Abstract
An aminopeptidase B (Ap-B) was previously purified to homogeneity from rat testis extracts and characterized. In the present work, by using oligonucleotides selected on the basis of partial amino acid microsequences of pure Ap-B and PCR techniques, the nucleotide sequence of a 2.2-kb cDNA was obtained. The deduced amino acid sequence corresponds to a 648-residue protein (72.3 kDa) containing the canonical “HEXXHX18E” signature, which allowed its classification as a member of the M1 family of metallopeptidases. It exhibits 33% identity and 48% similarity with leukotriene-A4 hydrolase, a relation further supported by the capacity of Ap-B to hydrolyze leukotriene A4. Both enzymes also were closely related to a partially sequenced protein from Dictyostelium discoideum, which might constitute the putative common ancestor of either aminopeptidase or epoxide hydrolase, or both. Ap-B and its mRNA were detected in the germ line and in the Sertoli and peritubular cells of the seminiferous tubules. Because the enzyme was found in the medium conditioned by spermatocytes and spermatids and in the acrosome during spermatozoa formation, together these observations suggested an involvement of this exometallopeptidase in the secretory pathway. It is concluded that this ubiquitous enzyme may be involved in multiple processing mechanisms.
Proteolytic activation of peptide precursors occurs, in most cases, at Arg and Lys residues and involves the sequential action of an endoprotease, cleaving on the C terminus of basic residues and subsequently of a carboxypeptidase of the B-type (1–3). This scheme was supported by the characterization of a number of subtilisin-like serine proteases, related to Kex2 (4, 5) and by the isolation of carboxypeptidase E (or H) (3, 6, 7). However, complete removal of the basic residues of a doublet in a precursor can, in principle, proceed alternatively by three different pathways, of which two may involve an aminopeptidase-B (Ap-B) (1). Several reports showed the existence of endoproteases with a cleavage specificity for the N terminus of basic residues present in the doublet (8–12) and of poorly characterized Ap-B-like activities (9, 13–15). An Ap-B (EC 3.4.11.6) activity initially was identified in several rat tissues (13) and was defined as an exopeptidase able to remove basic residues from the N terminus of l-amino acid-β-naphthylamide, dipeptides, and kallidin 10 (13, 14). More recent work has provided information about the biochemical and structural properties of the enzyme (16, 17). Purification and characterization of an Ap-B from rat testis (18) demonstrated the strict specificity of Ap-B for Arg and/or Lys residues at the N terminus of various peptides and allowed its definition as a putative Zn2+-exopeptidase. A 72-kDa immunologically related form of Ap-B detected in brain cortex and in other rat tissues indicated an ubiquitous distribution of the enzyme (18, 19).
The testis contains precursors for a number of peptide hormones, and Ap-B is abundant at late stages of spermatogenesis (18). It was used as a source for pure Ap-B and to determine its cDNA nucleotide sequence. The deduced primary structure demonstrates that the enzyme is a member of the M1 family of Zn2+-dependent peptidases (20) and that it is closely related to leukotriene-A4 (LTA4) hydrolase. Ap-B is detected in most seminiferous tubule cell types, in the acrosome of spermatids, and in the germinal cell conditioned medium. It is hypothesized that Ap-B may be involved in processing mechanisms during spermatid differentiation and participate in inflammatory processes.
MATERIALS AND METHODS
RNA Extraction.
Total RNAs extracted from rat testes, total germinal cells, isolated testis cells, and residual bodies were prepared using TRIzol reagent (GIBCO/BRL). Poly(A)+ mRNAs were purified from total testes RNA using the Oligotex mRNA kit (Quiagen, Hilden, Germany).
cDNA Cloning of Ap-B.
Ap-B was purified from rat testes (18), and the amino acid sequence of endolysine C-digested peptides was determined. Two amino acid microsequences (P1: NDHQEEFWK; P2: TYQLVYFLDK) were used to design the following degenerate oligonucleotides: A1a, 5′-GATCAT/CCAA/GGAA/GGAA/GTTT/CTGG-3′; A1b, 5′-GACCAT/CCAA/GGAA/GGA A/GTTT/CTGG-3′; A3a, 5′-CCAAAAT/CTCT/CTCT/CTGA/GTGATC-3′; A3b, 5′-CCAAAAT/C TCT/CTCT/CTGA/GTGGTC-3′; A2a, 5′-ACITAT/CCAA/GC/GTIGTITA T/CTTT/CCTIGA-3′; A2b, 5′-ACITAT/CCAA/GC/GTIGTITAT/CTTT/CTTIGA-3′; A4a, 5′-TCIAGA/GAAA/GTAIACIA G/AT/GTGA/GTAIGT-3′; A4b, 5′-TCIAAA/GAAA/GTAIACIAG/AT/GTGA/GTAIGT-3′ (Eurogentec, Brussels). Reverse transcriptions of poly(A)+ mRNA (0.2 μg) were performed with the SuperScript RNaseH− RT (GIBCO/BRL) using 400 pmol of the appropriate antisense primers. PCR experiments were carried out using different combinations of the above primers and Taq DNA polymerase (GIBCO/BRL). The 3′-end of the Ap-B cDNA was obtained using the 3′-rapid amplification of cDNA ends (RACE) system (GIBCO/BRL). The 5′-end of the Ap-B cDNA was obtained by successive 5′-RACE experiments (5′-RACE system, GIBCO/BRL) and the 5’ (single-strand ligation of cDNA) procedure (21). All the amplified fragments were cloned into the PCRII vector (Invitrogen) and sequenced by the dideoxynucleotide chain termination method using modified phage T7 DNA polymerase (Sequenase 2.0, United States Biochemical). PCR products were sequenced on both strands, and each base was sequenced at least five times.
