Abstract
Mutation of the adenomatous polyposis coli (APC) gene is an early step in the initiation of colon cancer. Because the distribution pattern of a protein within the cell can provide important clues as to function, we have used a combination of immunofluorescence microscopy and biochemical fractionation to determine the location of APC protein in epithelial cells. Immunofluorescence microscopy placed full-length APC protein in both the nucleus and the cytoplasm. The nuclear APC protein was concentrated in discrete subnuclear regions, including nucleoli, whereas the cytoplasmic APC protein concentrated at the leading edge of migrating cells. Colocalization of APC protein with rRNA confirmed a nucleolar localization. These immunocytochemical findings have been supported by cell fractionation, which demonstrated that full-length APC protein was located in both the membrane/cytoskeletal and the nuclear fractions.
Keywords: tumor suppressor, immunofluorescence microscopy, cellular fractionation, colon cancer
Inherited tumor predispositions provide an opportunity to define critical early genetic events in the development of tumors. An inherited colon cancer predisposition, familial adenomatous polyposis, is caused by mutant alleles of the adenomatous polyposis coli (APC) gene (1–4). The early appearance of hundreds or thousands of colon polyps in this inherited disorder indicates that mutations in the APC gene can be rate-limiting in polyp development. The majority of sporadic colon polyps and carcinomas also carry mutated APC genes, indicating that somatic mutations in the APC gene are an early event in the development of most colon polyps and carcinomas.
The distribution pattern of a protein within the cell can provide important clues as to its function. Previous studies of the intracellular distribution of endogenous APC have focused on APC protein in the cytoplasm (5–7). Importantly, endogenous APC protein has recently been shown to localize near the ends of microtubules that extend into actively migrating regions of MDCK or IEC-6 cell membranes (10). A recent examination of APC protein overexpressed in mouse colonocytes, however, also suggested that APC may be present in the nucleus (11).
To provide a more detailed characterization of the intracellular distribution of APC protein in epithelial cells, in particular to determine whether APC protein might, in fact, be present in the nucleus, we have examined the subcellular distribution of endogenous APC protein in epithelial cells in culture. We have used three commercially available mAbs and one polyclonal antiserum for cell staining experiments. Immunofluorescence microscopy revealed APC protein in both the cytoplasmic and the nuclear regions of the epithelial cells. As seen by others in different cells (10), we have seen a punctate cytoplasmic pattern and a more intense particulate labeling of the leading edges of migrating cells. In addition to the cytoplasmic pattern, we have also observed a distinct pattern of APC protein within the nucleus, with focal staining of the nucleoli. These immunocytochemical findings were supported by cell fractionation, demonstrating that full-length APC protein is found in the nucleus as well as the membrane/cytoskeletal fraction.
Most disease-causing mutations in the APC gene result in a truncated protein product (12). In contrast to wild-type APC protein, we found no truncated APC protein in the nuclear fraction of colon cancer cells containing only mutant APC. This result indicates that such truncations abrogate nuclear localization of APC protein, suggesting that elimination of the nuclear location and function of this protein may be important in colon cancer progression.
MATERIALS AND METHODS
Cell Lines and Tissue Culture.
Cell lines used in these experiments were maintained at 37°C in CO2 (5%) incubators. 184A1 cells are an immortalized human mammary epithelial cell line (13) (a gift from Martha Stampfer, Lawrence Berkeley Laboratory, Berkeley, CA). They were grown in MCDB 170 media (Clonetics, San Diego) supplemented as described (14). Primary epithelial or fibroblast outgrowths from normal breast tissue were grown in MCDB 170 media or MEM, 10% fetal bovine serum (FBS), respectively. Other cells were obtained from the American Type Culture Collection and were maintained in the following growth media: HCT116 (McCoy’s 5a medium/10% FBS), LS174T (Eagle’s MEM/10% FBS/1% nonessential amino acids), DLD-1 (RPMI medium 1640/10% FBS), T47D (RPMI medium 1640/10% FBS), and MDCK (MEM/10% FBS). HCT116 and LS174T are colon cancer cell lines that both express only full-length APC protein. DLD-1 is a colon cancer cell line that expresses no full-length APC protein but, rather, APC protein truncated at amino acid 1427. T47D is a breast cancer cell line that expresses full-length APC protein.
Immunofluorescence Microscopy.
