Abstract
Signaling by a variety of receptor and nonreceptor tyrosine kinases is mediated by Ras, a membrane-associated GTPase. Expression of v-Src, a transforming nonreceptor tyrosine kinase, results in Ras activation, and inhibition of Ras function in NIH 3T3 cells suppresses transformation by v-Src, indicating that in these cells Ras-dependent signaling pathways are required for v-Src to exert its biological effects. However, we show here that Ras was not activated in Rat-2 fibroblasts transformed by wild-type v-Src, or in chicken embryo fibroblasts transformed by SRX5, a v-Src mutant with a linker insertion at the major site of autophosphorylation. Expression of a dominant-negative mutant of Ras completely inhibited the ability of v-Src to activate the mitogen-activated protein kinase ERK2, which is downstream of Ras. However, dominant-negative Ras did not suppress transformation by v-Src as judged by a variety of criteria. Thus, v-Src can transform at least some cell types in the absence of Ras activation or Ras-stimulated ERK2 activity, and in these cells activation of Ras-independent signaling pathways must therefore be sufficient for transformation.
Expression of the transforming tyrosine kinase v-Src activates a number of signaling proteins, including the small GTPase Ras (1). Ras is a 21-kDa guanine nucleotide binding protein that functions as a molecular switch linking upstream activators, such as growth factor receptor and nonreceptor tyrosine kinases, to several downstream effectors (2). Ras transmits a signal when bound to GTP, and its biological activity is regulated by proteins that stimulate the exchange of GDP and GTP, as well as by proteins that stimulate its intrinsic GTPase activity. When Ras is in the GTP-bound state, its effector domain forms a high affinity binding site that interacts with specific effector molecules. The most well characterized effector of Ras is the protein serine/threonine kinase Raf (2), which functions in a kinase cascade that results in the activation of the mitogen-activated protein (MAP) kinases ERK1 and ERK2 (3). Once activated, these MAP kinases transit into the nucleus where they phosphorylate and activate transcription factors that regulate specific genes, such as c-fos (4). Thus, Ras provides a link between activated tyrosine kinases such as v-Src and the transcription of sets of genes that are important for regulating cell proliferation.
In invertebrate model systems, Ras and MAP kinase functions are necessary for the induction of gene expression in response to activation of receptor tyrosine kinases (5, 6). Similarly, Ras has been shown to be required for the malignant transformation of NIH 3T3 cells by v-Src: transformation and DNA synthesis induced by v-Src are suppressed by microinjection of neutralizing antibodies against Ras (7), by microinjection of a dominant-negative mutant Ras protein (8), or by overexpression of p120GAP, a negative regulator of Ras (9, 10). However, v-Src also activates a number of other signaling proteins, including phosphatidylinositol 3-kinase (PI 3-K) (11, 12), protein kinase C (13), and signal transducers and activators of transcription (STATs) (14), raising the possibility that other signaling pathways may also contribute to transformation by v-Src.
We report here that, in contrast to what has been observed in NIH 3T3 cells (15), Ras was not activated in Rat-2 fibroblasts transformed by wild-type v-Src or in primary chicken embryo fibroblasts (CEF) transformed by a v-Src mutant with a linker insertion at the major site of autophosphorylation. We also demonstrate that a dominant-negative mutant of Ras, while completely inhibiting the activation of ERK2, did not suppress transformation of either cell type by v-Src. These results indicate that, in these cells, activation of Ras-independent signaling pathways is sufficient for transformation.
METHODS
Cell Culture, Vector Construction, and Virus Production.
Primary cultures of CEF were prepared from 10-day-old embryos as described (16). The Rat-2-derived cell lines fpGV1-1-1, v-Src-A4, SRX5-C20, and MS1 have been described (16, 17). Focus assays, soft agar colony assays, and hexose uptake assays were performed as described (16, 18). Soft agar colonies were stained for photography by adding onto the agar 1 ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (Sigma; 0.5 mg/ml in PBS) and incubating the plates for 4–6 hr at 37°C.
To express dominant-negative Ras in CEF, a cDNA encoding the N17 mutant of H-ras (kindly provided by C. Der, Chapel Hill, NC) was subcloned into the helper-independent retroviral vector RCAS(B)-BP (19, 20), which encodes an envelope subgroup B virus. Wild-type Schmidt–Ruppin A v-src, the host-range mutant SRX5 (21), and the temperature-sensitive mutant tsUP1src (22) (kindly provided by J. Brugge, Cambridge, MA) were subcloned into the vector RCAN(A)-BP, which encodes an envelope subgroup A virus. CEF were transfected with these plasmids by polybrene/dimethyl sulfoxide shock, and virus stocks were harvested 5 or 6 days posttransfection, as described (16). To coexpress v-Src and N17Ras, cells were infected with the Ras virus 2 days prior to infection with the Src virus. Infections with subgroup B viruses were carried out in the presence of polybrene (2 μg/ml).
