Strapline
The National Center for Biotechnology Information has created the dbGaP public repository for individual-level phenotype, exposure, genotype, and sequence data, and the associations between them. dbGaP assigns stable, unique identifiers to studies and subsets of information from those studies, including documents, individual phenotypic variables, tables of trait data, sets of genotype data, computed phenotype-genotype associations and groups of study subjects who have given similar consents for use of their data.
Introduction
The technical advances and declining costs for high-throughput genotyping afford investigators fresh opportunities to do increasingly complex analyses of genetic associations with phenotypic and disease characteristics. The leading candidates for such genome wide association studies (GWAS) are existing large-scale cohort and clinical studies that collected rich sets of phenotype data. To support investigator access to data from these initiatives at the National Institutes of Health (NIH) and elsewhere, the National Center for Biotechnology Information (NCBI) has created a database of Genotypes and Phenotypes (dbGaP) with stable identifiers that make it possible for published studies to discuss or cite the primary data in a specific and uniform way. dbGaP provides unprecedented access to the large-scale genetic and phenotypic datasets required for GWAS designs, including public access to study documents linked to summary data on specific phenotype variables, statistical overviews of the genetic information, position of published associations on the genome, and authorized access to individual-level data.
The purposes of this description of dbGaP are three-fold: (1) to describe dbGaP's functionality for users and submitters; (2) to describe dbGaP's design and operational processes for database methodologists to emulate or improve upon; and (3) to reassure the lay and scientific public that individual-level phenotype and genotype data are securely and responsibly managed.
dbGaP accommodates studies of varying design. It contains four basic types of data: (1) Study documentation, including study descriptions, protocol documents, and data collection instruments, such as questionnaires; (2) Phenotypic data for each variable assessed, at both an individual level and in summary form; (3) Genetic data, including study subjects' individual genotypes, pedigree information, fine mapping results, and resequencing traces; and (4) Statistical results, including association and linkage analyses, when available.
To protect the confidentiality of study subjects, dbGaP accepts only de-identified data and requires investigators to go through an authorization process in order to access individual-level phenotype and genotype datasets. Summary phenotype and genotype data, as well as study documents, are available without restriction.
Phenotypes are described in a structured narrative framework
Traditionally, clinical phenotype data have only been shared among a limited group of collaborating researchers — a model that supports documentation of protocols, phenotypes, variables and analysis through paper-based manuals and forms, personal communication or, more recently, electronic access privileges to a project data coordinating center. A centralized resource model, designed specifically for broad data distribution, requires electronically accessible explanations of variables tied closely with the data tables. dbGaP addresses that goal by compiling all available descriptive metadata from participating clinical studies — documentation of protocols, data collection forms, manuals of procedure, and the corpus of statistical analyses — putting it in electronic form, and creating explicit links between measured phenotypic variables and related documentation (e.g., the measurement for blood pressure at time point 1 is linked to protocol instructions on how and when to take the measurement).
Tiered access for public and authorized users
dbGaP is divided into public and authorized-access sections for aggregate and individual-level data, respectively. The public dbGaP website provides a rich set of descriptive statistics for each phenotype variable, allowing users to assess whether the individual-level data may be relevant to their research. Users with required bona fides may download individual-level phenotypic data and genotype files, as well as genotype chip intensity files and pedigree information, for use in approved research. Authorization for access to clinical data obligates the investigator and her institution to obey data use restrictions dictated by participant informed consent agreements and to comply with requirements detailed in a governing Data Use Certification (DUC) document. Policy details for the GAIN project (1) nicely illustrate the process for GWAS studies in general.
Public Access
The public interface at http://view.ncbi.nlm.nih.gov/dbgap allows users to browse and search study metadata, phenotype variable summary information, documentation, and those association analyses that are in the public domain. It is hoped that this data will stimulate new hypotheses and help investigators identify those datasets that are suitable for research. Users can quickly navigate to any study and browse or search its components; those interested in specific diseases can browse dbGaP using disease terms linked to the Medical Subject Headings vocabulary. Users interested in a particular phenotype can use the search interface to query text words against phenotypic variable names and descriptions or against protocols and data collection forms, which contain explicit links to phenotypes.