Cells were seeded onto tissue culture chamber slides (25–50% confluency) and allowed to grow for 36–48 hr before manipulation. Cells were rinsed in PBS (10 mM phosphate, pH 7.5/100 mM NaCl) then fixed with 2% paraformaldehyde in PBS for 30 min at 4°C. Following two PBS rinses, cells were permeabilized with 0.2% Triton X-100 in TBS (10 mM Tris, pH 7.5/100 mM NaCl/5 mM KCl) for 5 min at room temperature. Following two TBS washes, cells were incubated with 0.5% Na2BH3 in water for 10 min at room temperature. Cells were rinsed with TBS and then incubated with primary antibody diluted in antibody buffer (1% BSA/3% normal goat sera/0.2% Triton X-100 in TBS) for 90 min at room temperature. Cells were rinsed three times with TBS prior to incubation with secondary antibody conjugated to fluorescein isothiocyanate or Texas Red for 30 min at room temperature. Cells were rinsed three times with TBS and mounted with Pro Long antifade (Molecular Probes) for immunofluorescence microscopy. Antibodies and dilutions used for the experiments are as follows: APC (mouse IgG1, Ab-2, Ab-4, or Ab-6) 1:150 (Oncogene Science) or APC64 (rabbit sera, a gift from the Arnold J. Levine laboratory, Princeton University, Princeton) 1:1000, α-tubulin (mouse IgG1, DM-1A) 1:200 (ICN), goat anti-mouse IgG1-fluorescein isothiocyanate and goat anti-mouse IgG1-Texas Red 1:200 (Southern Biotechnology Associates), goat anti-rabbit-Texas Red 1:200 (Accurate Chemical and Scientific, Westbury, NY), and goat anti-rabbit-fluorescein isothiocyanate 1:200 (Boehringer Mannheim). Nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI) following secondary antibody incubation. APC polyclonal serum was preadsorbed to DLD-1 cells that had been fixed and permeabilized as described for immunofluorescence staining. For the APC antibody blocking experiment, APC antibody was incubated with a peptide corresponding to APC protein amino acids 2717–2844 at 10-fold molar excess, in PBS for 12 hr at 4°C. Any precipitant protein was pelleted by centrifugation for 15 min prior to dilution of the peptide/antibody mixture in antibody buffer. Stained cells were examined using an Axioplan microscope (Zeiss) with a ×63 objective. Images of APC-stained cells and APC controls were captured using 400 ASA slide film (Eastman Kodak) and 2- to 4-min exposure times.
Cell Fractionation.
Cell fractionation was performed essentially as described (15). For cell fractionation procedures, protease inhibitors (Boehringer Mannheim) were added to the buffers in the following concentrations: pefablock, 0.2 mg/ml; aprotinin, 0.01 mg/ml; pepstatin, 0.01 mg/ml; and leupeptin, 0.01 mg/ml. Cells grown in 150-cm2 flasks were harvested by scraping into ice cold PBS. Cells were rinsed two times with cold PBS prior to lysis with detergent. Cells were resuspended in L-buffer (PBS/0.1% Triton X-100/0.1% Nonidet P-40) and incubated on ice for 10 min, or until they were determined to be >99% lysed using trypan blue exclusion. Nuclei were pelleted by centrifugation at 1000 × g for 10 min at 4°C. Supernatant was further fractionated by centrifugation at 100,000 × g for 60 min at 4°C. The supernatant fraction was collected and classified as cytoplasm. The pellet was resuspended in L-buffer and was classified as the membrane/cytoskeletal fraction. The nuclear pellet was purified from membrane contaminants by two rinses in L-buffer, passage through a 0.22-gauge needle three times, and passage through a 0.85 M sucrose cushion (15,000 rpm, microfuge, 15 min). Nuclei in the pellet were lysed by sonication (30 sec) in PBS prior to DNase (100 units/200 μl) treatment (45 min, 4°C). Nuclei were further sonicated two times, for 30 sec at 4°C to make a nuclear lysate. For nuclear scaffold/matrix isolation, nuclear pellets purified through the sucrose cushion were washed once, then resuspended in nuclei buffer (10 mM Tris, pH 7.4/20 mM KCl/0.125 mM spermidine/0.05 mM spermine/1% thiodiglycol). DNase I (100 units) and MgCl2 (5 mM final concentration) were added prior to incubation on ice for 30 min. CuSO4 (1 mM final concentration) was added, and the nuclei were incubated for 10 min at 37°C. Nuclear scaffold proteins were precipitated on ice by addition of an equal volume of 0.4 M (NH4)2SO4 in 10 mM Tris·HCl/0.2 mM MgCl2. Precipitate was raised in 15 ml TM-0.2 buffer [10 mM Tris·HCl, pH 7.4/0.2 mM MgCl2/0.2 M (NH4)2SO4] and pelleted at 1500 rpm for 15 min at 4°C. The pellet was washed three times with nuclei buffer and 70 mM NaCl prior to resuspension in L-buffer. Protein concentration was determined in all cell fractions using a Bio-Rad protein assay reagent as per manufacturer’s instructions.
