Abstract
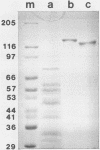
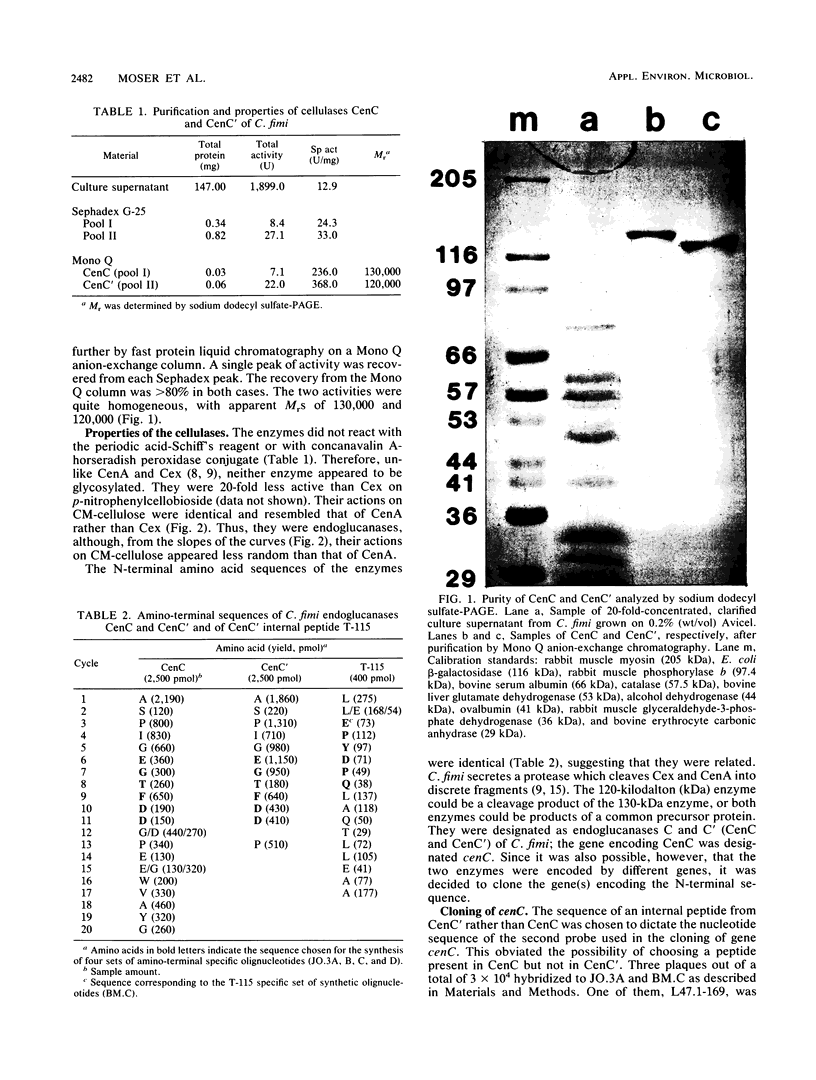
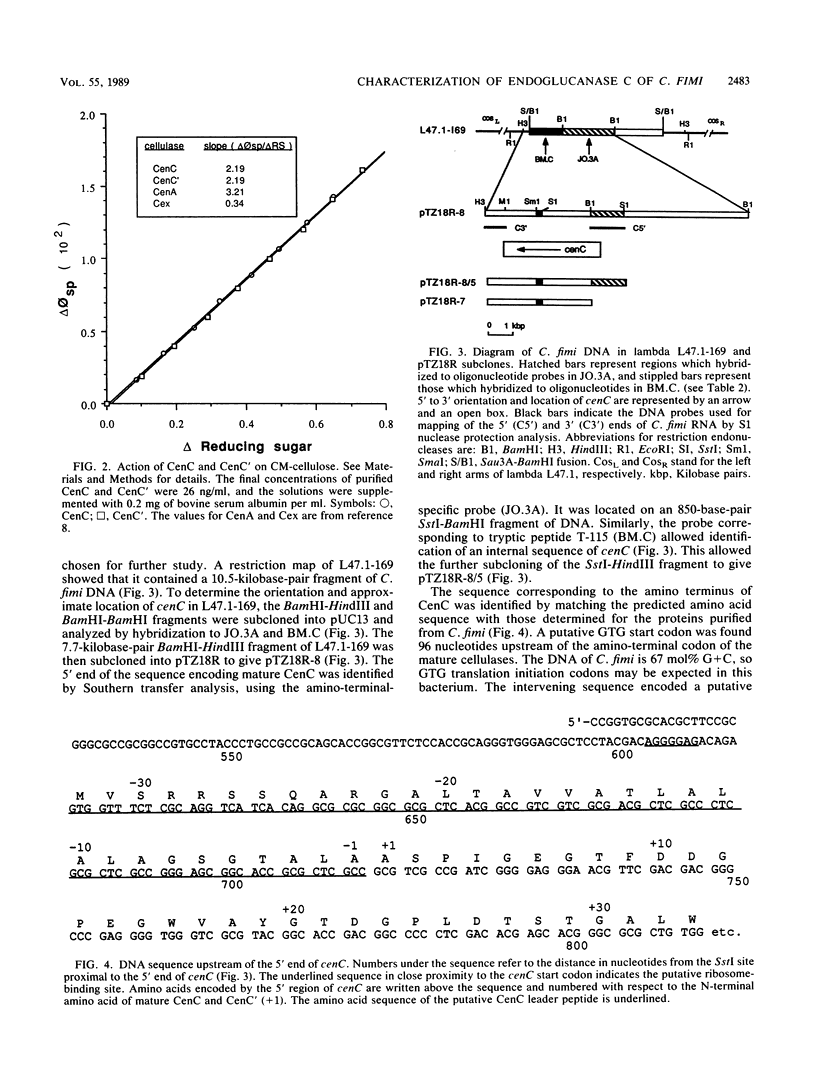
Two nonglycosylated endoglucanases which bind to Sephadex were purified from culture supernatants of Cellulomonas fimi grown on microcrystalline cellulose. Their Mrs were 120,000 and 130,000. The N-terminal amino acid sequences of the enzymes were identical, suggesting that the enzymes were related. A DNA fragment encoding this N-terminal sequence was cloned in Escherichia coli. The nucleotide sequence corresponding to the N-terminal amino acid sequence was preceded by a sequence encoding a typical leader peptide. Transcripts hybridizing to the cloned fragment were detected in total RNA isolated from C. fimi cells grown on carboxymethyl cellulose but not from cells grown on glycerol or glucose. Transcription started at a cluster of sites 53 to 59 nucleotides upstream of a GUG translation initiation codon and terminated at either of two closely spaced C residues immediately downstream of a region of potential secondary structure. The size of the transcript was approximately 3.5 kilobases, sufficient to encode a polypeptide of 130 kilodaltons. The 130-kilodalton polypeptide is designated endoglucanase C (CenC), and the gene encoding it is designated cenC.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Béguin P., Eisen H. Purification and partial characterization of three extracellular cellulases from Cellulomonas sp. Eur J Biochem. 1978 Jul 3;87(3):525–531. doi: 10.1111/j.1432-1033.1978.tb12403.x. [DOI] [PubMed] [Google Scholar]
- Dale R. M., McClure B. A., Houchins J. P. A rapid single-stranded cloning strategy for producing a sequential series of overlapping clones for use in DNA sequencing: application to sequencing the corn mitochondrial 18 S rDNA. Plasmid. 1985 Jan;13(1):31–40. doi: 10.1016/0147-619x(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Gilkes N. R., Langsford M. L., Kilburn D. G., Miller R. C., Jr, Warren R. A. Mode of action and substrate specificities of cellulases from cloned bacterial genes. J Biol Chem. 1984 Aug 25;259(16):10455–10459. [PubMed] [Google Scholar]
- Gilkes N. R., Warren R. A., Miller R. C., Jr, Kilburn D. G. Precise excision of the cellulose binding domains from two Cellulomonas fimi cellulases by a homologous protease and the effect on catalysis. J Biol Chem. 1988 Jul 25;263(21):10401–10407. [PubMed] [Google Scholar]