Rat-2 cells were transfected by electroporation with pM2N/N17ras (23) (kindly provided by G. Martin, Richmond, CA), which encodes the mutant Ras under the control of the metal-inducible metallothionein promoter, or with the empty expression vector pM2N. G418-resistant cell lines were isolated and expanded for further analysis. To express v-Src in these cells, the v-src gene was subcloned into the mammalian expression vector pBabe-Hygro (24). A helper-free ψ-2/HygroR/v-src virus was generated by transfecting ψ-2 cells (25) by electroporation with pBabe-Hygro/v-src, and harvesting virus from hygromycin-resistant clones. Rat-2 cell lines were infected with ψ-2/HygroR/v-src virus for focus assays, and hygromycin-resistant cell lines expressing v-Src were isolated for morphological analysis and soft agar colony assays.
For expression in MS1 cells, N17ras was subcloned into the mammalian expression vector pcDNA3 (Invitrogen). MS1 cells were transfected by electroporation with this construct (or the empty expression vector), and G418-resistant cell lines were isolated and expanded for further analysis.
Ras Assay.
The analysis of guanine nucleotides bound to Ras in vivo was carried out essentially as described (15), with the following modifications. Cells (≈5 × 106/100-mm dish) were serum-starved for 18 hr and then incubated for 4 hr in 4 ml phosphate-free DMEM containing 1 mCi [32P]orthophosphate (1 Ci = 37 GBq). The cells were lysed in 800 μl Ras lysis buffer (RLB: 1% Nonidet P-40/50 mM Tris, pH 7.5/10 mM MgCl2/1 mM phenylmethylsulfonyl fluoride/10 μM benzamidine/5 μM phenanthroline) with or without 1 μg anti-Ras mAb Y13–259 (Santa Cruz Biotechnology). Lysates were first cleared of free nucleotides by incubating 5 min with 0.1 ml activated charcoal (10% in PBS) that had been pre-equilibrated with 1% BSA. The cleared supernatant was then mixed with 20 μl protein A Sepharose beads [10% (wt/vol) in RLB] that had been coated with rabbit anti-rat IgG antibody. The beads were washed twice with RLB, once with 1% Nonidet P-40/0.1% SDS/20 mM Tris, pH 8.3/250 mM NaCl/ 10 mM MgCl2, once again with RLB, and once with 10 mM Tris, pH 7.5/10 mM MgCl2. Associated nucleotides were eluted by adding 20 μl elution buffer (1% SDS/20 mM Tris, pH 7.5/10 mM EDTA/100 μM GTP/100 μM GDP) and incubating at 65°C for 5 min. Eluted nucleotides were centrifuged through a 10,000 Mr cutoff membrane (UFC3 LGC; Millipore), separated on polyethyleneimine-cellulose plates (EM Science) that were developed with 1.3 M LiCl, and quantitated with a PhosphorImager (Molecular Dynamics).
Map Kinase Assay.
The activity of ERK2 was measured essentially as described (26), with the following modifications. Cells were serum-starved for 24 hr, in the absence or presence of 100 μM ZnCl2 and 2 μM CdCl2 in the case of Rat-2 cells, and lysed with lysis buffer (1% Nonidet P-40/0.5% deoxycholate/50 mM Hepes, pH 7.4/150 mM NaCl/1 mM Na3VO4/50 mM NaF/40 mM NaP2O7/5 mM EDTA/5 mM EGTA/1 mM phenylmethylsulfonyl fluoride/10 μM benzamidine/5 μM phenanthroline). The lysates were centrifuged at 14,000 × g for 10 min, and the supernatants (400 μg protein) were preincubated with 60 μl protein A Sepharose [10% (wt/vol) in lysis buffer] for 30 min. The samples were centrifuged again, and endogenous ERK2 was immunoprecipitated by incubating the supernatants with 2 μg anti-ERK2 (C-14) antibody (Santa Cruz Biotechnology) for 1 hr, and 60 μl protein A Sepharose for an additional hour. The immunoprecipitates were washed three times with lysis buffer, once with 100 mM NaCl/25 mM Tris (pH 7.5), and divided into two equal portions. One portion was resolved by SDS/PAGE and immunoblotted with the same anti-ERK2 antibody to confirm that equal amounts of ERK2 had been immunoprecipitated. The other portion was resuspended in 25 μl kinase buffer (20 mM Tris, pH 7.5/20 mM MgCl2/2 mM DTT) containing 10 μg myelin basic protein (Upstate Biotechnology, Lake Placid, NY), and reactions were initiated by adding 10 μl of 50 μM [γ-32P]ATP (10 Ci/mmol). Reactions were incubated at room temperature for 20 min and terminated by the addition of SDS/PAGE sample buffer. The reaction products were resolved on a gel containing 15% polyacrylamide, and quantitated by PhosphorImager analysis.
Immunoblots.
Cells were lysed in 1% SDS/10 mM Tris (pH 7.5) and boiled for 5 min. Equal amounts of protein were resolved by SDS/PAGE on 13% gels and transferred to polyvinylidene difluoride membranes (Millipore). The membranes were cut in half, and the appropriate segments were immunoblotted with either 1 μg/ml anti-Pan-Ras mAb (Santa Cruz Biotechnology) or 3.5 μg/ml anti-Src2–17 mAb (Microbiological Associates). The blots were then incubated with peroxidase-conjugated secondary antibody, and the immunoblotted proteins were detected by enhanced chemiluminescence (Renaissance; DuPont).