Western Immunoblot Analysis.
Proteins (70 μg/lane; 35 μg/lane for scaffold fractions) were separated electrophoretically using either 6% or 4–12% gradient acrylamide Tris tricine gels (NOVEX, San Diego) and Laemmli buffer. Gels were run for 2 hr at 125 V with cold circulating water. Proteins were transferred to nitrocellulose (Schleicher & Schuell) for 16 hr at 30 V in transfer buffer (192 mM glycine/20% methanol/25 mM Tris base/0.1% SDS) with circulating cold water. Rainbow molecular weight markers (Amersham) were loaded in one lane of each gel for size standardization. Nitrocellulose membranes containing transferred proteins were blocked with 5% BSA in TBST (TBS/0.1% Tween 20) then incubated with primary antibody diluted in 1% BSA in TBST for 1 hr at 20°C. Following three 10-min rinses with TBST, blots were incubated with an appropriate secondary antibody conjugated to horseradish peroxidase in 1% BSA/TBST. Blots were rinsed three times with TBST and then probed using an enhanced chemiluminescence detection system (Amersham) as per manufacturer’s instructions. Antibodies used for Western immunoblot analysis were as follows: APC (mouse IgG1, Ab-1, or Ab-2) 1:200 (Oncogene Science), α-tubulin 1:200 (ICN), lamin A/C (mouse IgG1, X-67), or lamin B (mouse IgG1, X233) 1:10 (American Research Products, Beltsville, MD), horseradish peroxidase–rabbit anti-mouse IgG1 1:20,000 (Zymed), β1,β2-adaptin (mouse IgG1) 1:2000 (Sigma), horseradish peroxidase–goat anti-rabbit IgG, and horseradish peroxidase–sheep anti-mouse 1:40,000 (Sigma).
RESULTS
Location of Full-Length APC Protein in Epithelial Cells.
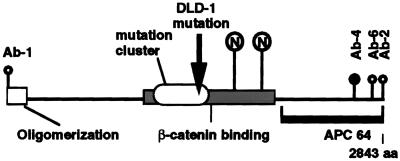
As a first step toward determining the distribution pattern for endogenous, full-length APC protein, we screened commercially available APC antibodies for their ability to stain epithelial cells using indirect immunofluorescence microscopy. Fig. 1 shows a schematic of the APC protein modified from ref. 12 showing the various epitopes recognized by the APC antibodies used in the present study. Ab-4 is an mAb made against the C-terminal 300 aa of APC protein. The precise location of its epitope has not been mapped. Additional features of the APC protein include a mutation cluster region, where 94% of all somatic and ≈62% of all inherited, disease-causing APC mutations occur (16), a β-catenin-binding region (17, 18), and an oligomerization region (19).
Figure 1.
Antibodies recognize different epitopes of the APC protein. Schematic representation of the APC protein as adapted from (12). The mutation cluster region is indicated by the oval. β-catenin-binding region is indicated by a shaded rectangle. □, Oligomerization region; ○, mAbs that recognize distinct APC epitopes used in the present study. Antibody Ab-4, whose epitope has not been mapped precisely, was made against a 300-amino acid peptide beginning at • and proceeding to the end of the APC protein. The solid line represents the APC region that was used to produce the polyclonal rabbit sera APC64. The arrow marks the point of APC protein truncation in the colon cancer cell line DLD-1. The circled Ns·· indicate two potential nuclear localization sequences beginning at amino acids 1773 and 2054.
