Abstract
Oxidative stress has been implicated to play a crucial role in the pathogenesis of a number of diseases, including neurodegenerative disorders, cancer, and ischemia just to name a few. Alzheimer's disease (AD) is an age-related neurodegenerative disorder that is recognized as the most common form of dementia. AD is histopathologically characterized by the presence of extracellular amyloid plaques, intracellular neurofibrillary tangles, the presence of oligomers of amyloid β-peptide (Aβ), and synapse loss. In this review we discuss the role of Aβ-peptide in the pathogenesis of AD and also the use of redox proteomics to identify oxidatively modified brain proteins in AD and mild cognitive impairment (MCI). In addition, redox proteomics studies in in vivo models of AD centered around human Aβ(1-42) are discussed.
Keywords: Alzheimer's disease, mild cognitive impairment, oxidative stress, amyloid β-peptide, redox proteomics
Introduction
Oxidative stress has been implicated to play a crucial role in the pathogenesis of a number of diseases including neurodegenerative disorders, cancer, ischemia [1]. Among all the body organs, brain is particularly vulnerable to oxidative damage because of its high utilization of oxygen, increased levels of polyunsaturated fatty acid (that are readily attacked by free radicals), and relatively high levels of redox transition metal ions; in addition, brain has relatively low levels of antioxidants [2-5]. The presence of iron ion in an oxygen-rich environment can further lead to enhanced production of hydroxyl free radicals and ultimately lead to a cascade of oxidative events.
Oxidative stress occurs due to an imbalance in the pro-oxidants and anti-oxidant levels. Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are highly reactive with biomolecules including proteins, lipids, carbohydrate, DNA and RNA [6]. Oxidative damage to these moieties lead to cellular dysfunctions [1, 2, 5, 7-9]. The markers of oxidative stress that are commonly used in biological samples include: protein carbonyls and 3-nitrotyrosine for protein oxidation; thiobarbituric acid reactive substance (TBARS), free fatty acid release, iso- and neuroprostane formation, 2-propen-1-al (acrolein) and 4-hydroxy-2-trans-nonenal (HNE) for lipid peroxidation; advanced glycation end products for carbohydrates; and 8-OH-2'-deoxyguanosine and 8-OH-guanosine and other oxidized bases, and altered DNA repair mechanisms for DNA and RNA oxidation. Among the earliest of these changes following an oxidative insult are increased levels of toxic carbonyls, 3-nitrotyrosine (3-NT), and HNE [2, 4, 7, 10-13].
Protein carbonyl groups are generated by direct oxidation of certain amino acid side chains (i.e., Lys, Arg, Pro, Thr, and His), peptide backbone scission, Michael addition reactions of His, Lys, and Cys residues with products of lipid peroxidation, or glycol-oxidation of Lys amino groups [6, 14-17]. Protein carbonyls are stable and hence are widely used as markers to assess the extent of oxidation of proteins both in in vivo and in vitro conditions [14, 15, 17, 18]. The levels of protein carbonyls can be determined experimentally by derivatization of the carbonyl groups with 2,4-dinitrophenylhydrazine (DNPH), followed by spectroscopic or immunochemical detection of the resulting hydrazone product [6, 15, 19].
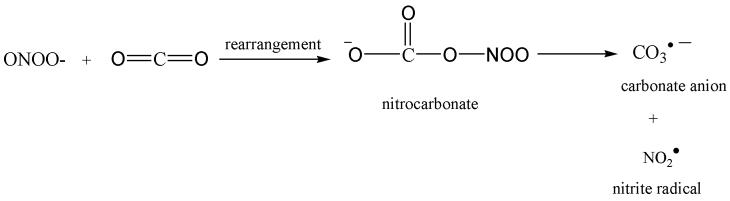
In addition to direct effects, oxidative stress could also stimulate additional damage in brain via the overexpression of inducible nitric oxide synthase (NOS: iNOS) and the action of constitutive neuronal NOS (nNOS) that increase the production of nitric oxide (NO.) via the catalytic conversion of arginine to citrulline. Nitric oxide reacts with superoxide anion (O2.−) at a diffusion controlled rate to produce peroxynitrite (ONOO−). Peroxynitrite is highly reactive with a half-life of less than a second, and can undergo a variety of chemical reactions depending upon its cellular environment, the presence of CO2, and the availability of reactive targets forming modifications such as 3-NT (Figure 1) [20, 21]. 3-Nitrotyrosine is a covalent protein modification that has been used as a marker of nitrosative stress under in a variety of disease conditions [22, 23]. Peroxynitrite can also react with sulfhydryl compounds intracellularly due to the high concentration of free thiols within the cell [24]. Sulfhydryls can also react via S-nitrosylation with NO to form a nitrosothiol (RSNO). Cysteine residues are preferentially nitrosylated due to favorable reaction kinetics [25, 26].
