluated by DNA-DNA solution hybridization studies. Restriction pattern analysis provided a distinctive reproducible fingerprint for each isolate. Isolates fell into nine separate groups (strain types). More than one strain type was isolated from most sites. Isolates from two different gel groups were found in 3 of 10 nodules examined. Of the 16 isolates, 10 contained extrachromosomal DNA. Six different extrachromosomal DNA banding patterns were found. Genomically similar isolates carried related, but different, banding patterns. DNA hybridization studies indicated that isolates from a single plant species can be minimally related as determined by total genome homology. Homology ranged from 12 to 99%. Highly divergent strains were isolated from the same plant and found to cohabit the same nodule. Thus, this study demonstrated that Frankia strains which infect the same host plant are not only phenotypically different but also genetically diverse.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloom R. A., Lechevalier M. P., Tate R. L., 3rd Physiological, chemical, morphological, and plant infectivity characteristics of Frankia isolates from Myrica pennsylvanica: correlation to DNA restriction patterns. Appl Environ Microbiol. 1989 Sep;55(9):2161–2166. doi: 10.1128/aem.55.9.2161-2166.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel K. D., Brill W. J. Diversity and Dynamics of Indigenous Rhizobium japonicum Populations. Appl Environ Microbiol. 1980 Nov;40(5):931–938. doi: 10.1128/aem.40.5.931-938.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normand P., Simonet P., Butour J. L., Rosenberg C., Moiroud A., Lalonde M. Plasmids in Frankia sp. J Bacteriol. 1983 Jul;155(1):32–35. doi: 10.1128/jb.155.1.32-35.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bodman S. B., Shaw P. D. Conservation of plasmids among plant-pathogenic Pseudomonas syringae isolates of diverse origins. Plasmid. 1987 May;17(3):240–247. doi: 10.1016/0147-619x(87)90032-1. [DOI] [PubMed] [Google Scholar]