Computer Analysis.
Sequence data were analyzed (BISANCE; ref. 22). Nucleotide sequences were translated in the six reading frames, and the deduced amino acid sequences were compared with protein sequences contained in the Genpro database (Release 96). blast and fasta programs were used for homology searches with the default parameters (22). The predicted secondary structures of Ap-B and LTA4 hydrolase proteins were determined with the Garnier and Robson algorithm (22).
LTA4 Hydrolase Activity Assays.
LTA4 hydrolase assays were performed using 300 ng of pure Ap-B and the Leukotriene B4 enzyme immunoassay kit (Cayman Chemicals, Ann Arbor, MI). A pure preparation of rat testis LTA4 hydrolase (300 ng), isolated as described for Ap-B in (18), was used as control.
Cell Isolation and Conditioned Media Preparation.
Pachytene spermatocytes, early spermatids, and residual bodies were prepared from adult Sprague–Dawley rat testes by centrifugal elutriation with a purity greater than 90% (23). Pachytene spermatocytes and early spermatids then were incubated at 32°C for 2, 4, 8, or 16 h, and 20 h for total germinal cells. At this time, 95% of cells were viable as shown by an erythrosine red dye exclusion test. Germinal cells-conditioned media were concentrated as described (24). Sertoli and peritubular cells were isolated from 20-day-old Sprague–Dawley rats, then cells and culture media were prepared as in ref. 24.
Northern Blot Analysis.
Total mRNAs were electrophoresed on 1% agarose/formaldehyde gel. Then, RNAs were transferred to Nytran (0.45 μm; Schleicher & Schuell, Dassel, Germany) and hybridized as in ref. 24. A 608-bp testis Ap-B cDNA (nucleotides 324–990) was cloned into EcoRI-digested PCRII vector (Invitrogen) and propagated in DH5α Escherichia coli strain. Plasmid was transcribed in vitro by using T7 RNA polymerase (Stratagene) and [α-32P]UTP (800 Ci/mmol; ICN). The blots were washed three times in 0.1× standard saline citrate (1× SSC = 0.15 M sodium chloride/0.15 sodium citrate, pH 7)/0.1% SDS/1 mM EDTA at 70°C for 2 h.
Immunoblot Analysis.
Tissues, residual bodies, and cells were prepared as in ref. 18. Samples of 3 μg of protein were run under denaturing conditions on 8% polyacrylamide electrophoresis gels. Electrotransfer of proteins to nitrocellulose membrane (0.45 μm; Schleicher and Schuell) was performed with a semidry blotting apparatus (Hoefer). Ap-B was detected using a rabbit polyclonal anti-Ap-B serum at 1:2000 dilution (18). The visualization of the antigen-antibody complex was carried out using alkaline phosphatase-coupled antibody and NBT/BCIP mixture (GIBCO/BRL).
Electron Microscopy.
Rat testis was fixed in situ at 4°C during 1 h in 0.1 M Sörensen phosphate buffer, pH 7.3, containing 4% paraformaldehyde (Merck). Tissues and ultrathin sections were prepared as in ref. 25. Grids bearing Lowicryl sections of rat testis were incubated for 30 min at room temperature on a 10-μl drop of a rabbit polyclonal anti-Ap-B serum (1:20 in PBS). After 15 min in PBS, the grids were floated for 30 min on a 10-μl drop of 1:25 dilution of goat anti-rabbit IgG-conjugated to gold particles of 10 nm in diameter (Biocell Laboratories). After a final PBS wash, grids were rinsed, air dried, and stained for 10 min with 5% aqueous uranyl acetate (25). For controls, a 1:10 dilution of preimmune serum was used.
RESULTS
After synthesis of rat testes cDNA using antisense degenerate primers (A3a, A3b, A4a, A4b; see Materials and Methods), PCR resulted in the amplification of a 173-bp (A2a/A3b) Ap-B cDNA fragment. Specific internal oligonucleotides then were used in successive 5′ and 3′ RACE experiments. In addition, several PCR were carried out to check the arrangement of the PCR products and the nucleotide sequence.