Figure 1.
Tyrosine nitration. ‘A’ represents reaction of peroxynitrite and carbon dioxide. ‘B’ represents formation of 3-nitrotyrosine.
Protein oxidation could lead to aggregation or dimerization of proteins; in addition, protein oxidation can also lead to unfolding or conformational changes in the protein thereby exposing more hydrophobic residues to an aqueous environment. This exposure may lead to loss of structural or functional activity and protein aggregation and subsequent accumulation of the oxidized proteins as cytoplasmic inclusions, such as tau aggregation in the form of tangles and amyloid-β aggregation as senile plaques, as observed in Alzheimer's disease (AD) [27, 28]. The accumulation of oxidatively modified proteins may disrupt cellular functions by alterations in protein expression and gene regulation, protein turnover, modulation of cell signaling, induction of apoptosis and necrosis, etc., which suggest that protein oxidation could have both physiological and pathological significance [6, 29-31]. In our laboratory, we used redox proteomics analyses to identify specific oxidatively modified brain proteins carbonylated proteins in neurodegenerative diseases and models thereof [10, 12, 13, 32-41].
Mild cognitive impairment is considered an intermediate phase between normal aging and Alzheimer's disease. Some researchers believe that MCI is in fact the earliest from of AD [42]. Persons with MCI have cognitive complaint, decline in cognition compared to previous years, and most notably no signs of dementia. Additionally, activities of daily living are not affected. These characteristics require informant confirmation for diagnosis of MCI. Accurate diagnosis can only be confirmed by medical examination to establish a level of cognitive decline [43-45]. Pathologically, MCI has also been characterized using magnetic resonance imaging technology to show measurable atrophy in the hippocampus and entorhinal cortex [46, 47]. Alzheimer's disease patients have considerable neurodegeneration in these aforementioned areas. Since the hippocampus is the region of the brain primarily responsible for processing of memory, atrophy in this brain region is consistent with memory loss in AD and MCI.
An age-related neurodegenerative disorder, AD is recognized as the most common form of dementia. AD is clinically associated with cognitive impairment, loss of language and motor skills, and changes in the behavior. AD is pathologically characterized by the presence of extracellular senile plaques, which consist of a core of Aβ, and intracellular neurofibrillary tangles (NFT), and loss of synaptic connections within entorhinal cortex and progressing into the hippocampus and cortex. NFTs are composed of paired helical filaments (PHF) that consist of aggregates of the hyperphosphorylated microtubule associated protein tau [48], while senile plaques (SP) are rich in Aβ [49].
In this paper, we review the involvement of Aβ and other sources of oxidative stress in AD brain and the use of redox proteomics to identify oxidatively modified brain proteins in AD and MCI. New insights into potential oxidative mechanisms underlying molecular processes in these disorders and progression from MCI to AD have emerged.
Amyloid β-peptide
Aβ, a 40-42 amino acid peptide, is derived by proteolytic cleavage of an integral membrane protein known as amyloid precursor protein (APP) by the action of beta- and gamma-secretases. Aβ(1-40) and Aβ(1-42) constitute the majority of the Aβ peptide found in human brain and has been considered to play a role in the development and progression of AD [49]. Aβ(1-42) is the more toxic of these species both in vitro and in vivo. Further, a number of studies suggest that the small oligomers of Aβ are the actual toxic species of this peptide rather than Aβ fibrils [50-53]. In addition, genetic mutations in genes for APP or presenilin-1 or presenilin-2, which lead to familial AD, have been reported to increase the production of Aβ(1-42) and, consequently, to early onset of AD [49]. Presenilin-1 and presenilin-2 comprise the catalytic element of gamma-secretase [54]. Another critical component of gamma-secretase is presenilin enhancer 2 (Pen-2). Pen-2 binds to presenilins and subsequently enhances gamma-secretase activity, thereby accumulating high levels of toxic Aβ (1-42) [55, 56].