RESULTS
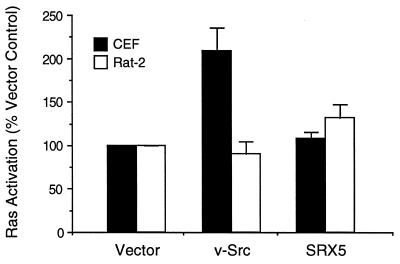
We have previously hypothesized that host-range mutants of v-Src, which are transformation-defective only in certain host cells, are defective in the activation of specific signaling pathways (16, 21). The host-range mutant SRX5, in which the major site of autophosphorylation (Tyr-416) is replaced by the sequence Ser-Arg-Asp, is fully transforming in CEF but not in Rat-2 fibroblasts, even though the mutant kinase is active in the latter cell type (16, 21). To determine if SRX5 is defective in activating Ras, we measured the fraction of GTP in the guanine nucleotides bound to Ras in both CEF and Rat-2 cells expressing wild-type v-Src or SRX5. Transformation of CEF by wild-type v-Src resulted in a 2-fold activation of Ras, whereas transformation by SRX5 did not activate Ras (Fig. 1). Transformation of Rat-2 fibroblasts by v-Src did not elevate Ras-GTP levels (Fig. 1). In control experiments, serum-starved Rat-2 cells carrying the empty vector were stimulated with serum for 5 min; this resulted in a 2-fold activation of Ras (data not shown). These experiments indicate either that activation of Ras is not necessary for the transformation of CEF or Rat-2 cells by v-Src or that only very low levels of activation are sufficient.
Figure 1.
Activation of Ras in CEF and Rat-2 cells expressing v-Src or SRX5. CEF (open bars) were infected with vector control, v-Src, or SRX5 virus 2 days before serum starvation. The Rat-2 cell lines (solid bars) fpGV1-1-1 (vector control), v-Src-A4, and SRX5-C20 were plated 24 hr before serum starvation. The activation state (% GTP-bound) of endogenous Ras was assayed as described and is expressed relative to the level observed in vector control cells. The data are expressed as the mean ± SEM of three independent experiments.
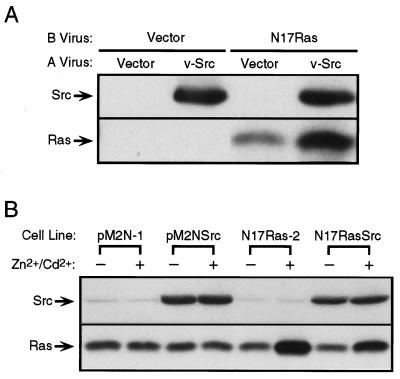
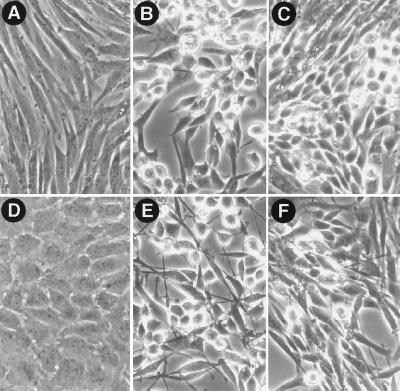
To determine if CEF can be transformed by v-Src when the activation of Ras is blocked, CEF were infected with a retrovirus encoding the dominant-negative Asn-17 mutant of H-ras, N17ras (27), and then superinfected with a retrovirus encoding v-src. Infection of CEF with the N17ras virus resulted in a high level of expression of dominant-negative Ras (Fig. 2A). The level of Ras increased further when the cells were superinfected with the v-src virus, consistent with the ability of v-Src to stimulate the Rous sarcoma virus long terminal repeat that drives expression of N17Ras in the retroviral construct (28). Expression of N17Ras slowed the growth rate of normal CEF but did not alter their morphology. The inhibition of MAP kinase activity observed in these cells (see below) confirmed that the block in Ras signaling was complete. Dominant-negative Ras did not block morphological transformation by v-Src, although some cells expressing both v-Src and N17Ras did not display the fully rounded, nonadherent morphology exhibited by the majority of cells expressing v-Src alone (Fig. 3 A–C). Immunofluorescent staining of Src and Ras in these cells indicated that the mutant Ras was expressed in >90% of the infected cells; the highest levels of Ras expression were observed in rounded, refractile cells also expressing v-Src (data not shown). Expression of N17Ras in CEF did not significantly alter the ability of v-Src to induce foci in monolayer cultures, or colonies in agar suspension cultures (Table 1), indicating that v-Src can transform CEF when the activation of endogenous Ras is blocked. In addition, v-Src stimulated the uptake of 2-deoxyglucose 5-fold in control CEF, and 4-fold in CEF expressing N17Ras (Table 1). Thus, dominant-negative Ras did not block the increase in hexose uptake induced by v-Src, although a slight decrease in the level of stimulation was observed; this correlates with our observation that CEF coexpressing v-Src and N17Ras displayed a 12- to 24-hr delay in acidification of the culture medium compared with CEF expressing v-Src alone.