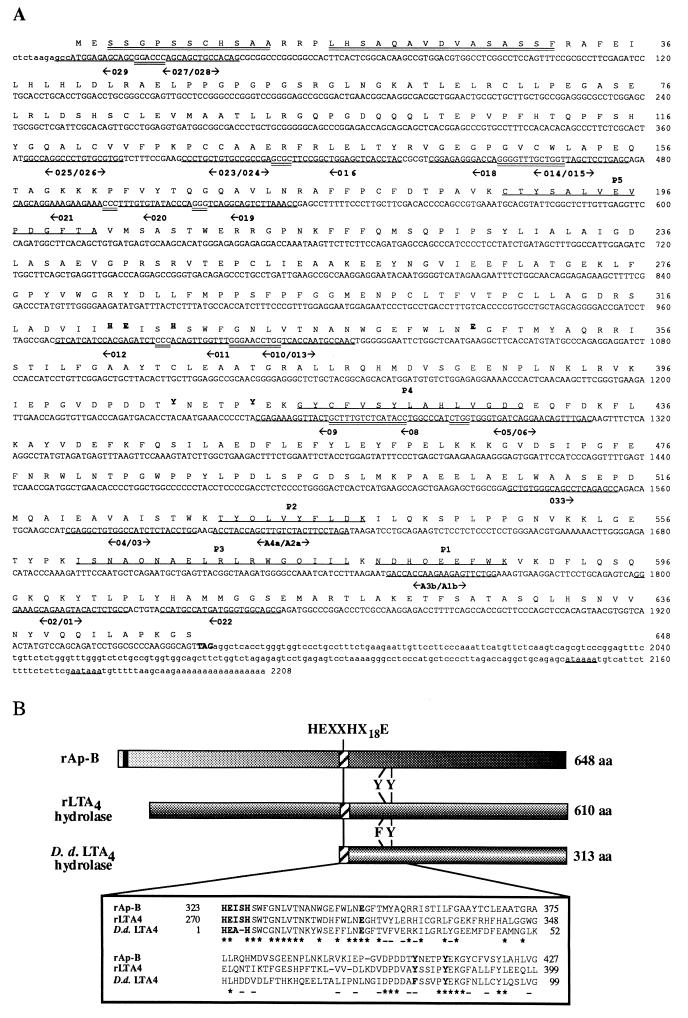
The Ap-B cDNA sequence is 2208 bp in length (55, 3% G+C). Although primer extension experiments showed the existence of a 40-nt-long additional sequence in the 5′ Ap-B cDNA, this region could not be obtained using two different methods of 5′-RACE. The high (G+C) content of this region (nucleotides 1–240: 71.2% G+C) could explain this problem and those encountered during the cloning of the 5′-Ap-B cDNA fragments. The whole Ap-B cDNA sequence should be about 2,250 bp long, which is similar to the estimated size of the Ap-B mRNA (2.2 kb; vide infra). The sequence consists of a short 11-base 5′ untranslated region (UTR), an ORF encoding a 648-amino acid protein, a 236-bp-long 3′ UTR, and a 17-bp-long poly(A)+ tail. No consensus regulation signals were found in the 3′ UTR. Two putative polyadenylylation signals are located in positions 2146 and 2172. The first ATG (nucleotide 12) in the sequence is assumed to be the initiation codon of the protein. The nucleotides flanking the putative AUG initiator codon are in agreement with Kozak’s rule (26). The ORF starting from this methionine and ending at the TAG terminator codon (nucleotide 1956) encodes a 72.3-kDa protein in agreement with the estimated mass on SDS/PAGE. The ORF translation product (nucleotides 12–1955) is in keeping with the rat or mouse codon usage, except for the first 18-amino-acid-long region. Sequencing of several independent Ap-B cDNA confirmed the nucleotide sequence of this fragment. The deduced amino acid sequence consists of 648 residues and contains the three other oligopeptide sequences of pure enzyme endolysine C fragments (P3: ISNAQNAELRLRWG QIIL, P4: GYCFVSYLAHLVGDQ, P5: CTYSALVEVPDGFTA). Two main features emerged from examination of Ap-B sequence. First, two mildly hydrophobic domains at the N terminus might constitute a putative signal peptide (Fig. 1). However, Von Heijne’s predictions (27) indicated that these domains probably are not very efficient for the translocation of the enzyme into the endoplasmic reticulum. The second feature is a typical consensus zinc binding motif HEXXHX18E in position 323 (Fig. 1). This confirms the reported metallopeptidase character of the enzyme and allows its classification under the M1 family of metalloexopeptidases (20). Neither consensus N-glycosylation sites nor consensus targeting signals were found in the sequence. On the other hand, several putative phosphorylation sites are present in Ap-B primary structure.
Figure 1.