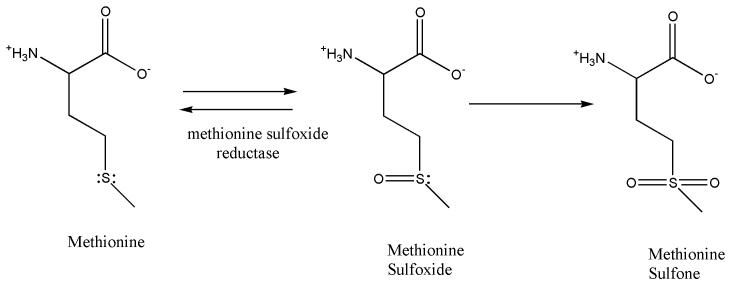
Aβ (1-42) has a critical methionine residue at position 35, which is believed to be associated with the toxicity of Aβ peptide [57-61]. Oxidation of the Met35 produces methionine sulfoxide (MetO) or through further irreversible oxidation, methionine sulfone (Figure 2) [62]. Methionine sulfoxide can be reduced back to methionine by the enzyme, methionine sulfoxide reductase. This stereospecific reaction is facilitated by thioredoxin and is NADPH-driven [63]. Methionine sulfoxide modulates oxidative stress and neurotoxic properties of Aβ (1-42), and studies have shown the activity of methionine sulfoxide reductase is reduced in AD brain [64, 65].
Figure 2.
Methionine chemistry. Formation of methione sulfoxide and methionine sulfone.
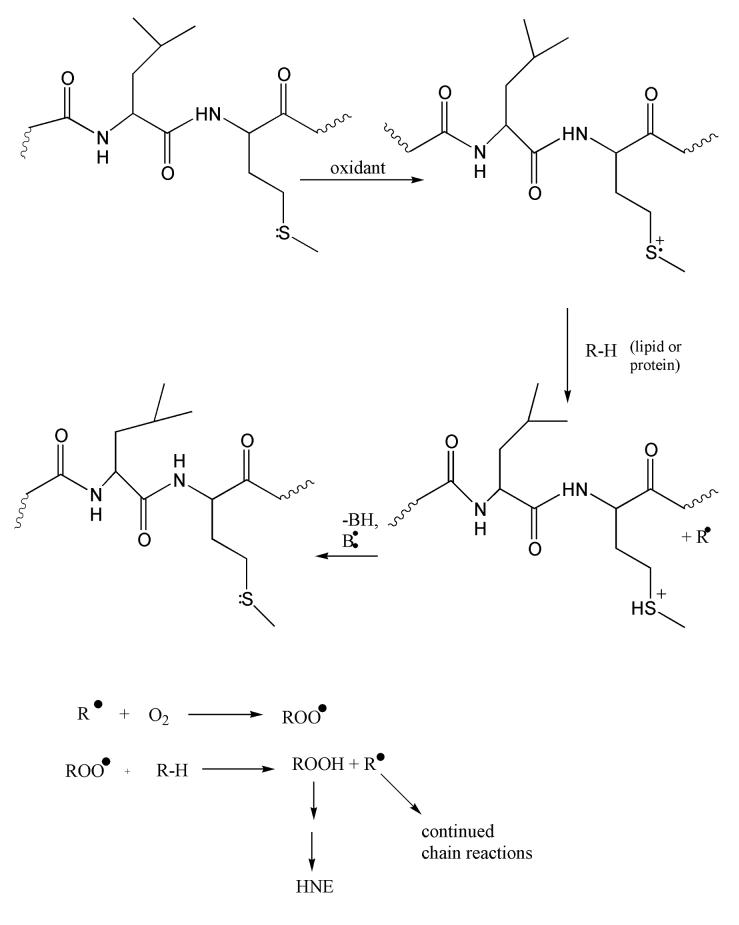
This Met35 residue has been shown to be critical for Aβ(1-42) toxicity and oxidative stress [57]. The secondary structure of Aβ(1-42), which is helical as a small oligomer in membrane lipid bilayers, contributes to the oxidative stress and neurotoxicity of this peptide [66]. When Ile31 of Aβ(1-42) is substituted by the helix-breaking amino acid, proline, in the “i + 4” α-helix conformation, no protein oxidation occurs [66]. A sulfuranyl radical (MetS·+) results as the lone pair of electrons on the S atom undergo a one-electron oxidation (Figure 3) [57, 67, 68]. The sulfuranyl radical can initiate free radical chain reactions with allylic H-atoms on unsaturated acyl chains of lipids until a termination step is reached. Sulfuranyl radicals can react with molecular oxygen to produce sulfoxide and superoxide [69]. Similarly, MetS·+ can react with oxygen to form MetO [70], making various proteins inactive [71]. Aβ (1-40) is a shorter peptide fragment produced from APP, which is also neurotoxic [72]. There is an abundance of Aβ(1-40) MetO in senile plaques [73]. The oxidation of this methionine is reported to prevent Aβ(1-40) from forming beta sheets during fibril formation [74]; therefore, Aβ(1-42) could be more toxic by this mechanism. With the I31P substitution of Aβ(1-42), the interaction of the carboxyl oxygen of Pro31 with the S-atom of Met35 is disrupted, yielding no neurotoxicity or protein oxidation in neurons [66]. The sulfuranyl radical can react with the methylene moieties of Met to form the α-(alkylthio)alkyl radical of methionine (-CH2- CH2-S- CH2). Others showed, using computer modeling, that the β-sheet conformation of this toxic peptide contains the sulfur-centered radical cation of Met35, which bolsters the creation of an α-carbon centered radical on the Gly33 residue. This gives rise to a hydrophobic environment that is ideal for lipid peroxidation in the lipid bilayer [75, 76]. The sulfuranyl radical (a positively charged radical) on the Met35 can in principle be stabilized by the helix-associated dipole of the C-terminal end of Aβ(1-42). Since the dipole of Aβ(1-42) would be greater than that of Aβ(1-40), the differential toxicity of Aβ(1-42) and Aβ(1-40) may be related to this characteristic. The importance of the hydrophobic environment of the Met35 residue was evaluated. By substituting aspartic acid for Gly37, the critical methionine residue no longer remained in a hydrophobic environment, and Aβ(1-42) oxidative stress and neurotoxicity was abrogated [76]. This result suggests that Aβ(1-42)-mediated lipid peroxidation is an early and crucial step in the toxicity of this peptide.