- Greenberg N. M., Warren R. A., Kilburn D. G., Miller R. C., Jr Regulation and initiation of cenB transcripts of Cellulomonas fimi. J Bacteriol. 1987 Oct;169(10):4674–4677. doi: 10.1128/jb.169.10.4674-4677.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg N. M., Warren R. A., Kilburn D. G., Miller R. C., Jr Regulation, initiation, and termination of the cenA and cex transcripts of Cellulomonas fimi. J Bacteriol. 1987 Feb;169(2):646–653. doi: 10.1128/jb.169.2.646-653.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Hazlewood G. P., Barker P. J., Gilbert H. J. Conserved reiterated domains in Clostridium thermocellum endoglucanases are not essential for catalytic activity. Gene. 1988 Sep 15;69(1):29–38. doi: 10.1016/0378-1119(88)90375-7. [DOI] [PubMed] [Google Scholar]
- Kotoujansky A., Diolez A., Boccara M., Bertheau Y., Andro T., Coleno A. Molecular cloning of Erwinia chrysanthemi pectinase and cellulase structural genes. EMBO J. 1985 Mar;4(3):781–785. doi: 10.1002/j.1460-2075.1985.tb03697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Langsford M. L., Gilkes N. R., Singh B., Moser B., Miller R. C., Jr, Warren R. A., Kilburn D. G. Glycosylation of bacterial cellulases prevents proteolytic cleavage between functional domains. FEBS Lett. 1987 Dec 10;225(1-2):163–167. doi: 10.1016/0014-5793(87)81150-x. [DOI] [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- MacKay R. M., Lo A., Willick G., Zuker M., Baird S., Dove M., Moranelli F., Seligy V. Structure of a Bacillus subtilis endo-beta-1,4-glucanase gene. Nucleic Acids Res. 1986 Nov 25;14(22):9159–9170. doi: 10.1093/nar/14.22.9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGavin M., Forsberg C. W. Isolation and characterization of endoglucanases 1 and 2 from Bacteroides succinogenes S85. J Bacteriol. 1988 Jul;170(7):2914–2922. doi: 10.1128/jb.170.7.2914-2922.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai R., Horinouchi S., Beppu T. Cloning and nucleotide sequence of a cellulase gene, casA, from an alkalophilic Streptomyces strain. Gene. 1988 May 30;65(2):229–238. doi: 10.1016/0378-1119(88)90459-3. [DOI] [PubMed] [Google Scholar]
- O'Neill G., Goh S. H., Warren R. A., Kilburn D. G., Miller R. C., Jr Structure of the gene encoding the exoglucanase of Cellulomonas fimi. Gene. 1986;44(2-3):325–330. doi: 10.1016/0378-1119(86)90197-6. [DOI] [PubMed] [Google Scholar]