(A) Nucleotide sequence of the Ap-B cDNA. The 5′ and 3′ untranslated regions are in lowercase letters. The stop codon (TAG) is boldface, and putative polyadenylylation signals are underlined. The oligonucleotides used are underlined and numbered. Their orientation is indicated by arrows. The inverse and complementary primers are separated by a slash. Overlapping regions between oligonucleotides are double underlined. The Ap-B proteolytic peptides identified by microsequencing are underlined (P1 to P5). The potential Zn2+ binding motif (HEXXHX18E) is boldface. Two putative signal sequences are double underlined. Two tyrosine residues (Y407 and Y412) potentially involved in the catalytic mechanism are boldface. (B) Schematic representation of primary structures and alignment of partial amino acid sequences of rat Ap-B, rat LTA4 hydrolase, and the C-terminal domain of Dictyostelium discoideum putative LTA4 hydrolase. The putative signal sequence (black box), the potential Zn2+ binding motif (HEXXHX18E) and the two tyrosine residues (LTA4 hydrolase: Y379 and Y384; Ap-B: Y407 and Y412) potentially involved in the catalytic mechanism are indicated. A conserved region of the three enzyme sequences is boxed. Identical residues (*) and similar residues (−; according to the rule T:A:S, I:L:V:M, K:R:H, D:E:Q:N, F:Y, G, P, W, C) are indicated.
A database search identified LTA4 hydrolase, another member of the M1 family, as the most closely related protein to Ap-B. Indeed, the putative Ap-B translation product exhibits 33% and 48%, respectively, amino acid identity and similarity with LTA4 hydrolase. This extends to the entire primary structures with some divergence in the N and C termini. Both sequences exhibit the putative canonical zinc binding site and two important conserved tyrosine residues (LTA4 hydrolase: Y379 and Y384; Ap-B: Y407 and Y412; Fig. 1). Interestingly, this databank search also revealed that a partial amino acid sequence of Dictyostelium discoideum (313 amino acids; accession number: V27538V27538, Jho and Kopachik, 1995, unpublished data), presumed to be a LTA4 hydrolase, showed important homology with rAp-B (31, 5% identity; 46, 9% similarity) and rLTA4 hydrolase (32, 7% identity, 51% similarity). This structural similarity overlapped the entire C terminal domains of the three enzymes (Fig. 1). However, it should be noted, on the one hand, that the putative zinc binding site of D. discoideum protein lacked one of the residues (HEXHX18E; Fig. 1). On the other hand, one of the two tyrosine residues, Y379 of LTA4 hydrolase (Y407 of Ap-B), known to be involved in the covalent binding of LTA4, is replaced by a phenylalanine (ref. 28; Fig. 1).
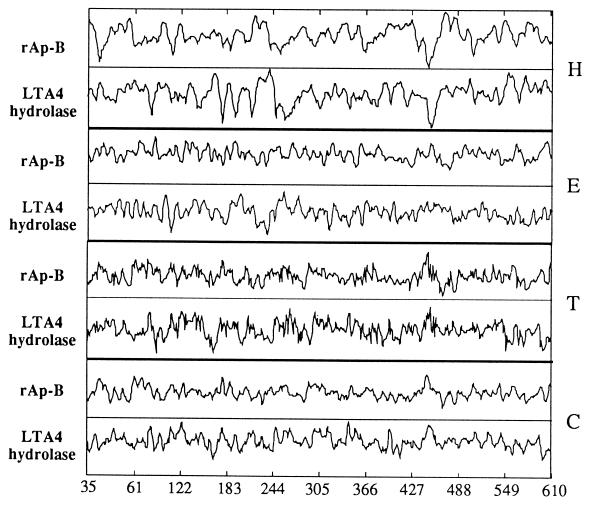
Moreover, secondary structure predictions (Garnier and Robson algorithm, ref. 22) underlined a strong homology supporting the evolutionary affinities of Ap-B and LTA4 hydrolase (Fig. 2). This clearly indicated LTA4 hydrolase as a potential homolog of Ap-B. Functional homology was supported by tests of LTA4 hydrolase activity with pure Ap-B shown to be free of any contaminating LTA4 hydrolase by Western blot analysis, using a pure testis LTA4 hydrolase enzyme as control. These preliminary assays demonstrated that Ap-B transformed LTA4 into proinflammatory LTB4. However, this second activity exhibited by Ap-B appeared to be 10-fold lower than the corresponding activity of LTA4 hydrolase (not shown).
Figure 2.
Schematic representation of secondary structure predictions for rat Ap-B and LTA4 hydrolase. Predictions for α-helix (H), β-sheet (E), reverse turns (T), and coil (C) structures are shown. Calculations were made using the Garnier and Robson algorithm.