Figure 3.
Aβ(1-42) as a small oligomer is postulated to reside in the lipid bilayer. One-electron oxidation of the S-atom on Met-35, facilitated by the α-helical i + 4 interaction of the backbone carbonyl of Ile-31 with the S-atom of Met-35, forms a positively charged sulfuranyl radical that is stabilized in part by the helical dipole of Aβ(1-42) located in the lipid bilayer. This radical can abstract a lipid acyl allyllic H-atom, forming a lipid carbon radical that immediately binds paramagnetic O2. This peroxyl radical can abstract an allylic H-atom making the lipid hydroperoxide and propagating the chain reaction. The lipid hydroperoxide on arachidonic acid can decompose to HNE, which can subsequently bind to proteins by Michael addition, resulting in oxidative damage to the protein. The SH+ acid on Met has a pKa of minus 5, so loses the H+ easily to reform Met. That is, the reaction is catalytic. This mechanism is consistent with an amplification of damage by a relatively minor degree of conversion of Met to the sulfuranyl free radical of Aβ(1-42).
If Gly33 residue is substituted by the hydrophobic amino acid, valine, neurotoxicity is minimal and protein oxidation is significantly reduced in comparison to the wildtype Aβ(1-42) peptide [77]. Several groups [58, 59, 61] have confirmed our findings that substitution of the S-atom of Met35 by CH2 (to make the R-group, norleucine, Nle) abrogates the oxidative stress and neurotoxicity of the resulting peptide [68]. Other researchers point to the role of Tyr10 as a source of free radicals of Aβ(1-42). This free radical is proposed to reduce Cu2+ bound to His6, His13, and His14, leading to Cu+-mediated chemistry to produce OH· [78, 79]. These researchers point out that rat Aβ(1-42) lacks Tyr10 and is not toxic. However, substitution of Tyr10 by Phe (to keep aromaticity present but to prevent any possible electron flow in contrast to this possibility with Tyr), as well as substitution of three His residues by Asn (which binds Cu2+ at least 100-fold less than His) led to a toxic peptide similar to native human Aβ(1-42) [57]. Also rat Aβ(1-42) was shown to be toxic upon longer incubation with neurons [80]. The p3 fragment of human Aβ(1-42), Aβ(17-42), which does not contain Tyr10 or His6, His13, and His14, was reported to be toxic [60]. This Met-containing peptide, which is present in AD brain, was no longer toxic if the Met35 was substituted by Nle [60]. The dissociation constant of Cu2+ binding to Aβ(1-42) was resigned to be attomolar [79]. While some of these researchers propose an AD therapy based on Cu2+ chelation using clioquinol, which has a Kd of nanomolar [81], the nine orders of magnitude difference in affinity of Cu2+ between Aβ(1-42) and clioquinol make it unlikely that the effectiveness of this agent is correctly ascribed to its chelating ability. However, we propose that clioquinol could chelate relevant weakly bound Cu2+, for example if Cu2+ were loosely bound to Met35 of Aβ(1-42). This scenario would account for the oxidation of Met to a sulfuranyl radical while Cu2+ would be reduced to Cu+, from whence further oxidative chemistry conceivably could ensue. Further investigation will be required to test this notion. Thus, while a role for Cu2+/Cu+ in the oxidative stress and neurotoxicity of Aβ(1-42) can not be excluded, it appears that there is a paramount role of Met35 in these properties.