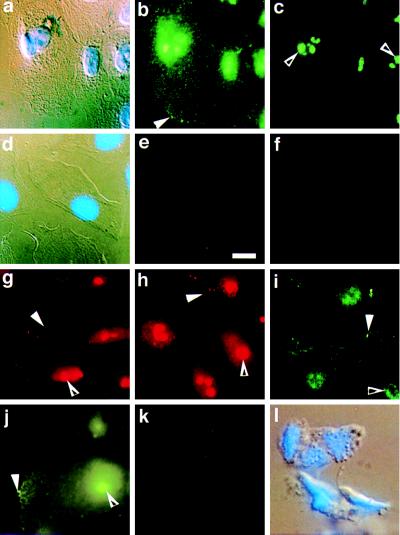
A panel of antibodies raised against various APC epitopes was used to stain 184A1 cells. 184A1 is an immortalized human mammary epithelial cell line that displays keratin staining patterns and phenotypic characteristics very similar to normal human mammary epithelial cells (13). Of seven mAbs and three polyclonal antibodies tested in immunofluorescence assays with the 184A1 cell line, the three mAbs raised against the C terminus of APC protein (Ab-2, Ab-4, and Ab-6), and the polyclonal antibody APC64, also raised against the C terminus, gave similar and reproducible staining patterns, as seen in Fig. 2 b, c, and g–i. These antibodies localized full-length APC protein to two distinct compartments of the epithelial cell. A punctate staining pattern is visible in the cytoplasm, with especially prominent staining at cell edges (Fig. 2 b and g–i, solid arrowheads). In migrating cells, the APC staining concentrated at the leading edge (data not shown). This cytoplasmic distribution of APC protein provides additional support for the very similar results reported recently in MDCK cells (10). In addition to the cytoplasmic staining, however, there is significant nuclear staining, shown most clearly in Fig. 2c (open arrowhead), where the same cells viewed in Fig. 2 a and b are photographed at a slightly different focal distance. Much of the total staining seems to be concentrated in discrete nuclear regions (open arrowheads). In Fig. 2a, the cells shown in Fig. 2 b and c were stained with DAPI and viewed using differential interference contrast (DIC) optics to distinguish cell nuclei, cell edges, and general morphology. In addition, a similar APC protein location was seen in several other cells tested, including T47D (Fig. 2j), normal human fibroblasts, MDCK, mink lung epithelial cells, primary outgrowth from normal breast tissue, and Cos7 (data not shown). The remaining four mAbs (Ab-1, Ab-3, Ab-5, and Ab-7; Oncogene Science) and one of the polyclonal antibodies (N-15; Santa Cruz Biotechnology) showed essentially no staining. Interestingly, each of these antibodies had been raised against the N terminus of the APC protein. The other polyclonal serum raised against the C terminus of APC protein (C-20; Santa Cruz Biotechnology) also showed only minimal staining.
Figure 2.
Localization of APC protein in 184A1 cells using immunofluorescence microscopy. 184A1 cells were grown on glass slides prior to fixation and immunofluorescence microscopy using Ab-4, an antibody specific for APC protein (b and c) or using Ab-4 preincubated with an APC peptide (e). b and c are photographs of the same group of cells taken at two focal distances to more clearly capture cell edge staining (b, solid arrowhead) and nuclear staining (c, open arrowhead). APC protein appears in a punctate pattern throughout the cytoplasm with areas of protein concentration at one edge (solid arrowhead). In addition, APC protein appears throughout the nuclei with a few areas of concentration (c). a and d are DIC and DAPI views of the fluorescence views shown in b, c, and e, respectively. Controls include: f, 184A1 cells stained with nonspecific antibody IgG1; g–i, 184A1 cells stained for APC using antibodies Ab-2 (g), Ab-6 (h) or APC64 (i); j, APC staining of T47D cells. For each antibody, both edge staining (solid arrowhead) and nuclear staining (open arrowhead) are apparent. In k, DLD-1 cells that express only truncated APC protein were stained using the C terminal antibody Ab-4 to demonstrate staining specificity. l is the corresponding DIC and DAPI view of the cells shown in k. (Bar = 10 μm.)
In addition to the observations that four independent antibodies give the same staining pattern, this result was further examined by several criteria. APC staining was eliminated by preincubation of the primary antibody with an APC peptide (Fig. 2e), while the same APC peptide did not affect immunofluorescence with an α-tubulin antibody (data not shown). In the absence of primary antibody (data not shown), or in the presence of a nonimmune mouse mAb of the same isotype, IgG1 (Fig. 2f), only a diffuse, very faint staining was seen, without the characteristic pattern obtained with APC antibody.
To further demonstrate the dependence of the staining pattern on the presence of full-length APC protein, cells from the colon cancer cell line DLD-1, which lack full-length APC protein, were used for immunofluorescence microscopy with each of the four antibodies directed against the C terminus of APC. DLD-1 cells express no full-length APC protein since they carry a mutation that truncates APC at amino acid 1427. This truncation deletes the C-terminal portion of the APC protein, thereby eliminating all APC epitopes recognized by antibodies directed toward APC protein’s C terminus. The distinctive staining pattern previously observed was not seen in the DLD-1 cells stained with any of the C-terminal antibodies [Ab-4 (Fig. 2k with corresponding DAPI/DIC in Fig. 2l), Ab-2, Ab-6, and APC64; data not shown]. This finding supports the interpretation that the characteristic staining pattern we observe in cells carrying normal APC protein is due to epitopes within the C terminus of full-length APC protein.