Authorized Access
Authorization for access to individual-level data files is obtained via the dbGaP authorized access portal at http://view.ncbi.nlm.nih.gov/dbgap-controlled. Entry into the system requires that investigators have either an NIH eRA Commons Account (extramural researchers) or an NIH log in (intramural researchers) and that they be classified as Principal Investigators. Non-PI's cannot independently apply for access to individual-level data; however, they can be approved for local access to downloaded data files within the PI's lab if they are listed as collaborating investigators on a PI's application. Although NCBI provides the interface for requests and downloads, data authorization decisions are made by the NIH institute that sponsors each study in dbGaP. The dbGaP authorized access portal authenticates users requesting access against the NIH eRA Commons system, provides the necessary forms, and forwards completed requests to the appropriate NIH Data Access Committee (DAC) for review. DAC review criteria varies by program; however, all reviews seek to ensure that the stated research purposes are compatible with participant consent and that the PI and her institution will abide by the study's data use certification and terms of use. Once access has been granted, researchers can log into the system and download the de-identified individual-level data files for which they have approval.
Publication
For many studies in dbGaP, the investigators who contributed the data will retain the exclusive right to publish analyses of the dataset for a defined period of time following release in dbGaP, typically 9 or 12 months. The date that the exclusive publication rights expire is called the embargo release date in dbGaP. During this period of exclusivity, other investigators may be granted access to download and analyze the data, but they are expected not to seek publication of their analyses or conclusions until the embargo release data. Embargo release dates are provided in several places: (1) the public dbGaP home page's list of studies; (2) the public dbGaP summary pages for each study; and (3) file download manifests that accompany each authorized-use data download.
Data Security
High density genomic data, even when de-identified, remain unique to the individual and could potentially be linked to a specific person if used in conjunction with other databases, hence the need for security around the storage, distribution and use of these data. Lowrance and Collins (2) provide a detailed exposition of the identifiability issues surrounding these data and conclude that protecting the privacy and confidentiality of participant research data is a shared responsibility between submitters, repositories and authorized users. These data must be managed with care to maintain public trust.
In accord with these principles, NCBI only releases de-identified data as encrypted files to authorized users. It is the responsibility of each PI to establish a secured computing facility for her local use of the data. Best practices for configuring a secure network are available at http://www.ncbi.nlm.nih.gov/projects/gap/pdf/dbgap_2b_security_procedures.pdf.
The goal of this process is to ensure that data provided by the NIH is kept sufficiently secure and not released to any person not permitted to access the data, either through malicious or inadvertent means. To accommodate these requirements, systems housing authorized-access data must not be directly accessible from the internet, and the data must not be posted on any web or ftp server. Data placed on shared systems must be secured and access must be limited to those involved in the research for which the data has been requested. If data is stored on laptops or removable devices, those devices must be encrypted.
Authorized data includes all instances of individual-level data. When specific studies are subject to an embargo release date, authorized data also includes certain summary data, such as the results of pre-computed association or genetic linkage tests. These summary components are distributed through the authorized access mechanism until the embargo release date has passed. Afterwards, summary data will be freely available through the public dbGaP FTP site.
dbGaP accessioned objects
The accessioned objects of dbGaP include studies, phenotype trait tables, phenotype variables, documents, genotypes and analyses (Figure 1).
Figure 1. Accession numbers in dbGaP.