Finally, several researchers employ the 11-mer, Aβ(25-35), as a model for the AD-relevant (but more costly) Aβ(1-42). Even in the case of the 11-mer, substitution of the C-terminal Met by Nle abrogated the free radical oxidative stress induced by this shorter peptide as did use of the 10-mer Aβ(25-34) [68]. Moreover, because Met is C-terminal in Aβ(25-35), while Met is intrachain in Aβ (1-42), the mechanism by which the sulfuranyl free radical on Met35 is generated is different in these two peptides [68]. This fact, coupled with the absence of evidence for Aβ(25-35) in AD brain, suggests to us that use of Aβ(25-35), though of possible academic interest, is of no relevance for gaining insight into AD.
Oxidative Stress in AD Brain
Oxidative stress is observed in the AD brain [1-5]. This increase has been well documented with markers for protein, DNA, and RNA oxidation as well as lipid peroxidation [4, 6, 11, 82, 83].
Protein Oxidation
Protein oxidation is indexed in the AD brain by an increase in carbonylated [82], HNE- [84], and 3-NT- [11] modified proteins. The initial origin of AD pathogenesis has not been determined though it has become evident that oxidative stress is implicated in the development of this disease [1, 5, 85]. Studies have shown an increase in protein carbonyls in the hippocampus and parietal cortex, but not in the cerebellum where there is less significant AD pathology [82].
Protein Nitration
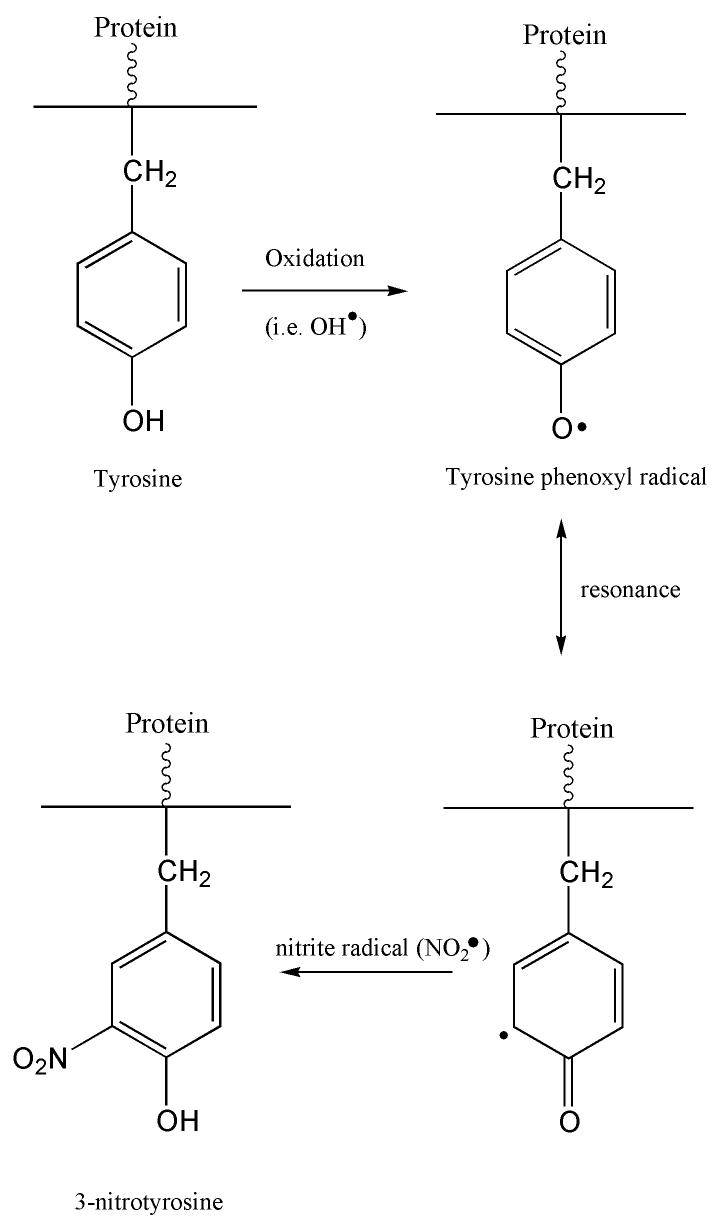
Another common marker of protein oxidation is the addition of a nitro group to tyrosine residues forming 3-NT. Increased protein nitration in the AD brain supports the notion that nitrosative stress also contributes to neurodegeneration in AD [10, 11, 13, 16, 86]. Protein nitration also increases the susceptibility of brain proteins to proteosomal degradation [26]. The overexpression of iNOS and nNOS could be responsible for increased levels of RNS. Previous studies have reported mitochondrial abnormalities in the AD brain [87], which could lead to superoxide (O2−·) leakage. As shown in Figure 1, superoxide then reacts with nitric oxide (NO·) to form peroxynitrite (ONOO −).