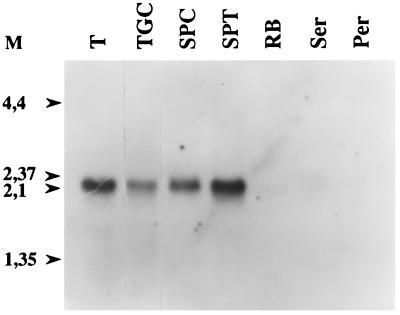
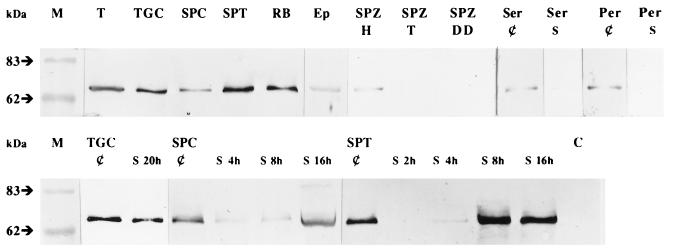
A single 2.2-kb Ap-B transcript was detected by Northern blot, in rat testis and in isolated testis components (Fig. 3). Pachytene spermatocytes and particularly round spermatids appeared to be the main Ap-B mRNA expressing cells. A weak signal was observed in residual bodies and a very faint one in the Sertoli and peritubular somatic cells, corroborated by Western blot of the same samples (Fig. 4). In contrast to its mRNA, the protein was very abundant in residual bodies as observed by immunofluorescence (18). This cytoplasmic structure is shed by elongated spermatids just before leaving the seminiferous tubules. The difference between the amounts of mRNA and protein suggested a concentration effect of Ap-B in this particular structure subsequently eliminated by the Sertoli cells. Ap-B also was expressed in the epididymis and in the spermatozoa from the head of this organ, but no signal was detected in those from the tail of epididymis or from the deferent duct.
Figure 3.
Northern blot analysis of Ap-B mRNA in rat testes, isolated testis cells, and residual bodies. Each lane contains 10 μg of total mRNA. M, molecular size markers expressed in kilobases; T, testes; TGC, total germinal cells; SPC, spermatocytes (pachytene stage); SPT, round spermatids; RB, residual bodies; Ser, Sertoli cells; Per, peritubular cells.
Figure 4.
Western blot analysis of the expression of Ap-B in rat testis, isolated testis components, epididymis, and cultured or conditioned media. M, prestained molecular mass markers; T, testes; TGC, total germinal cells; SPC, spermatocytes (pachytene stage); SPT, round spermatids; RB, residual bodies; Ep, epididymis; spermatozoa isolated from the head (SPZ-H) and from the tail of epididymis (SPZ-T); spermatozoa isolated from the deferent duct (SPZ-DD); Ser, Sertoli cells; Per, peritubular cells; ¢, cellular protein extract; S, cultured or conditioned media collected at the indicated time; C, control medium.
Because Ap-B might be secreted, the enzyme in the culture or incubation media of germinal and somatic cells was evaluated (Fig. 4). Only traces of Ap-B were detectable in Sertoli and peritubular cell culture media, in agreement with the low level of expression of the enzyme in these somatic cells. In spermatocytes and in spermatids conditioned media, the immunoreactive protein increased with time, and spermatids released Ap-B in much higher quantity than spermatocytes. This extracellular Ap-B could not result from a cell lysis process, because 95% of them were viable after 20 h. Molecular mass of the secreted enzyme appeared unchanged, suggesting the absence of either major posttranslational events or degradation. Lower amounts of Ap-B in germinal cells-conditioned medium versus spermatocytes and spermatids media suggested a lower rate of secretion of the enzyme in the other germinal cell types such as spermatogonia and prepachytene spermatocytes. Together, these data argued in favor of the passage of Ap-B via the germinal cell secretory route.
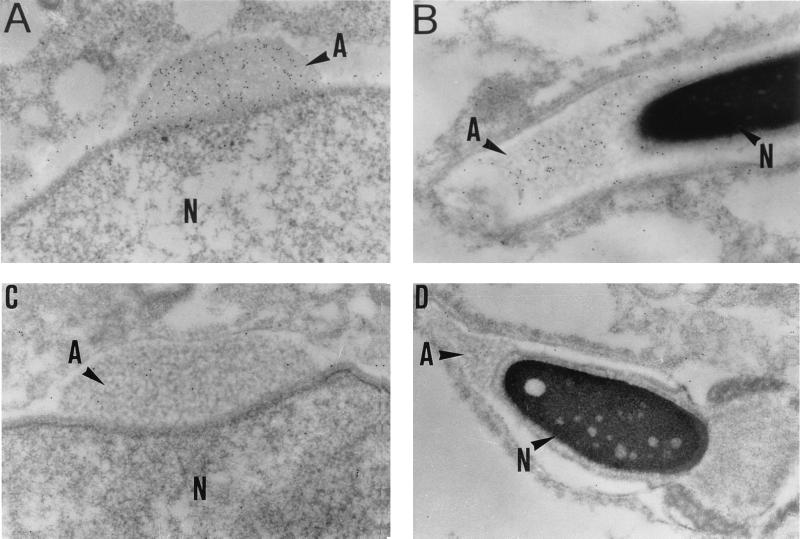
Electron microscopy examination of Ap-B subcellular localization in germinal cells showed that it was present in the trans-Golgi network around the apical pole of the nucleus of round spermatids (data not shown) and concentrated in the proacrosomic granule of these cells (Fig. 5A). At a later step of spermatid differentiation, it was observed in the acrosome of the elongated spermatids, but to a lower extent (Fig. 5B). No signal was detected using the preimmune serum (Fig. 5 C and D). Because the acrosome is generated from saccules of the Golgi apparatus, this confirmed the presence of Ap-B in the secretory apparatus of spermatids during their transformation into spermatozoa.