APC Protein Within the Nucleus Localizes to the Nucleoli.
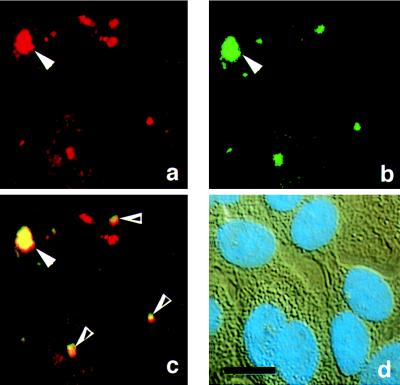
Nuclei were identified with DAPI stain as shown in Fig. 2a, with the blue DAPI signal overlying the DIC image. The size and shape of the concentrated staining suggested that APC protein might be in nucleoli. This suggestion was confirmed using a fluorescein-conjugated DNA probe that recognizes rRNA, a major component of nucleoli. As shown in Fig. 3, most regions with concentrated nuclear APC protein, identified using a Texas Red-conjugated secondary antibody (Fig. 3a), overlapped with the nucleoli, visualized with a green, fluorescein-conjugated, antisense oligonucleotide rRNA probe (Fig. 3b). The micrographs shown in this figure were taken with the nuclear region of the cell in focus, eliminating most of the cytoplasmic and edge staining. The overlap of APC protein and rRNA is shown in Fig. 3c, where a double exposure shows the coincidence of red APC signal and green rRNA signal as yellow. In Fig. 3, solid arrows designate nuclear staining in a single cell, and open arrows indicate additional regions of staining overlap. Some regions that stained positive for APC protein did not stain positive for rRNA. The DIC view with DAPI signal overlay is shown in Fig. 3d. A fluorescein-conjugated oligonucleotide probe corresponding to the rRNA sense strand sequence did not show the nucleolar staining pattern (data not shown).
Figure 3.
Nuclear APC protein co-localizes with rRNA. 184A1 cells were stained simultaneously for APC protein (a) and rRNA (b) using antibodies specific for the APC protein and a fluorescein isothiocyanate-conjugated DNA oligo complementary to rRNA. The yellow staining in c reflects the colocalization of rRNA and APC protein. Solid arrowheads designate nuclear staining in a single cell (a–c). Open arrowheads designate additional regions of staining overlap. d is a DIC and DAPI view of the fluorescence views shown in a–c. (Bar = 10 μm.)
Biochemical Fractionation Locates Full-Length APC Protein to Both Membrane/Cytoskeletal and Nuclear Cell Fractions.
Biochemical fractionation was employed as an independent test of the two cellular locations of APC protein. Because the colon cancer cell lines HCT116 and LS174T both carry normal APC alleles, and express 5–10 times more full-length APC protein than 184A1 cells, they were used to optimize the biochemical fractionation procedure. Both cell lines were subjected to lysis by detergent (>99% cell lysis as determined by trypan blue exclusion) followed by purification of the nuclei from the cytoplasmic fraction. The cytoplasmic fraction was further separated into a membrane/cytoskeletal fraction and a soluble fraction. Nuclear matrix/scaffold proteins were purified from the nuclear fraction by DNAse treatment, stabilization with CuSO4, precipitation in 0.2 M ammonium sulfate, and washes. Because similar results were obtained with both LS174T and HCT116 cell lines, only data from fractionation of LS174T is shown. The fractions were characterized by Western immunoblot analysis using antibodies directed against compartment-specific proteins: α-tubulin fractionated with the cytoplasm and cytoskeleton, β-adaptins with cytoplasm and membranes, and lamins A, B, and C with the nucleus and nuclear matrix/scaffold.