Tyrosine is particularly susceptible to nitration but tryptophane and phenylalanine can also be nitrated, albeit at lower rates. Tyrosine residues are important to redox cell signaling and are often the site of phosphorylation, a prominent regulation function [88]. The nitration of tyrosine at the 3-position sterically hinders the phosphorylation and also may change the structure of proteins, thereby rendering a protein dysfunctional, and could lead to cell death [6, 89].
Increased levels of nitrated proteins have been reported to be present in AD brain and cerebrospinal fluid CSF implying a role for RNS in AD pathology [10, 11, 13, 90]. An increase in 3-NT immunoreactivity in neurons from AD brain was observed [11], as well as elevated dityrosine and 3-NT levels in hippocampus, inferior parietal lobule (IPL), and neocortical regions of the AD brain and in ventricular cerebrospinal fluid (VF) when compared with aged matched controls. [11, 86]. The demonstration of nitrated tau in pretangles, tangles, and tau inclusions in AD brain, suggests that tau nitration may be an early event in AD [91]. This notion is strengthened by our finding of elevated 3-NT in brain of subjects with MCI (see below).
Lipid Peroxidation
Amplified lipid peroxidation has been described in several neurodegenerative diseases including AD [7, 83, 84, 92]. Analysis of AD brains demonstrates an increase in free HNE in amygdala, hippocampus, and parahippocampal gyrus of the AD brain when compared with age-matched controls [83]. This increased alkenal concentration corresponds with the regions showing the most striking histopathologic alterations in AD. A significant elevation of free HNE in ventricular cerebrospinal fluid (CSF) and serum provides a potential biomarker for AD [93, 94]. Protein-bound HNE, which is indication of Michael addition of HNE to proteins, is elevated in AD [2, 4, 84, 95]. HNE is elevated in neurons treated with Aβ (1-42) [8, 84, 96]. Protein–bound HNE alters conformation and function of proteins [84, 97] . In AD brain protein–bound HNE is found on the glutamate transporter (Glt-1 or EAAT), glutathione–S-transferase (GST), and mulit-drug resistant protein (MRP-1) [84, 98]. Thus processes related to excitotoxicity may be facilitated, while processes related to removal of HNE from neurons (via GST and MRP1) may be compromised.
DNA Oxidation
More than four decades ago, it was suggested that DNA is the primary target of ROS leading to cellular aging [99]. Due to the high oxygen consumption rate by the brain, ROS may contribute to neuronal damage in aging and neurological disorders [5]. Oxidative damage to DNA by ROS results in strand breaks, DNA–DNA and DNA–protein cross-linking, and sister chromatid exchange and translocation [100, 101]. DNA bases are also attacked by lipid peroxidation products HNE and acrolein, which lead to formation of bulky exocyclic adducts. This modification can cause inappropriate base pairing that alters protein synthesis. DNA oxidation by ROS also produces oxidized base adducts, such as 8-hydroxy-2-deoxyguanine (8-OHdG) [102, 103]. Guanine, because it has the lowest oxidation potential of the four DNA bases, is the most readily oxidized base and therefore the mostly commonly used analysis of DNA oxidation.
Previous studies have demonstrated a two-fold increase in DNA strand breaks in AD brain that consequently results in depletion of energy stores and cell death [104]. DNA oxidation has been shown to escalate with age, shown by an increase in 8-OHdG in the cerebral cortex and cerebellum brain regions [105]. Similarly both mitochondrial DNA (mtDNA) and nuclear DNA (nDNA) have increased 8-hydroxyguanine (8-OHG), 8-hydroxyadenine (8-OHA), and 5-hydroxyuracil (5-OHU) in temporal, parietal, and frontal lobes in AD [105, 106]. Overall, mtDNA shows an approximately 10-fold higher intensity of oxidized bases than nDNA. This result demonstrates that nDNA and mtDNA undergo extensive oxidative damage in AD, which may contribute to the neurodegenerative pathology of this disorder.
RNA Oxidation
Studies have shown that 30–70% oxidation of the mRNAs in the frontal cortex of the AD brain in comparison to only 2% oxidation age-matched controls [107]. A specific increase in rRNA oxidation has also been shown in the AD IPL when compared to age-matched controls [108]. Increased levels of 8-hydroxyguanine (8-OHG) have also been reported in the hippocampus and cerebral neocortex of the AD brain, while the 8-OHG level in the cerebellum was not significantly altered when compared with controls [108-110]. An increase in 8-OHdG has been identified not only in brain tissue but also in CSF from AD patients [111]. RNA oxidation in the AD brain could render the cell incapable of initiating protein synthesis, hindering the cell's defense against further oxidative damage an effect observed in AD [108].