Figure 5.
Subcellular localization of Ap-B in germinal cells by electron microscopy. (A) Young spermatid at stage VII of the rat seminiferous cycle. (B) Elongated spermatid at stage III-IV of the rat seminiferous cycle. (C and D) Controls run with the preimmune serum. A, acrosome; N, nucleus. (×24,000.)
DISCUSSION
Trimming of extra amino acids at both the N and C termini of peptides are crucial activation reactions in proteolytic processing. Therefore, identification of exopeptidases with either carboxyl- or amino-peptidase activity is essential for understanding the mechanisms leading to functional peptides and for the modulation of their properties. Moreover, in the case of the endoproteolytic processing of basic amino acid doublets, exopeptidases of the B-type are needed (2, 3, 7). In contrast to Cp-E (6, 7), Ap-B is relatively poorly understood. The present and previous reports (13, 18, 19) unequivocally demonstrate the existence in several tissues of an Ap-B with a strict selectivity for the removal of Arg and/or Lys residues at the N terminus of peptides. Ap-B can be considered, on the basis of the presence of the HEXXHX18E zinc binding motif in its amino acid sequence, as a metalloexoprotease of the M1 family (20).
This family is composed of aminopeptidase A, aminopeptidase N, cysteine aminopeptidase, and LTA4 hydrolase. The latter initially was identified as an hydrolase implicated in the biosynthesis of the proinflammatory substance LTB4 (29). Moreover, LTA4 hydrolase exhibits an aminopeptidase activity (30, 31) and was considered as an arginine aminopeptidase despite its wide cleavage specificity (32). This data probably led to the hypothesis that LTA4 hydrolase and Ap-B might be identical (33), a classification inappropriate in light of the present evidence. Ap-B exhibits in vitro an epoxide hydrolase activity in good agreement with the degree of homology between both enzymes. In LTA4 hydrolase, two tyrosine residues (Y379 and Y384) were implicated in the suicide inactivation of the enzyme and in catalysis, respectively. Indeed, the first one covalently binds the LTA4 substrate to inactivate the enzyme (28), and the latter is necessary to the peptidase activity as a proton donor in a general base mechanism (34). The conservation of these two residues in Ap-B further supports its in vitro, and probably in vivo, bifunctionality. This also could explain why some atypical LTA4 hydrolase and Ap-B activities have been reported (35–39).
The structural relationships between rat Ap-B, LTA4 hydrolase, and a D. discoideum protein is intriguing. The observed similarities between the three proteins indicate they are related by evolution. Biochemical studies on this protein will clarify which kind of activities this protein exhibits. Indeed, D. discoideum is an amoebae that assembles, via cAMP-dependent chemotaxis, into a multicellular organism in case of starvation. As LTB4 induces chemotaxis of leukocytes and their aggregation in vitro (40), this could indicate that this slime mold ancestral putative aminopeptidase/epoxide hydrolase protein could participate in a primitive intercellular communication system. In that case, the question about a similar function for Ap-B remains open.
Concerning the participation of Ap-B in the secretory pathway, the determination of its subcellular distribution clearly identified Ap-B in compartments derived from the Golgi apparatus and in cell culture or incubation media. These are strong arguments in favor of its passage through this membrane network. The predicted inefficiency of the putative Ap-B signal sequence (27) and the invariant molecular mass of the intra- or extracellular enzyme raise the question of functionality of this signal peptide. However, several proteins such as ovalbumin and the plasminogen activator inhibitor-2 (PAI-2; ref. 41) are secreted without previous cleavage of their signal sequence (42, 43). Moreover, PAI-2 is translocated in the secretory pathway despite a weak recognition of its mildly hydrophobic bipartite signal sequence (43). Noticeably, Ap-B also was found in the secretory pathway and in the culture medium of neuroendocrine cells, and its secretion was dependent upon the cAMP pathway. Finally, electron microscopy examination of Ap-B producing cells by peroxidase labeled antibodies confirmed a marked association of the protein to the plasma membrane (unpublished data).
At the present time, the physiological relevance of the bifunctionality of Ap-B in vitro remains an open question and more precise in vitro and in vivo determinations might clarify this issue. The broad pH dependence of the enzyme (18) and its ubiquitous presence (19) argue in favor of its adaptability to various cellular subcompartments and its implication in a broad spectrum of physiological phenomena, including processing, but also inflammatory processes in which some potential peptide substrates of Ap-B such as kallidin (14), enkephalins (15, 44), and somatostatin (45) might play a central role.