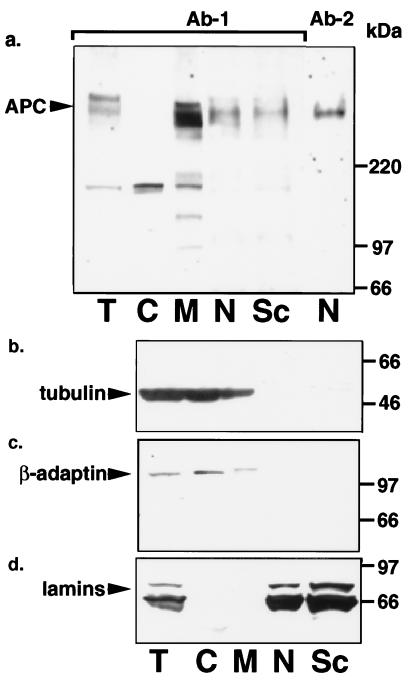
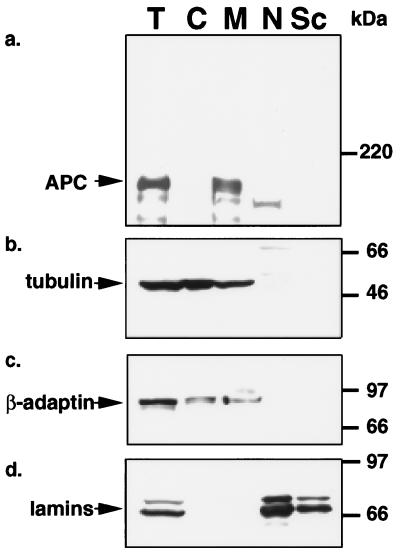
As seen in Fig. 4a, LS174T cells contain full-length APC protein in both the membrane/cytoskeletal (lane M) and the nuclear (lane N) fractions with ≈one-sixth of the total APC protein residing in the nucleus. The majority of the nuclear APC protein further fractionated with the nuclear matrix (lane Sc). The slowest band recognized by the APC antibody migrated at the molecular size predicted for full-length APC protein relative to the molecular weight markers (Fig. 4a, lanes T, M, N, and Sc) and was recognized by antibodies raised to both N-terminal (Ab-1) and C-terminal (Ab-2) regions of APC protein. Band intensities were greatly diminished by preincubation of the APC antibody with an APC peptide (data not shown). The various cell fractions were characterized by stripping and re-probing the blot for the marker proteins α-tubulin (Fig. 4b, lanes C and M), β-adaptin (Fig. 4c, lanes C and M), and lamins (Fig. 4d, lanes N and Sc).
Figure 4.
Full-length APC protein locates to both membrane/cytoskeletal and nuclear cell fractions in LS174T cells. Proteins within the various fractions of LS174T cells were analyzed by SDS/PAGE and immunoblotting. Fractions are labeled at the figure bottom as follows: T, total; C, cytoplasm; M, membrane/cytoskeleton; N, nucleus; Sc, nuclear scaffold. The antibodies used for the Western immunoblots are as follows: a, APC;, b, tubulin as a cytoskeletal marker; c, β-adaptin as a membrane marker; and d, lamins as nuclear and nuclear matrix scaffold markers.
Once fractionation conditions were optimized, they were applied to 184A1 cells, used previously for the immunofluorescence microscopy analysis. Although these cells express much lower levels of APC protein, we were able to show full-length APC protein in the membrane/cytoskeletal, the nuclear, and the nuclear matrix fractions (data not shown). The 184A1 cells showed a more even distribution of APC protein between the nuclear and the membrane/cytoskeletal fractions, consistent with the ratio seen using immunofluorescence microscopy (Fig. 2). The various cell fractions were again characterized by stripping and reprobing the blot for the marker proteins α-tubulin, α-adaptin, and lamins (data not shown).
Truncated APC Protein from a Colon Cancer Cell Line Does Not Locate to the Nuclei.
Cells from the DLD-1 colon carcinoma cell line, which carries only a mutant allele of the APC gene, were fractionated by the above procedure. As seen in Fig. 5a, the truncated APC peptide is found in the membrane/cytoskeletal fraction. Longer exposure times reveal truncated APC protein in the cytoplasmic fraction but not the nuclear or nuclear scaffold fractions. The band appearing slightly below that of truncated APC protein represents cross-reactivity of the secondary antibody with a protein in the DLD-1 cell line as it is present in blots probed with an antibody to APC protein’s C terminus and also with secondary antibody alone (data not shown). Compartment-specific proteins α-tubulin (Fig. 5b), β-adaptin (Fig. 5c), and lamins A, B, and C (Fig. 5d), had the expected distribution patterns.
Figure 5.
Truncated APC protein does not fractionate with the nucleus of DLD-1 cells. Proteins within the various fractions of DLD-1 cells were analyzed by SDS/PAGE and immunoblotting. Fractions are labeled as follows: T, total; C, cytoplasm; M, membrane/cytoskeleton; N, nucleus; Sc, nuclear scaffold. The antibodies used for the Western immunoblots include: a, APC; b, tubulin as a cytoskeletal marker; c, β-adaptin as a membrane marker; and d, lamins as nuclear and nuclear matrix scaffold markers. Truncated APC protein is present in the membrane/cytoskeletal fraction, but not the nuclear or nuclear scaffold fractions.