Mitochondrial Dysfunction
Mitochondrial dysfunction in AD is central to the development of oxidative stress since it is primary source of cellular oxidants [112, 113]. In vivo positron emission tomography (PET) has provided specific evidence of brain metabolism abnormalities associated with AD [114], which precede neuropsychological impairment and visual atrophy [115, 116]. Frontal cortex and middle temporal gyrus show a significant decrease in metabolism as well as synaptic dysfunction [117-121]. Studies of autopsied AD brain tissue also revealed decreased pyruvate dehydrogenase activity in the parietal, temporal, and frontal cortex [122-124]. Cytochrome c oxidase (COX), Complex IV in the electron transport chain (ETC), consistently shows decreased activity in AD brain [125]. Decreased α-ketoglutarate dehydrogenase activity in the parietal and temporal cortex of AD patients has also been reported that could be related to the HNE binding to α-KG [126-130] , though this could be affected by longer postmortem intervals [131]. All the above-mentioned mitochondrial changes would limit ATP production and increases ROS production and suggests possible abnormalities in mitochondrial functions in AD. ROS production is directly related to mitochondrial membrane potential (Δψ) such that hyperpolarization (high Δψ) promotes ROS production [132-134]. In addition, several in vitro studies of Aß and mitochondrial function have reported that Aß affects mitochondrial DNA and proteins, leading to impairments of the electronic transport chain (ETC) and ultimately mitochondrial dysfunction [135, 136]. Dysfunction of mitochondria is reported to alter APP metabolism, enhancing intraneuronal accumulation of amyloid beta-peptide and enhancing neuronal vulnerability [137]. Consistent with the importance of Aβ(1-42) in AD pathogenesis, recent research has suggested the presence of intracellular Aβ (1-42) in mitochondria from brains of transgenic mice and AD patients [138, 139], and showed increased expression of mitochondrial genes before Aβ deposition in APP transgenic mice, and suggested that the up regulation of mitochondrial genes may be a compensatory response because of mitochondrial oxidative damage caused by the over-expression of mutant APP and/or Aß [139] . Further, the accumulation of Aβ in the mitochondria may be associated with diminished enzymatic activity of mitochondria [140], where the peptide is proposed to disrupt energy production while promoting mitochondrially derived apoptotic processes via the intrinsic pathway. This claim of mitochondrial needs replication but is a provocative hypothesis that provides potential insight into metabolic alterations and oxidative stress in AD brain.
Vascular Factors in the Conversion from MCI to AD
Oxidative stress is also associated with vascular factors [141-144] that reportedly play a role in the conversion of MCI to AD [145]. Vascular factors include, but are not limited to, APOE4 allele, diabetes, smoking, hypertension and heart disease [146]. ApoE4 isoform is associated with increased peripheral lipid levels, glial cell activation, and decreased peripheral lipid metabolism, and cerebral glucose metabolism, that may eventually lead to inflammation, oxidative stress, and dementia. A recent study by Wolozin and Bednar (147) suggests that ApoE does not directly contribute to AD by increasing serum cholesterol, but rather ApoE might act via a mechanism involving Abeta [147]. From our laboratory we showed that increased oxidative stress in APP/PS-1 mice is independent of dietary cholesterol, which further supports the role of Abeta in the induction of oxidative stress rather than the direct effect of cholestrol [148] . In addition to ApoE other risk factors for AD are coronary artery bypass surgery, congestive heart failure, cerebrovascular-carotid atherosclerosis, atrial hypotension, and transient ischemia attacks [149]. In all these conditions, one common factor is the production of oxidative stress that often occurs by ischemic reperfusion injury. Patients with cardiovascular disease and MCI are reported to have increased short-term memory loss and decreased ability to learn and retain new material, and decreased recall and short-term memory compared to MCI patients without vascular risk factors [150]. Another risk factor for AD is diabetes mellitus in which increased levels of glucose and insulin may increase the levels of free radicals such as superoxide and hydrogen peroxide [151]. Monomeric Aβ is known to compete with insulin for insulin degrading enzyme, so diabetes may lead to elevated levels of Aβ, thereby posing a risk factor for development of AD [49]. Prevention or treatment of vascular risk factors can reduce oxidative stress [152]. Vascular factors are also associated with AD [153-156]. Therefore, it is possible that vascular factors contribute to the conversion of MCI to AD [157, 158].
Oxidative stress in MCI Brain
As described above, MCI is characterized by mild current memory loss without dementia or significant impairment of other cognitive functions or activities of daily living [159, 160]. As also noted above, a number of MCI subjects show neuropathological hallmarks similar to AD, including temporal lobe atrophy and low CSF Aβ levels [161].