Figure 2.
Expression of dominant-negative Ras in CEF and Rat-2 cells. (A) CEF were infected with an envelope subgroup B empty vector virus or a similar virus encoding N17ras, and 2 days later were superinfected with an envelope subgroup A empty vector virus or a similar virus encoding v-src. Lysates were prepared 3 days after infection with the subgroup A virus and were subjected to immunoblotting for Src and Ras as described. (B) Rat-2 cell lines stably transfected with a vector encoding N17ras under the control of a metal-inducible promoter were infected with a retrovirus encoding v-src. Infected cell lines (Rat-2/pM2NSrc and Rat-2/N17RasSrc) and uninfected control cell lines (Rat-2/pM2N-1 and Rat-2/N17Ras-2) were grown in serum-containing medium in the absence or presence of 100 μM ZnCl2 and 2 μM CdCl2 for 24 hr, and then lysed and subjected to SDS/PAGE and immunoblotting as described.
Figure 3.
Effect of N17Ras expression on morphological transformation of CEF and Rat-2 cells. (A–C) Morphology of CEF that were infected with empty vector viruses (A), empty vector subgroup B and v-src subgroup A viruses (B), or N17ras subgroup B and v-src subgroup A viruses (C). (D–F) Morphology of Rat-2 cell lines that were grown in serum-containing medium in the presence of 100 μM ZnCl2 and 2 μM CdCl2 for 24 hr: (D) Rat-2/pM2N-1, (E) Rat-2/pM2NSrc, and (F) Rat-2/N17RasSrc (see Fig. 2B legend for description of cell lines). Images were acquired using a Zeiss Axiovert microscope with a ×20 objective.
Table 1.
Effect of dominant-negative Ras on transformation of CEF by v-Src
| Cells* | Src virus† | Focus formation,‡ % | Soft agar colony formation,‡ % | Hexose uptake,§ % |
|---|---|---|---|---|
| CEF/Vec | − | 0 | 0 | 100 ± 9 |
| CEF/Vec | + | 100 ± 3 | 100 ± 10 | 536 ± 85 |
| CEF/N17Ras | − | 0 | 0 | 59 ± 5 |
| CEF/N17Ras | + | 110 ± 16 | 144 ± 23 | 414 ± 51 |
The data are expressed as the mean ± SEM (percent of control) of three replicate plates in a single experiment, and are representative of two to four independent experiments.
CEF/Vec, cells that were infected with the subgroup B empty vector virus; CEF/N17Ras, cells that were infected with the subgroup B N17Ras virus.
Cells were infected with the subgroup B virus 2 days before superinfection with the subgroup A empty vector (−) or v-Src (+) virus.
Cells were infected with the subgroup A virus at a low multiplicity of infection (<1) for focus and soft agar colony assays.
Cells were infected with the subgroup A virus at a high multiplicity of infection (≈10) for the hexose uptake assay.
To determine if activation of Ras is necessary for transformation of Rat-2 cells, we generated cell lines that express N17Ras under the control of a metal-inducible promoter and tested the ability of v-Src to transform these cells in the presence and absence of the inducing metals zinc and cadmium. In the absence of metal, cells transfected with the N17Ras expression construct contained the same amount of Ras protein as cells transfected with the empty expression vector pM2N (Fig. 2B), indicating that there was little basal expression of N17Ras. Exposure of the N17Ras cell lines to 100 μM ZnCl2 and 2 μM CdCl2 for 24 hr resulted in expression levels that were ≈2- to 4-fold higher than that of endogenous Ras in the same cell lines, whereas exposure of the vector control cells to metal did not alter the levels of expression of endogenous Ras (Fig. 2B). Expression of dominant-negative Ras did not alter the morphology of normal Rat-2 cells or cause reversion of the transformed morphology of cells expressing v-Src, although the transformed cells carrying the metal-inducible N17Ras appeared slightly flatter, with or without induction of the mutant Ras, than the transformed cells carrying the empty vector (Fig. 3 D–F). Infection of three different clones of Rat-2/N17Ras with a helper-free virus encoding v-src resulted in the formation of foci, although the N17Ras cell lines exhibited slightly decreased infection efficiencies and altered focus morphologies. Moreover, induction of N17Ras did not significantly alter the number of foci resulting from virus infection (Table 2). In soft agar colony assays, induction of dominant-negative Ras did not alter the cloning efficiency of Rat-2 cells expressing v-Src (Table 2).
Table 2.