Acknowledgments
We thank Drs. Bernard Jégou and Alain Dupaix (Institut National de la Santé et de la Recherche Médicale, Campus Beaulieu, Université de Rennes I, Rennes Cedex, France) for a kind gift of isolated germinal and somatic testes cells; Dr. Philippe Ravassard (Hopital de la Pitié Salpétrière, Paris, France) for the gift of 5′SLIC anchored primers; and Drs. Edmond Puvion (Centre National de la Recherche Scientifique, Villejuif, France) and Jean Charles Schwartz (Institut National de la Santé et de la Recherche Médicale, Paris, France) for helpful discussions.
ABBREVIATIONS
- Ap-B
aminopeptidase B
- LTA4 hydrolase
leukotriene- A4 hydrolase
- RACE
rapid amplification of cDNA ends
Footnotes
References
- 1.Cohen P. Biochimie. 1987;69:87–89. doi: 10.1016/0300-9084(87)90239-2. [DOI] [PubMed] [Google Scholar]
- 2.Darby N J, Smyth D G. Biosci Rep. 1990;10:1–13. doi: 10.1007/BF01116845. [DOI] [PubMed] [Google Scholar]
- 3.Fricker L D. Annu Rev Physiol. 1988;50:309–321. doi: 10.1146/annurev.ph.50.030188.001521. [DOI] [PubMed] [Google Scholar]
- 4.Julius D, Brake A, Blair L, Kunizawa R, Thorner J. Cell. 1984;37:1075–1089. doi: 10.1016/0092-8674(84)90442-2. [DOI] [PubMed] [Google Scholar]
- 5.Seidah N G, Chrétien M. Trends Endocrinol Metab. 1992;3:133–139. doi: 10.1016/1043-2760(92)90102-7. [DOI] [PubMed] [Google Scholar]
- 6.Fricker L D, Snyder S H. J Biol Chem. 1983;258:10950–10955. [PubMed] [Google Scholar]
- 7.Fricker L D. In: Peptide Biosynthesis and Processing. Fricker L D, editor. Caldwell, NJ: Telford; 1991. pp. 1–16. [Google Scholar]
- 8.Peng Loh Y, Parish D C, Tujeta R. J Biol Chem. 1985;260:7194–7205. [PubMed] [Google Scholar]
- 9.Gluschankof P, Gomez S, Morel A, Cohen P. J Biol Chem. 1987;262:9515–9520. [PubMed] [Google Scholar]
- 10.Azaryan A V, Hook Y H. FEBS Lett. 1994;341:193–196. doi: 10.1016/0014-5793(94)80456-7. [DOI] [PubMed] [Google Scholar]
- 11.Chesneau V, Pierotti A R, Barré N, Créminon C, Tougard C, Cohen P. J Biol Chem. 1994;269:2056–2061. [PubMed] [Google Scholar]
- 12.Pierotti A R, Prat A, Chesneau V, Gaudoux F, Leseney A M, Foulon T, Cohen P. Proc Natl Acad Sci USA. 1994;91:6078–6082. doi: 10.1073/pnas.91.13.6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopsu V K, Mäkinen K K, Glenner G G. Arch Biochem Biophys. 1966;114:567–566. doi: 10.1016/0003-9861(66)90380-8. [DOI] [PubMed] [Google Scholar]
- 14.Hopsu V K, Mäkinen K K. Nature (London) 1966;212:1271–1272. doi: 10.1038/2121271a0. [DOI] [PubMed] [Google Scholar]
- 15.Gainer H, Russel J T, Peng Loh Y. FEBS Lett. 1984;175:135–139. doi: 10.1016/0014-5793(84)80586-4. [DOI] [PubMed] [Google Scholar]
- 16.Yamada M, Sukenaga Y, Fujii H, Abe F, Takeuchi T. FEBS Lett. 1994;342:53–56. doi: 10.1016/0014-5793(94)80583-0. [DOI] [PubMed] [Google Scholar]
- 17.Belhacène N, Mari B, Rossi B, Auberger P. Eur J Immunol. 1993;23:1948–1955. doi: 10.1002/eji.1830230833. [DOI] [PubMed] [Google Scholar]
- 18.Cadel S, Pierotti A R, Foulon T, Créminon C, Barré N, Segrétain D, Cohen P. Mol Cell Endocrinol. 1995;110:149–160. doi: 10.1016/0303-7207(95)03529-g. [DOI] [PubMed] [Google Scholar]
- 19.Foulon T, Cadel S, Chesneau V, Draoui M, Prat A, Cohen P. Ann N Y Acad Sci. 1996;780:106–120. doi: 10.1111/j.1749-6632.1996.tb15115.x. [DOI] [PubMed] [Google Scholar]
