Abstract
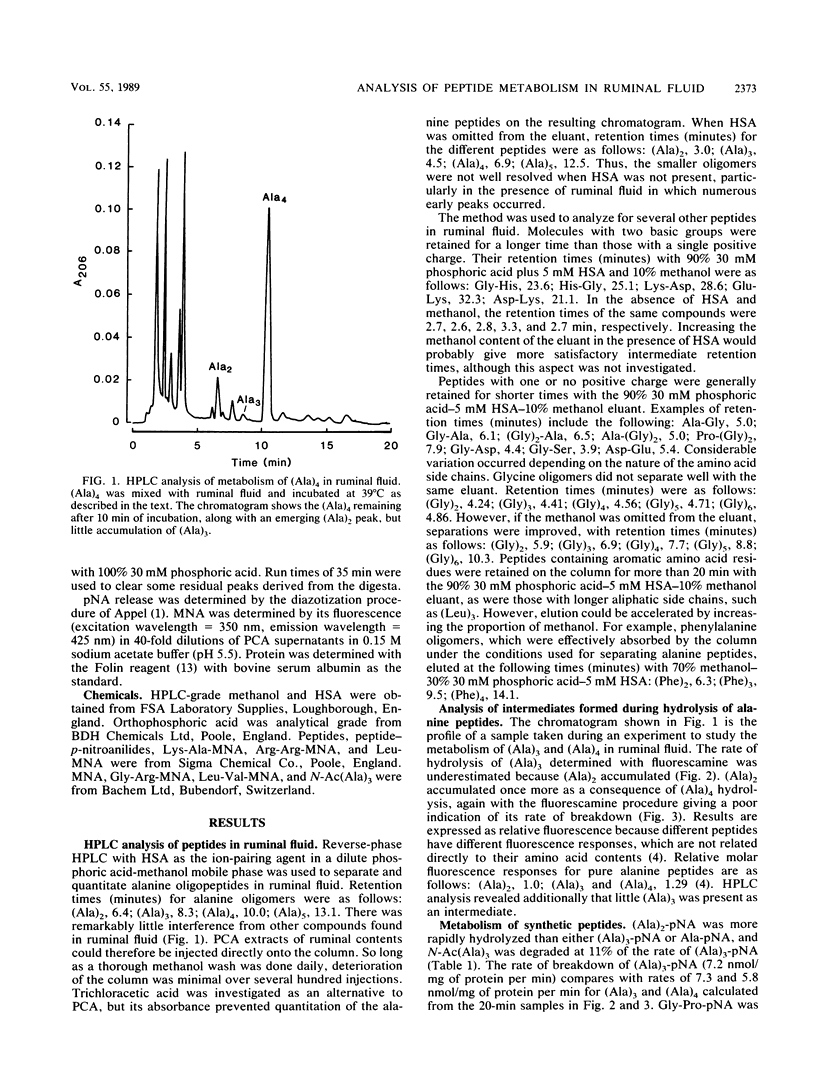
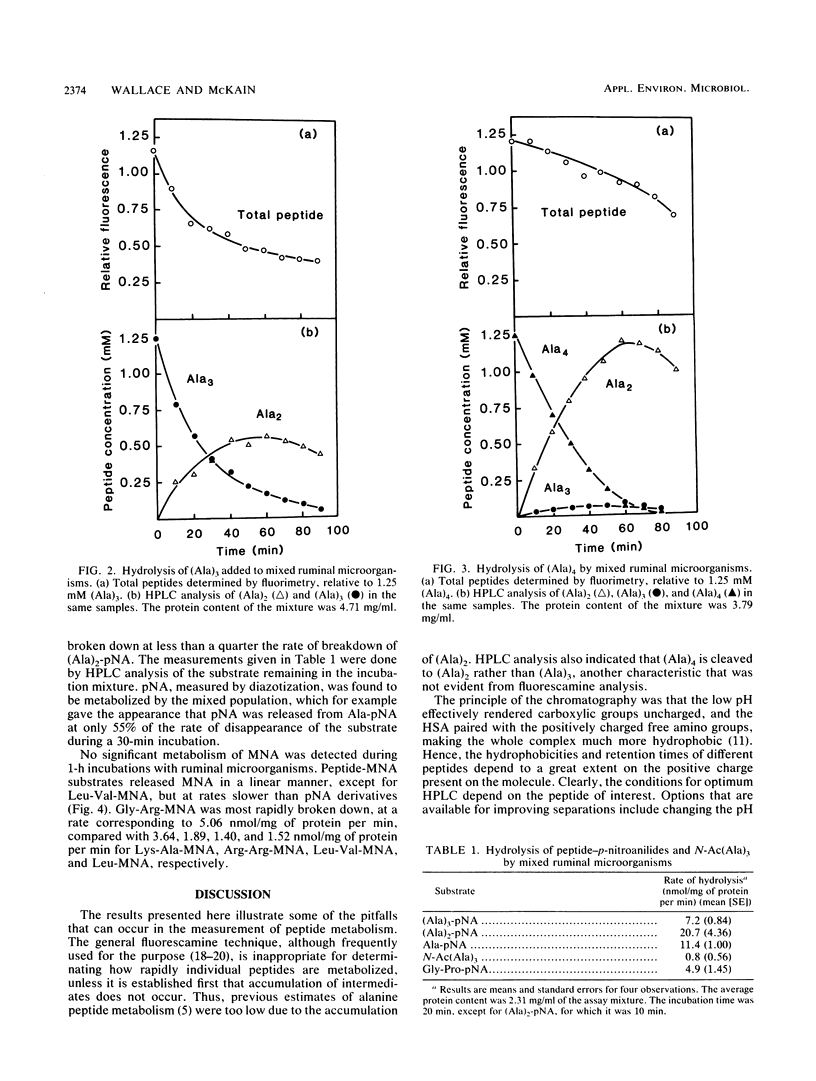
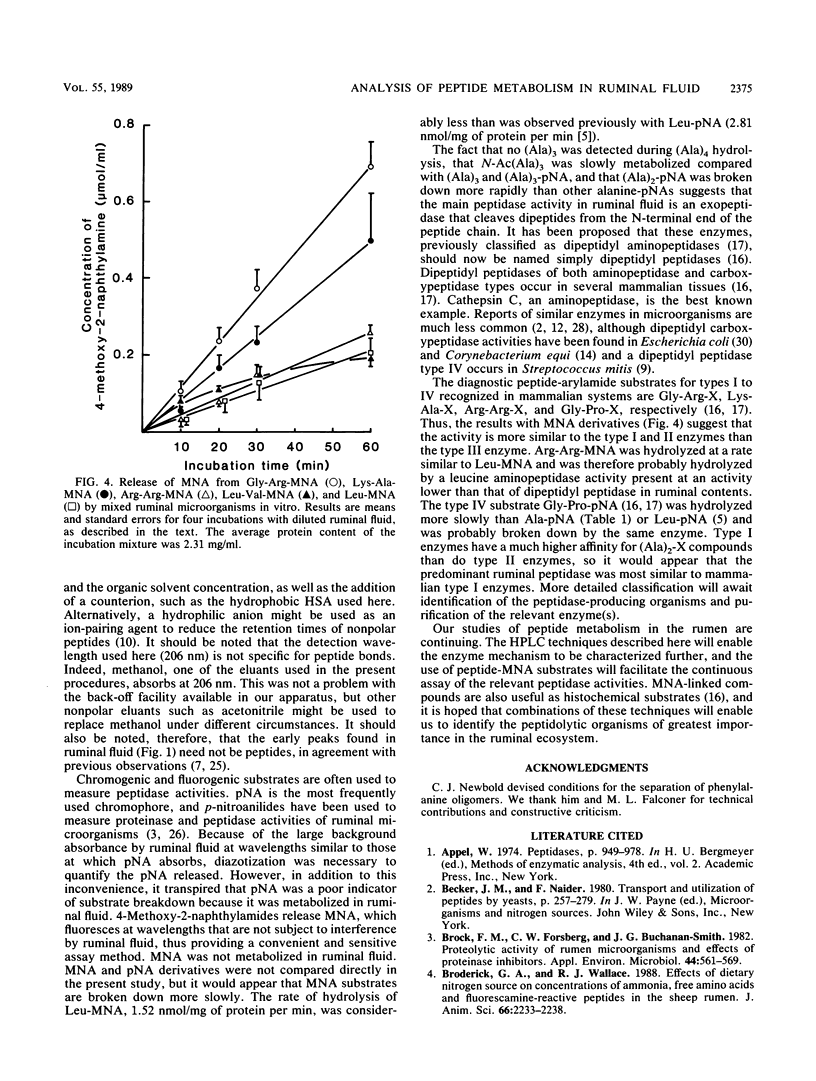
Methods were developed for the determination of oligoalanine and other short-chain peptides and peptide analogs in ruminal fluid by using reverse-phase high-pressure liquid chromatography. Chromatographic analysis of the breakdown of (Ala)3 and (Ala)4 in ruminal fluid in vitro revealed that the predominant mechanism of hydrolysis was a dipeptidyl peptidase-like activity. Hydrolysis proceeded from the N terminal of the peptide chain; N-acetyl-(Ala)3 was broken down at 11% of the rate of breakdown of (Ala)3 or (Ala)3-p-nitroanilide. (Ala)2-p-nitroanilide was hydrolyzed most rapidly of the arylamide substrates tested, but fluorogenic 4-methoxy-2-naphthylamide (MNA) compounds were more convenient and potentially more versatile substrates than p-nitroanilides. Gly-Arg-MNA was the most rapidly hydrolyzed dipeptidyl peptidase substrate, suggesting that ruminal peptidase activity was predominantly of a type I specificity.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brock F. M., Forsberg C. W., Buchanan-Smith J. G. Proteolytic activity of rumen microorganisms and effects of proteinase inhibitors. Appl Environ Microbiol. 1982 Sep;44(3):561–569. doi: 10.1128/aem.44.3.561-569.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Russell J. B., Sniffen C. J. A procedure for measuring peptides in rumen fluid and evidence that peptide uptake can be a rate-limiting step in ruminal protein degradation. J Dairy Sci. 1987 Jun;70(6):1211–1219. doi: 10.3168/jds.S0022-0302(87)80133-9. [DOI] [PubMed] [Google Scholar]