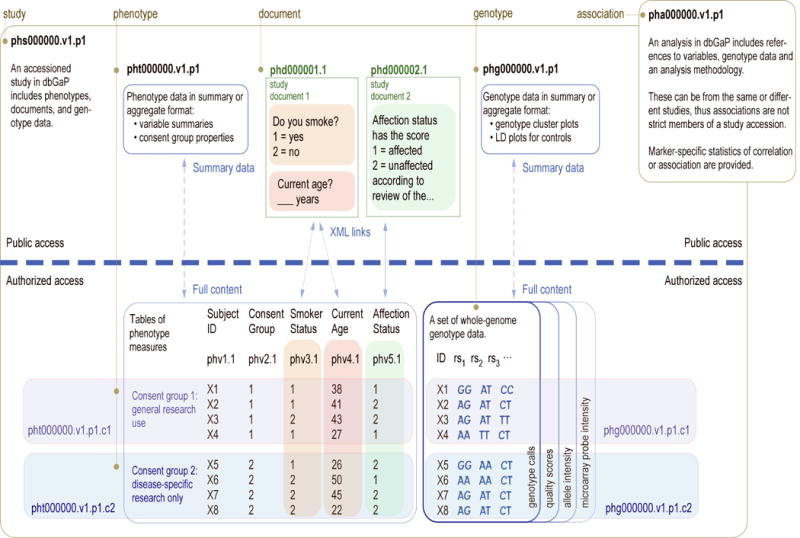
are created separately for a study and its subordinate objects: phenotype variables, phenotype trait tables, documents, and genotype datasets, prefixed a phs, phv, pht, phd, and phg, respectively. Accession numbers have suffixes “v” for data version, “p” for participant set version, and “c” for consent group version. Phenotype and genotype data are distributed as both public summary records and individual-level data that require authorization to use. Associations between genotypes and a phenotype trait of interest are not subordinate objects of a study, as an analysis may include data components from several studies simultaneously.
A Study is the largest unit of submitted content organized and accessioned by dbGaP. The organization of studies in the database can be hierarchical: a top-level study may contain sub-studies reflecting an intuitive or historical grouping of the data. dbGaP generally includes a high-level description of the study, a timeline or history, a bibliography of related published articles, and attribution for study investigators. In addition, studies can be browsed by tagged disease terms. The database is designed to accommodate studies with any number of sub-studies, phenotypic variables, documents, genotype batches, and analyses.
Each study is assigned a unique, stable, and versioned identifier prefixed by ‘phs,’ indicating a phenotype study. The ID is suffixed by both a version number (.v#) that increases when changes occur to data columns (phenotype values) and a participant set number (.p#) that increases when the number of individuals in a set changes due to alterations in informed consent status. An example of a study accession in dbGaP is phs000001.v1.p1, where v1 indicates the data version and p1 indicates the participant set version. To comply with current subject consents, authorized downloads of individual-level data will be limited to only the most recent participant set, both for current and previous versions.
Phenotype Variables
A generic database schema is used to describe measured traits from tabular submission files, where columns are phenotypic variables and rows are individuals. Every cell in a data table is stored in the database and mapped to the appropriate phenotype variable and individual. Public summaries of phenotypes and genotypes are prepared and displayed from these tables without exposing individual-level details. Phenotype variable summary descriptions provided by the submitter via a data dictionary include variable name, description, unit, and a list of any coded responses. Variable data type (text strings, integers, decimals, and dates) is automatically determined for each column by calculating what type is in the majority. Conflicts between submitted and calculated data types are reconciled by dbGaP curators in consultation with each submitter.
For each phenotype variable, several descriptive statistics are calculated: mean, median, standard deviation, minimum value, and maximum value. Categorical variables are reported with the frequency of each response. The number of missing (null) values for each phenotype measurement is listed along with the overall number of individuals in the dataset (n). Individuals in datasets are separately classified by sex, case-control status, and consent category. When appropriate, variable distributions are presented separately for cases and controls, with accompanying statistics for deviation of cases from controls. Phenotypic variables are assigned ‘phv’ (phenotype variable) identifiers along with increasing numeric suffixes for new versions and participant sets, similar to studies. Document parts, such as clinical protocols, play an integral role in defining phenotypic variables in dbGaP and aid a user's understanding of a variable. Thus, changes to documents or other metadata that significantly alter the meaning of a variable also result in a new variable version. Changes in participant sets will alter the statistical summary of values and prompt the regeneration of all versions of data files to reflect the new consent structure. Although individual-level data from obsolete participant sets will not be available for authorized download, prior summaries will be maintained on the public website.