- 20.Rawlings N D, Barrett A J. Biochem J. 1993;290:205–218. doi: 10.1042/bj2900205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumas Milne Edwards J B, Delort J, Mallet J. Nucleic Acids Res. 1991;19:5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dessen P, Fondrat C, Valencien C, Mugnier C. Comput Applied Biosci. 1990;6:355–356. doi: 10.1093/bioinformatics/6.4.355. [DOI] [PubMed] [Google Scholar]
- 23.Pineau C, Syed V, Bardin C W, Jégou B, Cheng C Y. J Androl. 1993;14:87–98. [PubMed] [Google Scholar]
- 24.Chesneau V, Prat A, Segretain D, Hospital V, Dupaix A, Foulon T, Jegou B, Cohen P. J Cell Sci. 1996;109:2737–2745. doi: 10.1242/jcs.109.11.2737. [DOI] [PubMed] [Google Scholar]
- 25.Puvion-Dutilleul F, Besse S, Chan E K L, Tan E M, Puvion E. J Cell Sci. 1995;108:1143–1153. doi: 10.1242/jcs.108.3.1143. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 27.Von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mueller M J, Blomster M, Oppermann U C T, Jörnvall H, Samuelsson B, Haeggström J Z. Proc Natl Acad Sci USA. 1996;93:5931–5935. doi: 10.1073/pnas.93.12.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radmark O, Shimizu T, Jörnvall H, Samuelsson B. J Biol Chem. 1984;259:12339–12345. [PubMed] [Google Scholar]
- 30.Haeggström J Z, Wetterholm A, Vallee B L, Samuelsson B. Biochem Biophys Res Commun. 1990;173:431–437. doi: 10.1016/s0006-291x(05)81076-9. [DOI] [PubMed] [Google Scholar]
- 31.Minami M, Ohishi N, Mutoh H, Izumi T, Bito H, Seyama Y, Toh H, Shimizu T. Biochem Biophys Res Commun. 1990;173:620–626. doi: 10.1016/s0006-291x(05)80080-4. [DOI] [PubMed] [Google Scholar]
- 32.Orning L, Gierse J K, Fitzpatrick F A. J Biol Chem. 1994;269:11269–11273. [PubMed] [Google Scholar]
- 33.NC-IUMB (Nomenclature Committee of the International Union and Molecular Biology) Eur J Biochem. 1996;237:1–5. doi: 10.1111/j.1432-1033.1996.t01-1-00001.x. [DOI] [PubMed] [Google Scholar]
- 34.Blomster M, Wetterholm A, Mueller M J, Haeggström J Z. Eur J Biochem. 1995;231:528–549. doi: 10.1111/j.1432-1033.1995.0528d.x. [DOI] [PubMed] [Google Scholar]
- 35.Orning L, Jones D A, Fitzpatrick F A. J Biol Chem. 1990;265:14911–14916. [PubMed] [Google Scholar]
- 36.Bigby T D, Lee D M, Minami M, Ohishi N, Shimizu T, Baker J R. Am J Respir Cell Mol Biol. 1994;11:615–624. doi: 10.1165/ajrcmb.11.5.7946391. [DOI] [PubMed] [Google Scholar]
- 37.Baker J R, Kilstra T A, Bigby T D. Biochem Pharmacol. 1995;50:905–912. doi: 10.1016/0006-2952(95)00210-q. [DOI] [PubMed] [Google Scholar]
- 38.Mäkinen K K, Virtanen K K. Clin Chim Acta. 1976;67:213–218. doi: 10.1016/0009-8981(76)90262-x. [DOI] [PubMed] [Google Scholar]
- 39.Knuuttila M, Virtanen K, Söderling E, Mäkinen K K. Biochem Biophys Res Commun. 1978;81:374–381. doi: 10.1016/0006-291x(78)91543-7. [DOI] [PubMed] [Google Scholar]
- 40.Ford-Hutchinson A W. Immunology. 1990;10:1–12. [PubMed] [Google Scholar]
- 41.Belin D, Bost S, Vassali J-D, Strub K. EMBO J. 1996;15:468–478. [PMC free article] [PubMed] [Google Scholar]
- 42.Tabe L, Krieg P, Strachan R, Jackson D, Wallis E, Colman A. J Mol Biol. 1984;180:645–666. doi: 10.1016/0022-2836(84)90031-7. [DOI] [PubMed] [Google Scholar]
- 43.Ye R D, Wun T-C, Sader J E. J Biol Chem. 1988;263:4869–4875. [PubMed] [Google Scholar]
- 44.Heagy W, Duca K, Finberg R W. J Clin Invest. 1995;96:1366–1374. doi: 10.1172/JCI118171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karalis K, Mastorakos G, Sano H, Wilder R L, Chrousos G P. Endocrinology. 1995;136:4133–4138. doi: 10.1210/endo.136.9.7544277. [DOI] [PubMed] [Google Scholar]