- Chen G., Strobel H. J., Russell J. B., Sniffen C. J. Effect of hydrophobicity of utilization of peptides by ruminal bacteria in vitro. Appl Environ Microbiol. 1987 Sep;53(9):2021–2025. doi: 10.1128/aem.53.9.2021-2025.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K. M., Harada M. Purification and properties of dipeptidyl peptidase IV from Streptococcus mitis ATCC 9811. Arch Biochem Biophys. 1981 Aug;210(1):230–237. doi: 10.1016/0003-9861(81)90184-3. [DOI] [PubMed] [Google Scholar]
- Lee H. J., LaRue J. N., Wilson I. B. Dipeptidyl carboxypeptidase from Coryne bacterium equi. Biochim Biophys Acta. 1971 Dec 15;250(3):608–613. doi: 10.1016/0005-2744(71)90266-x. [DOI] [PubMed] [Google Scholar]
- Leng R. A., Nolan J. V. Nitrogen metabolism in the rumen. J Dairy Sci. 1984 May;67(5):1072–1089. doi: 10.3168/jds.S0022-0302(84)81409-5. [DOI] [PubMed] [Google Scholar]
- Payne J. W. Peptide transport in bacteria: methods, mutants and energy coupling. Biochem Soc Trans. 1983 Dec;11(6):794–798. doi: 10.1042/bst0110794. [DOI] [PubMed] [Google Scholar]
- Perrett D., Webb J. P., Silk D. B., Clark M. L. The assay of dipeptides using fluorescamine and its application to determining dipeptidase activity. Anal Biochem. 1975 Sep;68(1):161–166. doi: 10.1016/0003-2697(75)90690-9. [DOI] [PubMed] [Google Scholar]