Effect of dominant-negative Ras on transformation of Rat-2 cells by v-Src
| Cells | Zn2+/Cd2+ | Src virus | Focus formation,* % | Soft agar colony formation,† % |
|---|---|---|---|---|
| Rat-2/pM2N-1 | − | + | 100 ± 12 | |
| Rat-2/pM2N-1 | + | + | 103 ± 6 | |
| Rat-2/pM2N-2 | − | + | 112 ± 6 | |
| Rat-2/pM2N-2 | + | + | 135 ± 3 | |
| Rat-2/N17Ras-1 | − | + | 124 ± 9 | |
| Rat-2/N17Ras-1 | + | + | 159 ± 18 | |
| Rat-2/N17Ras-2 | − | + | 126 ± 15 | |
| Rat-2/N17Ras-2 | + | + | 132 ± 18 | |
| Rat-2/N17Ras-3 | − | + | 88 ± 3 | |
| Rat-2/N17Ras-3 | + | + | 82 ± 3 | |
| Rat-2/pM2N-1 | − | 0 | ||
| Rat-2/pM2N-1 | + | 0 | ||
| Rat-2/pM2NSrc | − | 100 ± 3 | ||
| Rat-2/pM2NSrc | + | 99 ± 7 | ||
| Rat-2/N17Ras-2 | − | 0 | ||
| Rat-2/N17Ras-2 | + | 0 | ||
| Rat-2/N17RasSrc | − | 151 ± 8 | ||
| Rat-2/N17RasSrc | + | 164 ± 1 |
The data are expressed as the mean ± SEM (percent of control) of three replicate plates in a single experiment, and are representative of two independent experiments.
Focus assays with Rat-2-derived cell lines were conducted with two independent clones carrying the empty metal-inducible expression vector (Rat-2/pM2N-1 and -2) and three independent clones carrying metal-inducible N17Ras (Rat-2/N17Ras-1, -2, and -3), and were corrected for efficiency of infection by normalizing to hygromycin-resistant colony forming units.
Soft agar colony assays with Rat-2-derived cell lines were conducted with one clone transfected with the empty vector (Rat-2/pM2N-1), one clone transfected with the metal-inducible N17Ras (Rat-2/N17Ras-2), and single clones of these cells expressing v-Src (Rat-2/pM2NSrc and Rat-2/N17RasSrc).
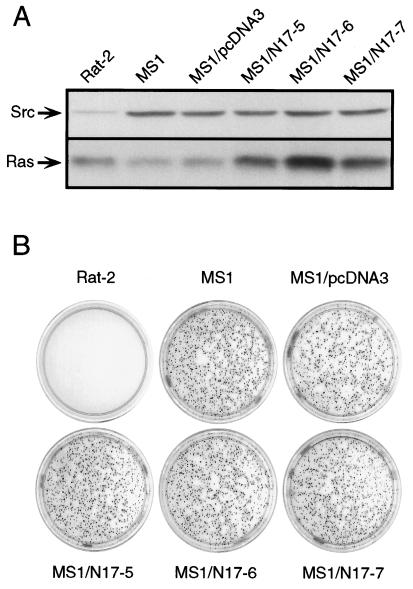
When v-Src expression is driven by the long terminal repeats of an avian or mammalian retrovirus, the level of expression is considerably higher than the threshold level required for transformation (17). To determine if a threshold dose of v-Src can transform cells independently of Ras, MS1 cells, a clone of Rat-2 cells transformed by a threshold dose of v-Src (17), were transfected with an N17Ras expression vector, and several clones that expressed the mutant Ras at levels 2- to 4-fold over the level of endogenous Ras were isolated (Fig. 4A). Dominant-negative Ras did not significantly alter the cloning efficiency of MS1 cells in soft agar (Fig. 4B), indicating that the continued function of Ras is not required for transformation even by a threshold dose of v-Src.
Figure 4.
Effect of dominant-negative Ras on transformation by a threshold dose of v-Src. (A) Expression of Src and Ras in untransfected Rat-2 and MS1 cells, MS1 cells stably transfected with the empty expression vector (MS1/pcDNA3), and three independent clones transfected with the N17Ras expression construct (MS1/N17Ras-1, -2, and -3). (B) Soft agar colony assays were performed with the cell lines described in A. Colonies were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) 16 days after plating.