DISCUSSION
Endogenous APC protein has been found in both the nucleus and the cytoplasm of cultured human epithelial cells. Immunocytochemistry revealed a particulate distribution of APC protein in distinct nuclear regions with foci in the nucleoli (Figs. 2c and 3a), and throughout the cytoplasm (Fig. 2b), with concentrations at the leading edge of migrating cells. Each of three mAbs and one polyclonal antibody directed against APC protein’s C terminus consistently showed the same staining pattern. In contrast, five antibodies directed against the APC protein’s N terminus failed to show significant staining, suggesting that the N-terminal epitopes might be masked in vivo, or that these antibodies might recognize denatured but not native protein.
Cell fractionation experiments provided further support for a nuclear as well as cytoskeletal/membrane localization for the APC protein. The relative cellular distribution of APC protein between nucleus and cytoskeletal/membrane fractions varied among cell types; 184A1 cells showed a high proportion of APC protein in the nucleus (up to half of the total), whereas the colon cancer cell line LS174T showed only ≈one-sixth of the total APC protein in the nucleus. Approximately 70% of the nuclear APC protein fractionated together with the nuclear matrix.
The observation that a significant fraction of APC protein localized to the nucleus of epithelial cells prompted a search for putative nuclear localization sequences within the APC amino acid sequence. As indicated in Fig. 1, APC protein has two stretches of basic amino acid residues, bordered by glycine or proline, typical of nuclear localization sequences. Interestingly, almost all mutations mapping to the APC gene in both familial adenomatous polyposis and sporadic colon cancer patients result in elimination of both putative nuclear localization sequences from the resulting APC protein.
Our findings extend previous studies that revealed primarily cytoplasmic components of the intracellular distribution of APC. Consistent with our results, a recent report using MDCK and rat intestinal epithelial cells has shown punctate cytoplasmic and leading edge localization of APC protein, although nuclear staining was not reported as significant (10). Previous studies differ from those reported here in fixation, staining, and in the cells examined, focusing on immunohistochemical analyses of frozen colon tissue sections (6, 20), overexpressed exogenous protein (8, 9), APC protein expressed in neural cells (21), canine and rat epithelial cells (10), or murine enterocytes (11). Our studies, focusing on human epithelial cells, indicate that the ability to clearly see details of the nuclear component of APC is highly dependent on protocol details: e.g., the use of paraformaldehyde vs. formaldehyde in cell fixation and DNAse treatment of nuclear lysates prior to immunoblot analysis.
Identification of β-catenin (17, 22), and plakoglobin (23, 24) as APC protein partners suggests that APC may modulate the WNT1 signaling pathway, perhaps by increasing the level of β-catenin turnover (18, 25). The threonine-serine kinase GSK3β was also recently found to bind and phosphorylate APC in vitro and this phosphorylation was necessary for β-catenin binding (25). Recent observations demonstrating the presence of β-catenin in the nucleus (26), in association with the transcription complex component, LEF-1 or XTcf-3 (27, 28), together with our observations reported here of APC protein in the cell nucleus, raise the possibility that APC protein interacts with β-catenin in both the cytoplasm and the nucleus.
It is intriguing to relate these observations to possible functional roles for APC in colonocyte development. The developing colonocyte must coordinate a number of cellular events as it migrates up the walls of the crypt. At early stages there are several cell divisions but at later stages, cell division ceases, with cessation of ribosomal synthesis, and specific differentiated functions such as mucin production are initiated, while cell migration continues up the crypt. Upon reaching the top of the crypt, the colonocyte initiates a program of apoptosis and is shed into the lumen of the colon. Several pieces of evidence point to a cell regulatory function for APC in this process. Removal of APC can initiate the deregulated growth of adenomatous colon polyps, characterized by continued cell division and a failure to complete the normal differentiation program. Localization of APC protein in the nucleus with foci in the nucleoli may suggest a regulatory role for APC in both mitosis and ribosomal RNA synthesis. Taken together, these results suggest that APC may be playing a role as a global regulator of colonocyte development, orchestrating the activities of multiple systems.
Acknowledgments
We are grateful to Dr. Martha Stampfer for 184A1 cells, Dr. Arnold Levine, A. A. Sayler, S. Hayashi, and A. K. Teresky for APC antisera, Ed Meenan for oligonucleotide synthesis, the University Hospital Tissue Core for human tissue specimens, the Huntsman Cancer Institute Tissue Culture Facility, and Drs. Mary Beckerle, Mina Bissell, Jerry Kaplan, Mark Noble, and Steven Prescott for useful comments on the manuscript. This work was supported by National Institutes of Health Grant P30 CA42014–10. K.L.N. is a postdoctoral fellow supported by National Institutes of Health Basic Cancer Research Training Grant CA09602.