Data Submission
There are three requirements that must be met before a study can be submitted to dbGaP: (1) PIs and their institutions must certify that all applicable laws and regulations have been followed; (2) the data submitted to dbGaP are compliant with U.S. federal privacy regulations, e.g. the Health Insurance Portability and Accountability Act (HIPAA) of 1996 and Title 45 of the Code of Federal Regulations; and (3) if the study is not sponsored by NIH, the investigators must reach an agreement with an NIH Institute to have a Data Access Committee manage data access requests for the study data.
After these assurances are met, dbGaP staff will work with the primary investigator to correctly reflect the study's logical organization, documentation, phenotype data sets and individual informed consent categories. While NCBI works with the primary investigator to identify and remove any detectable errors in the data, NCBI does not warrant the accuracy, quality or fitness of dbGaP data for any particular purpose.
Study Documents
The dbGaP document system formats, accessions, indexes, and displays all submitted study documentation in such a way that (1) links are created between the variable values in the database and their references in the documents; (2) document representation can be rendered in a web browser for easy navigation between variable summary report pages and their referencing documents; and (3) a framework is established that will support future generation of web-based forms and questionnaires that capture variable data directly into dbGaP.
To achieve these goals documents are encoded using XML (eXtensible Markup Language), which allows semantic annotation of documents and is used to link phenotypes with the parts of documents that describe how they were measured. This type of encoding lends itself naturally to creating multiple output streams (such as HTML and PDF), and allows for later reuse and repurposing of the documents. (3, 4)
Document models and markup
NCBI created and maintains the NLM Archiving and Interchange XML TagSet and the NLM Archiving and Interchange DTD, a schema defined using elements from the TagSet to markup journal articles in PubMed Central, NLM's electronic archive of full-text life sciences journal literature. Extensions to the NLM DTD that support study documents, like protocols and questionnaires, are available (see Supplementary Note). Study documents receive a unique stable identifier, along with the ‘phd’ prefix that indicates a phenotype document, and a version suffix (ie: phd000001.1); this full identifier should be cited in publications.
Genotypes
dbGaP archives genotype data from study samples and distributes these data, providing investigators with direct access to raw data on which to apply best practices and develop new techniques for analysis. Unfiltered genotype datasets include such raw data as microarray probe intensity data, normalized allele-specific signal intensities, and pre-generated genotype cluster plots, as well as vendor-supplied genotype calls. As investigators apply new techniques, such as improved genotype calling algorithms, these results can be incorporated into dbGaP as additional versions of the genotype data. Prior to release of genotype data several checks are applied to confirm sample tracking and family relationships, including Mendelian inheritance, gender agreement with the manifest file, comparison with previous genotypes, and checks for unexpected duplicate samples. Samples flagged as presenting genotypes from the wrong individual are excluded from the data release. Genotypes are provided in a compact matrix format, and may be available in a more expansive format with one genotype per row. All genotypes are reported in genomic orientation. Sets of genotypes on a particular platform across all individuals are assigned a unique and stable ‘phg’ identifier along with a version and participant set.
Data Cleaning and Quality Control
The thresholds used during the data cleaning process, and the resulting selection of which individuals and markers to use as input to downstream analysis, profoundly affect the results as well as the reproducibility of analyses. Thus, dbGaP works collaboratively with data submitters and the study advisory committee to create a documented process of data cleaning as well as the filtered dataset used to generate the analyses (discussed in the next section). Sample quality control metrics include genotype call rate, sex prediction, average heterozygosity, likelihood of observed genotypes, and Mendelian error rate. Marker-specific quality control metrics include Mendelian error rate per marker, Hardy-Weinberg Equilibrium test p-value, call rate, duplicate concordance rate, concordance rate with known genotypes, minor allele frequency, and vendor-supplied genotype quality score. Summary results and selected thresholds are provided publicly when available, while detailed and embargoed results are components of the authorized-access release.