- Pittman K. A., Lakshmanan S., Bryant M. P. Oligopeptide uptake by Bacteroides ruminicola. J Bacteriol. 1967 May;93(5):1499–1508. doi: 10.1128/jb.93.5.1499-1508.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prins R. A., van Hal-Van Gestel J. C., Counotte G. H. Degradation of amino acids and peptides by mixed rumen micro-organisms. Z Tierphysiol Tierernahr Futtermittelkd. 1979 Dec;42(6):333–339. doi: 10.1111/j.1439-0396.1979.tb01226.x. [DOI] [PubMed] [Google Scholar]
- Russell J. B. Fermentation of Peptides by Bacteroides ruminicola B(1)4. Appl Environ Microbiol. 1983 May;45(5):1566–1574. doi: 10.1128/aem.45.5.1566-1574.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell J. B., Sniffen C. J., Van Soest P. J. Effect of carbohydrate limitation on degradation and utilization of casein by mixed rumen bacteria. J Dairy Sci. 1983 Apr;66(4):763–775. doi: 10.3168/jds.S0022-0302(83)81856-6. [DOI] [PubMed] [Google Scholar]
- Wallace R. J., Kopecny J. Breakdown of diazotized proteins and synthetic substrates by rumen bacterial proteases. Appl Environ Microbiol. 1983 Jan;45(1):212–217. doi: 10.1128/aem.45.1.212-217.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw F. G., Bruce L. A., Eadie J. M., Shand W. J. 2-Aminoethylphosphonic acid concentrations in some rumen ciliate protozoa. Appl Environ Microbiol. 1983 Oct;46(4):951–953. doi: 10.1128/aem.46.4.951-953.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright D. E. Metabolism of peptides by rumen microorganisms. Appl Microbiol. 1967 May;15(3):547–550. doi: 10.1128/am.15.3.547-550.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron A., Mlynar D., Berger A. A dipeptidocarboxypeptidase from E. coli. Biochem Biophys Res Commun. 1972 May 26;47(4):897–902. doi: 10.1016/0006-291x(72)90577-3. [DOI] [PubMed] [Google Scholar]