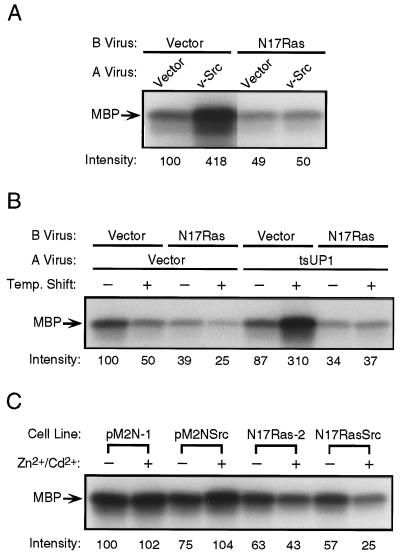
Ras is thought to signal to numerous effectors, the most well characterized of which is the protein kinase Raf. Activation of Raf by Ras leads to the activation of the MAP kinases ERK1 and ERK2, events that have been proposed to be necessary and sufficient for transformation by tyrosine kinases (26, 29, 30). We therefore examined the effect of dominant-negative Ras on the activity of endogenous ERK2 in CEF and Rat-2 cells transformed by v-Src. In CEF expressing v-Src alone, the activity of ERK2 was constitutively elevated 2- to 4-fold compared with normal serum-starved cells. When N17Ras was expressed, the ability of v-Src to activate ERK2 was completely inhibited (Fig. 5A). Moreover, whether or not v-Src was also expressed, the activity of ERK2 in CEF expressing N17Ras was inhibited to a level significantly below that in normal serum-starved CEF (Fig. 5A), confirming that the Ras block in these cells was complete. The complete inhibition of Src-stimulated ERK2 activity by N17Ras was also observed in cells that were not serum starved (data not shown). These findings indicate that constitutive activation of ERK2 is not required for maintenance of the transformed state. To determine if v-Src could transiently activate ERK2 in the presence of dominant-negative Ras, we examined the effect of N17Ras expression on the activation of ERK2 by a temperature-sensitive mutant of v-Src (tsUP1). When CEF expressing tsUP1 without N17Ras were maintained at the nonpermissive temperature, the activity of ERK2 was comparable to that observed in the vector control cells (Fig. 5B). When shifted to the permissive temperature for 30 min, a modest decrease in ERK2 activity was observed in the control cells, and a 3-fold stimulation of ERK2 was observed in cells expressing tsUP1 alone. However, coexpression of N17Ras completely inhibited the ability of tsUP1 to stimulate ERK2 (Fig. 5B). Expression of N17Ras did not inhibit the ability of tsUP1 to induce protein-tyrosine phosphorylation after a 30-min shift to the permissive temperature or to induce morphological transformation after further incubation at the permissive temperature (data not shown). These observations indicate that the transformation of CEF by v-Src is not dependent on a transient induction of ERK2 activity. The activity of ERK2 was not elevated in Rat-2 cells transformed by v-Src (Fig. 5C), which is consistent with the observations of others (31), and with our observation that Ras is not constitutively activated in these cells (Fig. 1). However, induction of N17Ras reproducibly inhibited ERK2 activity in the Src-transformed Rat-2/N17RasSrc cell line by ≈50%, to levels below those observed in control Rat-2 cells (Fig. 5C), confirming that even low levels of ERK2 activation are not required for transformation by v-Src.
Figure 5.
Effect of dominant-negative Ras on the activation of the MAP kinase ERK2 by v-Src. (A) CEF were infected as in Fig. 2A, and the activity of endogenous ERK2 was determined as described. (B) CEF were infected as in Fig. 2A, except that a temperature-sensitive mutant of v-Src (tsUP1) was used. Cells were maintained at the nonpermissive temperature (41°C) during the entire incubation period (Temp. Shift, −) or were shifted to the permissive temperature (36°C) 30 min prior to lysis (Temp. Shift, +). The activity of endogenous ERK2 was then determined as described. (C) Rat-2-derived cell lines carrying the empty metal-inducible expression vector (pM2N-1), metal-inducible N17Ras (N17Ras-2), or clones of these cells that also express v-Src (pM2NSrc and N17RasSrc) were assayed for the activity of endogenous ERK2 as described. Numbers below the autoradiograms refer to the radioactivity incorporated into the substrate (myelin basic protein, MBP) as determined by PhosphorImager analysis, and are expressed as percentages relative to the vector control lanes.
DISCUSSION
The current model for transformation by v-Src and other oncogenic tyrosine kinases posits Ras and its effectors as central and necessary components in the signaling pathways that lead to transformation (1). This model is based on reports that Ras is constitutively activated in v-Src-transformed cells (15) and that inhibition of Ras function suppresses the biological effects of v-Src (7–10). Our findings indicate that signaling through the Ras–MAP kinase pathway is not necessary for the malignant transformation of other cell types. It might be argued that in certain cells, the requirement for signals from Ras is overcome by the strength of the signal generated by the highly transforming v-src allele. However, this explanation appears to be excluded by our observation that expression of N17Ras does not affect transformation even by a threshold dose of v-Src. The results presented here are consistent with recent reports that MAP kinase activity is not constitutively elevated in certain cells transformed by v-Src (31, 32). Our findings are also consistent with a recent report that a primary role of c-Src in growth factor receptor signaling is to activate a Ras-independent pathway that leads to the induction of Myc (33). It is not yet clear why transformation by v-Src is Ras-dependent in NIH 3T3 cells, and Ras-independent in CEF and Rat-2 cells. The expression of dominant-negative Ras is lethal in NIH 3T3 cells (27, 34), but only decreases the growth rate of CEF or Rat-2 cells (D.T.A. and G.S.M., unpublished observations). These observations suggest that significant differences exist in the signaling programs of these different cell types.