The process of selecting quality control thresholds and analysis may be iterative. A shared pattern of quality control metrics among highly significant markers revealed by a preliminary association test may signal a common false-positive error mode. dbGaP provides several metrics to detect statistical inflation in the data, e.g. quantile-quantile plots of Cochran-Armitage test statistics (5) and genomic control lambda values (6), for consideration by the study investigators and advisory committee as they optimize data cleaning thresholds. These conclusions are documented as part of the genotype release, and the resulting dataset is provided as the filtered release set.
In addition to the filtered set of high-quality genotype data, all remaining genotype data that fails Hardy-Weinberg or Mendelian inheritance checks will be available for investigator use, with flags for the set of quality filters they failed to pass. These data that failed quality checks are provided because they may occasionally contain information of value, but they should be used with the requisite cautions.
Analyses
Analyses between genotypes and phenotypes can be calculated using different statistical methods and different underlying data, such as SNP markers or sequence traces. Each analysis, defined as the set of computed association statistics for a single phenotype variable against all genetic markers, is assigned a unique and stable accession.version (e.g. ., pha000001.1). dbGaP is designed to be flexible enough to represent any analysis done where the output is a set of statistics mapped to genomic coordinates. While dbGaP staff has the ability to do statistical computation, as was done at the request of the National Eye Institute for the AREDS study (http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/analysis.cgi?id=pha000001), we would prefer to store, represent, and distribute analyses done by study domain experts.
Analysis records directly link individual phenotype and genotype measures with a sufficiently detailed account of the statistical method used to allow others to reproduce the analysis. Secondary published analyses can also be posted and distinguished from the analysis of the original submitter, allowing for comparison of different analytical methods. dbGaP displays analyses on a new genome browser, Supplementary Figure 1, that facilitates filtering by various statistics and quick access to regions of possible significance. Multiple genome tracks can be overlaid within the browser to view analysis results in the same context, and individual marker statistics can be explored for numerical properties (genotype counts) and genotype quality (allele intensity cluster plots). Users can download analysis results for local use or for display in a different browser.
An example of an analysis between cases and controls (for the derived diagnosis of age-related macular degeneration (AMD) status in the NEI AREDS study) can be accessed at: http://www.ncbi.nlm.nih.gov/SNP/GaP.cgi?rm=genomeTraitView&test_id=1&method_id=3. For those studies with public analyses, the analysis browser is accessed through a link on the analysis report page, which is linked from the main study description page; the analysis report page describes the analysis and the statistical method used, and provides links to the phenotypic variable report page for the trait analyzed.
dbGaP Participating Studies
dbGaP is a general repository for studies examining the association between phenotype and genotype. At the time of writing, dbGaP has 12 public studies at various stages of completion. Several other genome-wide association studies are being processed and will be made public in the next several months. Table 1 lists the studies that are expected to be available in dbGaP through 2007 and into 2008; together they will include well over 100,000 individuals measured for thousands of distinct phenotypic variables. In addition to genome-wide association and linkage studies, dbGaP will store medical resequencing studies, fine-mapping studies, and literature and database reviews of disease-causing mutations. There are obvious links between these types of studies, some of which will be follow-up analyses of other studies in dbGaP.
Table 1.
dbGaP participating projects.