ABBREVIATIONS
- APC
adenomatous polyposis coli
- DAPI
4, 6-diamidino-2-phenylindole
- DIC
differential interference contrast
- FBS
fetal bovine serum
References
- 1.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, et al. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 2.Joslyn G, Carlson M, Thliveris A, Albertsen H, Gelbert L, et al. Cell. 1991;66:601–613. doi: 10.1016/0092-8674(81)90022-2. [DOI] [PubMed] [Google Scholar]
- 3.Kinzler K W, Nilbert M C, Su L-K, Vogelstein B, Bryan T M, et al. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 4.Nishisho I, Nakamura Y, Miyoshi Y, Miki Y, Ando H, et al. Science. 1991;253:665–668. doi: 10.1126/science.1651563. [DOI] [PubMed] [Google Scholar]
- 5.Smith S A, Easton D F, Ford D, Peto J, Anderson K, Averill D, Stratton M, Ponder M, Pye C, Ponder B A J. Genetics. 1993;52:767–776. [PMC free article] [PubMed] [Google Scholar]
- 6.Miyashiro I, Senda T, Matsumine A, Baeg G, Kuroda T, Shimano T, Miura S, Noda T, Kobayashi S, Monden M, et al. Oncogene. 1995;11:89–96. [PubMed] [Google Scholar]
- 7.Boman B, Lovas S, Abraham C, Adrian T, Murphy R, Marbello R, Bhattacharya G. Biochem Biophys Res Commun. 1995;206:909–915. doi: 10.1006/bbrc.1995.1129. [DOI] [PubMed] [Google Scholar]
- 8.Munemitsu S, Souza B, Muller O, Albert I, Rubinfeld B, Polakis P. Cancer Res. 1994;54:3676–3681. [PubMed] [Google Scholar]
- 9.Smith K, Levy D, Maupin P, Pollard T, Vogelstein B, Kinzler K. Cancer Res. 1994;54:3672–3675. [PubMed] [Google Scholar]
- 10.Nathke I S, Adams C L, Polakis P, Sellin J H, Nelson J. J Cell Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong M H, Hermiston M L, Syder A J, Gordon J I. Proc Natl Acad Sci USA. 1996;93:9588–9593. doi: 10.1073/pnas.93.18.9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polakis P. Curr Opin Genet Dev. 1995;5:66–71. doi: 10.1016/s0959-437x(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 13.Walen K H, Stampfer M R. Cancer Genet Cytogenet. 1989;37:249–261. doi: 10.1016/0165-4608(89)90056-3. [DOI] [PubMed] [Google Scholar]
- 14.Taylor-Papadimitriou J, Stampfer M R. In: Culture of Epithelial Cells. Freshney R I, editor. Vol. 1. New York: Wiley; 1992. pp. 107–133. [Google Scholar]
- 15.Sprott S, Hammond K, Savage N. Anal Biochem. 1991;194:407–412. doi: 10.1016/0003-2697(91)90249-s. [DOI] [PubMed] [Google Scholar]
- 16.Miyaki M, Konishi M, Kikuchi-Yanoshita R, Enomoto M, Igari T, K, Muraoka M, Takahashi H, Amada Y, Fukayama M. Cancer Res. 1994;54:3011–3020. [PubMed] [Google Scholar]
- 17.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 18.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joslyn G, Richardson D, White R, Alber T. Proc Natl Acad Sci USA. 1993;90:11109–11113. doi: 10.1073/pnas.90.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith K J, Johnson K A, Bryan T M, Hill D E, Markowitz S, Willson J K, Paraskeva C, Petersen G M, Hamilton S R, Vogelstein B, Kinzler K. Proc Natl Acad Sci USA. 1993;90:2846–2850. doi: 10.1073/pnas.90.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg G-H, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- 22.Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain S H, Masiarz F R, Munemitsu S, Polakis P. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 23.Rubinfeld B, Souza B, Albert I, Munemitsu S, Polakis P. J Biol Chem. 1995;270:5549–5555. doi: 10.1074/jbc.270.10.5549. [DOI] [PubMed] [Google Scholar]
- 24.Shibata T, Gotoh M, Ochiai A, Hirohashi S. Biochem Biophys Res Commun. 1994;203:519–522. doi: 10.1006/bbrc.1994.2213. [DOI] [PubMed] [Google Scholar]
- 25.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 26.Funayama N, Fagotto F, McCrea P, Gumbiner B M. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 28.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]