Our results support the idea that some Ras-dependent and Ras-independent signaling pathways are functionally redundant. For example, PI 3-K, which is regulated by Ras (12), may also be activated by the Src homology 3 (SH3) domain of v-Src (35, 36) or by tyrosine-phosphorylated Cbl (a substrate of v-Src) (37, 38). An avian sarcoma virus encoding a transduced PI 3-K gene can transform CEF (P. K. Vogt, personal communication), suggesting that elevated PI 3-K activity is sufficient for transformation, and that PI 3-K may contribute to Ras-independent transformation by v-Src. PI 3-K can activate Rac1, a member of the Rho family of small GTPases, which in turn can directly activate a kinase cascade leading to the activation of c-Jun N-terminal kinases (JNKs) (3). However, v-Src does not constitutively or transiently activate JNK1 in CEF (39) (D.T.A. and G.S.M., unpublished observations), suggesting that JNKs do not play a role in the Ras-independent transformation of cells by v-Src. Nevertheless, other PI 3-K-dependent pathways may contribute to Ras-independent transformation. Other signaling proteins implicated in transformation by v-Src include protein kinase C family members (13) and p130CAS (40). However, whether the function of these proteins is necessary for Ras-independent transformation by v-Src remains to be determined.
Another signaling pathway that is likely to play a role in the Ras-independent transformation of cells by v-Src is the STAT pathway. STATs are directly activated by tyrosine phosphorylation, which induces formation of dimeric complexes that are transported into the nucleus and stimulate transcription from specific response elements (41, 42). Although MAP kinases may also phosphorylate STAT1 and STAT3 on serine residues (43, 44), the function of Ras does not appear to be required for signaling through the STAT pathway (45, 46). STATs were initially characterized as transducers of interferon receptor signals, where tyrosine phosphorylation is mediated by members of the Janus kinase family. It is now known that some STATs (including STAT3) are phosphorylated by other tyrosine kinases such as the EGF receptor and v-Src (14, 47), and STAT activity is significantly increased in cells expressing v-Src (14). However, it is not yet known whether this activation is required for mitogenesis or transformation induced by these tyrosine kinases.
The results of this study indicate that transformation by an activated tyrosine kinase can be independent of Ras, and that the signaling pathways involved in transformation can be functionally redundant. It remains to be determined whether the Ras-independent pathways involved in transformation by v-Src are mediated by PI 3-K, isozymes of protein kinase C, p130CAS, STATs, or other as yet unidentified signaling proteins.
Acknowledgments
We thank M. Botchan, J. DeClue, G. Rubin, and members of the Martin Laboratory for advice and comments on the manuscript; L. England for purifying the Src2-17 mAb; and J. Brugge, C. Der, and G. Martin for providing plasmids. D.T.A. was supported by a postdoctoral fellowship from the American Cancer Society (PF-3791) and by a training grant from the National Cancer Institute (CA09041). J.K. was supported by a University of California President’s Undergraduate Research Fellowship. This work was supported by National Cancer Institute Grant CA17542 (to G.S.M) and by the facilities of the Cancer Research Laboratory of the University of California, Berkeley.
ABBREVIATIONS
- PI 3-K
phosphatidylinositol 3-kinase
- STATs
signal transducers and activators of transcription
- CEF
chicken embryo fibroblasts
- MAP
mitogen-activated protein
References
- 1.Erpel T, Courtneidge S A. Curr Opin Cell Biol. 1995;7:176–182. doi: 10.1016/0955-0674(95)80025-5. [DOI] [PubMed] [Google Scholar]
- 2.Marshall C J. Curr Opin Cell Biol. 1996;8:197–204. doi: 10.1016/s0955-0674(96)80066-4. [DOI] [PubMed] [Google Scholar]
- 3.Waskiewcz A J, Cooper J A. Curr Opin Cell Biol. 1995;7:798–805. doi: 10.1016/0955-0674(95)80063-8. [DOI] [PubMed] [Google Scholar]
- 4.Treisman R. Curr Opin Cell Biol. 1996;8:205–215. doi: 10.1016/s0955-0674(96)80067-6. [DOI] [PubMed] [Google Scholar]
- 5.Kayne P S, Sternberg P W. Curr Opin Genet Dev. 1995;5:38–43. doi: 10.1016/s0959-437x(95)90051-9. [DOI] [PubMed] [Google Scholar]
- 6.Wassarman D A, Therrien M, Rubin G M. Curr Opin Genet Dev. 1995;5:44–50. doi: 10.1016/s0959-437x(95)90052-7. [DOI] [PubMed] [Google Scholar]
- 7.Smith M R, DeGudicibus S J, Stacey D W. Nature (London) 1986;320:540–543. doi: 10.1038/320540a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacey D W, Roudebush M, Day R, Mosser S D, Gibbs J B, Feig L A. Oncogene. 1991;6:2297–2304. [PubMed] [Google Scholar]