| Projected Availability | # studies | Study Name / Disease Focus | Sponsor | Type | Number of Participants |
|---|---|---|---|---|---|
| Nov-06 | 1 | AREDS | NEI | Case-Control GWAS | 600 |
| Nov-06 | 1 | Parkinsonism | NINDS/NIA | Case-Control GWAS | 2,573 |
| Jun-07 | 1 | ADHD | GAIN | Trio GWAS | 2,874 |
| Aug-07 | 2 | Diabetic Nephropathy | GAIN | Case-Control GWAS | 1,835 |
| Sep-07 | 0 | GeneLink | NHLBI | Multipoint linkage analyses | n.d. |
| Sep-07 | 1 | Stroke | NINDS | Case-Control GWAS | 1,555 |
| Sep-07 | 1 | Motor Neuron Disease/ALS | NINDS | Case-Control GWAS | 1,876 |
| Sep-07 | 1 | LEAPS | MJFF | Tiered case-control GWAS | 886 |
| Sep-07 | 1 | Major Depression | GAIN | Case-Control GWAS | 3,720 |
| Oct-07 | 1 | Framingham SHARe | NHLBI | Family-Based Longitudinal GWAS | ∼9,500 |
| Oct-07 | 1 | Psoriasis | GAIN | Case-Control GWAS | 2,898 |
| Nov-07 | 2 | DCCT/ EDIC | NIDDK | Longitudinal GWAS | |
| Dec-07 | 1 | Schizophrenia | GAIN | Case-Control GWAS | 2,909 |
| Dec-07 | 1 | Bipolar Disorder | GAIN | Case-Control GWAS | 2,400 |
| Early 2008 | 1 | Alzheimer's Disease | NIA | Case-Control GWAS | 10,000 |
| Late 2008 | 8 | 8 GEI Studies | NHGRI | TBD | >30,000 |
| Late 2008 | 0 | Medical Resequencing, phase 1 | NHGRI | TBD | ∼15,000 |
| Late 2008 | 1 | MESA SHARe | NHLBI | Longitudinal GWAS | 8,000 |
| TBD | 0 | Women's Health Genome Study | Brigham and Women's Hospital, NHLBI, Amgen | Longitudinal GWAS | ∼28,000 |
| TBD | 0 | Cornelia de Lange Study | CETT | Clinical Diagnostic | TBD |
| TBD | 0 | Duchenne Muscular Dystrophy Study | CETT | Clinical Diagnostic | TBD |
| TBD | 0 | Kallman Syndrome | CETT | Clinical Diagnostic | TBD |
| TBD | 0 | TSC2 | CETT | Clinical Diagnostic | TBD |
| TBD | 0 | OMIM | Johns Hopkins University | Literature | TBD |
| TBD | 0 | Dystrophin Mutation Study | CETT | LSMDB | TBD |
|
| |||||
| 25 | |||||
Projected availability is for planning purposes only. Specific availability dates will be guided by the data access policy for each particular study. AREDS - Age-Related Eye Disease Study; CETT - Collaboration, Education and Test Translation Program; DCCT - Diabetes Control and Complications Trial; EDIC - Epidemiology of Diabetes Intervention and Complications; GAIN - Genetic Association Information Network; GEI - Genes, Health and Environment Initiative; GWAS -Genome-Wide Association Study; LEAPS - Linked Efforts to Accelerate Parkinson's Solutions; LSMDB - Locus-Specific Mutation Database; MESA - Multi-Ethnic Study of Atherosclerosis; MJFF - Michael J. Fox Foundation; NEI - National Eye Institute; NHGRI - National Human Genome Research Institute; NIA - National Institute of Aging; NIDDK - National Institute of Diabetes & Digestive & Kidney Disease; NINDS - National Institute of Neurological Disorders and Stroke; OMIM - Online Mendelian Inheritance in Man; SHARe - SNP Health Association Resource; TBD -To be determined.
Publication References to dbGaP Studies
dbGaP provides a critical contribution to the scientific community by establishing a central repository for uniform representation of clinical phenotype, genotype, and analysis data that can be accessed and browsed through a common interface. Published analyses of data from dbGaP should explicitly reference unique and stable accession numbers in method descriptions and an explicit acknowledgement of each utilized study as directed by the Data Use Certification. Publications that reference a dbGaP analysis data set should reference the unique and stable pha identifier assigned to that analysis. Embargoed data has an Embargo Release Date clearly indicated for each set of downloaded data. Studies subject to embargo release dates are clearly indicated on the public dbGaP search page, and each study's respective description page.