- Owolabi J. B., Beguin P., Kilburn D. G., Miller R. C., Warren R. A. Expression in Escherichia coli of the Cellulomonas fimi Structural Gene for Endoglucanase B. Appl Environ Microbiol. 1988 Feb;54(2):518–523. doi: 10.1128/aem.54.2.518-523.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ståhlberg J., Johansson G., Pettersson G. A binding-site-deficient, catalytically active, core protein of endoglucanase III from the culture filtrate of Trichoderma reesei. Eur J Biochem. 1988 Apr 5;173(1):179–183. doi: 10.1111/j.1432-1033.1988.tb13982.x. [DOI] [PubMed] [Google Scholar]
- Tomme P., Van Tilbeurgh H., Pettersson G., Van Damme J., Vandekerckhove J., Knowles J., Teeri T., Claeyssens M. Studies of the cellulolytic system of Trichoderma reesei QM 9414. Analysis of domain function in two cellobiohydrolases by limited proteolysis. Eur J Biochem. 1988 Jan 4;170(3):575–581. doi: 10.1111/j.1432-1033.1988.tb13736.x. [DOI] [PubMed] [Google Scholar]
- Warren R. A., Beck C. F., Gilkes N. R., Kilburn D. G., Langsford M. L., Miller R. C., Jr, O'Neill G. P., Scheufens M., Wong W. K. Sequence conservation and region shuffling in an endoglucanase and an exoglucanase from Cellulomonas fimi. Proteins. 1986 Dec;1(4):335–341. doi: 10.1002/prot.340010407. [DOI] [PubMed] [Google Scholar]
- Whittle D. J., Kilburn D. G., Warren R. A., Miller R. C., Jr Molecular cloning of a Cellulomonas fimi cellulose gene in Escherichia coli. Gene. 1982 Feb;17(2):139–145. doi: 10.1016/0378-1119(82)90066-x. [DOI] [PubMed] [Google Scholar]
- Wong W. K., Gerhard B., Guo Z. M., Kilburn D. G., Warren A. J., Miller R. C., Jr Characterization and structure of an endoglucanase gene cenA of Cellulomonas fimi. Gene. 1986;44(2-3):315–324. doi: 10.1016/0378-1119(86)90196-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zacharius R. M., Zell T. E., Morrison J. H., Woodlock J. J. Glycoprotein staining following electrophoresis on acrylamide gels. Anal Biochem. 1969 Jul;30(1):148–152. doi: 10.1016/0003-2697(69)90383-2. [DOI] [PubMed] [Google Scholar]
- Zappe H., Jones W. A., Jones D. T., Woods D. R. Structure of an endo-beta-1,4-glucanase gene from Clostridium acetobutylicum P262 showing homology with endoglucanase genes from Bacillus spp. Appl Environ Microbiol. 1988 May;54(5):1289–1292. doi: 10.1128/aem.54.5.1289-1292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]