- 9.DeClue J E, Zhang K, Redford P, Vass W C, Lowy D R. Mol Cell Biol. 1991;11:2819–2825. doi: 10.1128/mcb.11.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nori M, Vogel U S, Gibbs J B, Weber M J. Mol Cell Biol. 1991;11:2812–2818. doi: 10.1128/mcb.11.5.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukui Y, Hanafusa H. Mol Cell Biol. 1991;11:1972–1979. doi: 10.1128/mcb.11.4.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rodriguez-Viciana P, Warne P H, Dhand R, Vanhaesebroeck B, Gout I, Fry M J, Waterfield M D, Downward J. Nature (London) 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 13.Zang Q, Frankel P, Foster D A. Cell Growth Differ. 1995;6:1367–1373. [PubMed] [Google Scholar]
- 14.Yu C L, Meyer D J, Campbell G S, Larner A C, Carter-Su C, Schwartz J, Jove R. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 15.Gibbs J B, Marshall M S, Skolnick E M, Dixon R A F, Vogel U S. J Biol Chem. 1990;265:20437–20442. [PubMed] [Google Scholar]
- 16.Liebl E C, England L J, DeClue J E, Martin G S. J Virol. 1992;66:4315–4324. doi: 10.1128/jvi.66.7.4315-4324.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jakobovits E B, Majors J E, Varmus H E. Cell. 1984;38:757–765. doi: 10.1016/0092-8674(84)90271-x. [DOI] [PubMed] [Google Scholar]
- 18.Martin G S, Venuta S, Weber M, Rubin H. Proc Natl Acad Sci USA. 1971;68:2739–2741. doi: 10.1073/pnas.68.11.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hughes S H, Greenhouse J J, Petropoulos C J, Sutrave P. J Virol. 1987;61:3004–3012. doi: 10.1128/jvi.61.10.3004-3012.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petropoulos C J, Hughes S H. J Virol. 1991;65:3728–3737. doi: 10.1128/jvi.65.7.3728-3737.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeClue J E, Martin G S. J Virol. 1989;63:542–554. doi: 10.1128/jvi.63.2.542-554.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maroney A C, Qureshi S A, Foster D A, Brugge J S. Oncogene. 1992;7:1207–1214. [PubMed] [Google Scholar]
- 23.Cook S J, Rubinfeld B, Albert I, McCormick F. EMBO J. 1993;12:3475–3485. doi: 10.1002/j.1460-2075.1993.tb06022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mann R, Mulligan R C, Baltimore D. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 26.Troppmair J, Bruder J T, Munoz H, Lloyd P A, Kyriakis J, Banerjee P, Avruch J A, Rapp U R. J Biol Chem. 1994;269:7030–7035. [PubMed] [Google Scholar]
- 27.Feig L A, Cooper G M. Mol Cell Biol. 1988;8:3235–3243. doi: 10.1128/mcb.8.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dutta A, Stoeckle M Y, Hanafusa H. Genes Dev. 1990;4:243–254. doi: 10.1101/gad.4.2.243. [DOI] [PubMed] [Google Scholar]
- 29.Cowley S, Paterson H, Kemp P, Marshall C J. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 30.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 31.Stofega M R, Yu C L, Wu J, Jove R. Cell Growth Differ. 1997;8:113–119. [PubMed] [Google Scholar]
- 32.Greulich H, Reichman C, Hanafusa H. Oncogene. 1996;12:1689–1695. [PubMed] [Google Scholar]
- 33.Barone M V, Courtneidge S A. Nature (London) 1995;378:509–512. doi: 10.1038/378509a0. [DOI] [PubMed] [Google Scholar]
- 34.Chen S Y, Huff S Y, Lai C C, Der C J, Powers S. Oncogene. 1994;9:2691–2698. [PubMed] [Google Scholar]
- 35.Liu X, Marengere L E M, Koch C A, Pawson T. Mol Cell Biol. 1993;13:5225–5232. doi: 10.1128/mcb.13.9.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pleiman C M, Hertz W M, Cambier J C. Science. 1994;263:1609–1612. doi: 10.1126/science.8128248. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka S, Neff L, Baron R, Levy J B. J Biol Chem. 1995;270:14347–14351. doi: 10.1074/jbc.270.24.14347. [DOI] [PubMed] [Google Scholar]
- 38.Meisner H, Conway B R, Hartley D, Czech M P. Mol Cell Biol. 1995;15:3571–3578. doi: 10.1128/mcb.15.7.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bojovic B, Rodrigues N, Dehbi M, Bedard P A. J Biol Chem. 1996;271:22528–22537. doi: 10.1074/jbc.271.37.22528. [DOI] [PubMed] [Google Scholar]
- 40.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schindler C, Darnell J E. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 42.Winston L A, Hunter T. Curr Biol. 1996;6:668–671. doi: 10.1016/s0960-9822(09)00445-x. [DOI] [PubMed] [Google Scholar]
- 43.Wen Z, Zhong Z, Darnell J E. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 44.David M, Petricoin E, Benjamin C, Pine R, Weber M J, Larner A C. Science. 1995;269:1721–1723. doi: 10.1126/science.7569900. [DOI] [PubMed] [Google Scholar]
- 45.Silvennoinen O, Schindler C, Schlessinger J, Levy D E. Science. 1993;261:1736–1739. doi: 10.1126/science.8378775. [DOI] [PubMed] [Google Scholar]
- 46.Coffer P, Lutticken C, Puijenbroek A v, Jonge M K, Horn F, Kruijer W. Oncogene. 1995;10:985–994. [PubMed] [Google Scholar]
- 47.David M, Wong L, Flavell R, Thompson S A, Wells A, Larner A C, Johnson G R. J Biol Chem. 1996;271:9185–9188. doi: 10.1074/jbc.271.16.9185. [DOI] [PubMed] [Google Scholar]