Future directions
The dbGaP database schema is designed to generally store phenotype data that can be associated with other relevant data types, including genotype, sequence-derived genotypes and haplotypes, gene expression, proteomic, metabolomic, and epigenetic data. While current GWAS activity is focused on high-density micro array genotype data, new programs such as the National Human Genome Research Institute (NHGRI) Medical Resequencing initiative will couple phenotype data with new technologies such as short read sequencing. Additionally, the database is not restricted to human data or even to clinical data. In the future, the underlying architecture of dbGaP could be used as a repository for model organisms of human disease and for data on other non-clinical traits, such as agricultural phenotypes.
URLs
dbGaP public access homepage: http://view.ncbi.nlm.nih.gov/dbgap
dbGaP authorized access system: http://view.ncbi.nlm.nih.gov/dbgap-controlled
NIH GWAS policy: http://grants.nih.gov/grants/guide/notice-files/NOT-OD-07-088.html
Medical Subject Heading Vocabulary: http://www.nlm.nih.gov/mesh
NLM Archiving and Interchange DTD: http://dtd.nlm.nih.gov/
BOX 1.
dbGaP Key Features
The database of Genotypes and Phenotypes (dbGaP) was developed to archive and distribute the results of studies that have investigated the association between genotype and phenotype.
Content for each submitted study is comprehensive, including study documents (protocols, questionnaires, etc.), phenotype measures, genetic data derived from genotyping arrays or sequencing experiments, and details of statistical associations between phenotypes and genotypes.
Phenotype measurements are linked to the related study documentation. Measurement data are accepted in various formats, and are not further standardized by NCBI.
All content receives stable, unique public accession IDs, allowing specific data or studies to be cited in publications or pointed to from within other bioinformatics resources.
Data are distributed in a uniform download format across studies.
All individual-level data are de-identified by the submitter; individuals are represented by coded IDs, and NIH does not hold or have access to the keys.
Data Access Structure
Study protocols and summary phenotype and genotype data are available to the public without restrictions on use.
Access to individual-level data requires preauthorization from sponsoring NIH programs. Use of the data is limited to approved research activities, and must follow the basic principles set forth in the NIH policy for Genome-Wide Association Studies.
Association data may be limited to the authorized-access portion of dbGaP during an initial embargo period designed to give submitters exclusivity on publishing their data. After expiration of the embargo period, the association data will move to the public portion of the database.
Authorized-access data should be used in a secured computing environment and in accord with the applicable terms of use for the specific dataset(s), e.g. restrictions on use defined by participant informed consent agreements, prohibitions on redistributing the data, and complying with restrictions on the public dissemination of results during embargo periods.
Supplementary Material
Acknowledgments
Thanks to T. Manolio, J. Coleman, F Collins and C. O'Donnell for useful comments and discussion. This research was supported by the Intramural Research Program of the NIH, National Library of Medicine (NLM).
References
- 1.The GAIN Collaborative Research Group. New Models of Collaboration in Genome-Wide Association Studies: The Genetic Association Information Network (GAIN) Nat Genet. 2007;39:1045–1051. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 2.Lowrance WW, Collins FS. ETHICS: Identifiability in Genomic Research. Genomic data are unique to the individual and must be managed with care to maintain public trust. Science. 2007;317:600–602. doi: 10.1126/science.1147699. [DOI] [PubMed] [Google Scholar]
- 3.Harold ER. XML Bible. 2. Hungry Minds; New York, Indianapolis, Cleveland: 2001. [Google Scholar]
- 4.World Wide Web Consortium (W3C) Bray Tim, Paoli Jean, Sperberg-McQueen CM, Maler Eve, Yergeau Francois., editors. Extensible Markup Language (XML) 1.0. (Fourth) 2006 http://www.w3.org/TR/REC-xml/
- 5.Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, Smink LJ, Lam AC, Ovington NR, Stevens HE, Nutland S, Howson JM, Faham M, Moorhead M, Jones HB, Falkowski M, Hardenbol P, Willis TD, Todd JA. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 6.Devlin B, Roeder K. Genomic Control for Association Studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