Plasma of MCI patients is reported to have decreased levels of non-enzymatic antioxidants and decreased activity of antioxidant enzymes compared to those of controls, while there were no alterations of protein levels [162]. This diminution of antioxidants could possibly increase the production of free radicals during the progression of the disease. Previous studies reported increased oxidative damage in nuclear and mitochondrial DNA in MCI, as indexed by increased levels of 8-OHdG, 2,6-diamino-4-hydroxy-5-formamidopyrimidine (fapyguanine), 8-hydroxyadenine, 4,6-diamino-5-formamidopyrimidine (fapyadenine) and 5-hydroxycytosine [163, 164]. Markesbery and co-workers showed that subjects with MCI have higher levels of isoprostanes (F2isoP) in the plasma, urine and cerebrospinal fluid compared to that of the healthy subjects [165]. Lipid peroxidation markers such as free HNE, protein-bound HNE, TBARS, and MDA were reported to be elevated in MCI brain [166-168]. Our laboratory showed that protein-bound HNE levels and 3-NT are elevated in the MCI brain compared to control IPL and hippocampus [166, 169]. Protein carbonyls are also elevated in MCI brain [36, 167]. These results suggest the accumulation of oxidative stress [36, 108, 166, 167] in MCI brain and are consistent with the notion that oxidative stress could be an early event in the progression of MCI to AD.
Redox Proteomics identification of oxidatively modified brain proteins in AD
Redox Proteomics
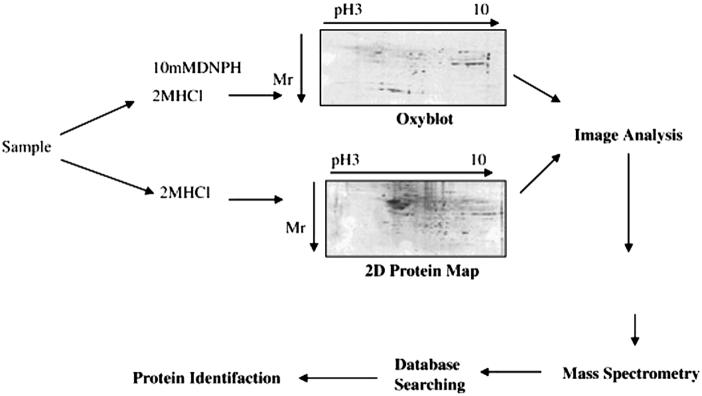
Redox proteomics used in our laboratory to identify specifically oxidized proteins in Alzheimer's disease brain involves coupling two-dimensional-polyacrylamide gel electrophoresis-mediated separation of proteins to mass spectroscopic analysis (Figure 4). Two-dimensional (2D) gel electrophoresis allows the analyses of complex protein mixtures based on two important physicochemical properties, i.e., isoelectric focusing (IEF) and relative mobility (Mr) [170]. The 2-D gel and blot maps obtained from proteomics provide information about the pI, molecular weight, expression, and post-and co-translational modifications of a protein of interest. Usually a single spot on the 2-D gel corresponds to a single protein [171]. 2D-PAGE for separation of proteins has some important limitations, such as solubilization of membrane proteins [172], highly basic proteins, and inability to detect low-abundance proteins.
Figure 4.
Diagrammatic representation of redox proteomics.
To use the 2D for maximum optimization, chaotropic agents, such as urea and thiourea, coupled with nonionic or zwitterionic detergents, are used to solubilize proteins and avoid protein precipitation during the IEF and the SDS gel electrophoresis. To avoid cathodic drift, immobilized pH IEF strips are used instead of tube gels, which improve protein map reproducibility between samples. Further, the use of narrow range IEF strips can expand the area of interest in a map of a proteome to give high-resolution separation.
In addition to the often-used method of 2D-PAGE, other separation methods, including 2D-HPLC and isotopically coded affinity tags (ICAT), are also used for protein separation. The ICAT method is used to analyze protein expression in two different sets of samples together by labeling the samples with different isotopes that bind to cysteine residues [173].
In our laboratory we used redox proteomics techniques to identify oxidatively modified proteins in oxidative stress-related diseases and their models. In this method we use a parallel analysis that couples 2D-PAGE with 2D-immunochemical detection of protein carbonyls derivatized by DNPH, nitrated proteins indexed by 3-NT, or protein adducts of HNE, on 2D Western blots, followed by MS analysis of the corresponding gel spot, as shown in Fig. 4. Proteins containing reactive carbonyl groups/3-NT/HNE in AD and control brain samples are detected by 2D Western blot analysis using specific antibodies. The 2D Western blots and 2D gel images are matched by computer-assisted image analysis, and the anti-DNP/nitrotyrosine/HNE immunoreactivity of individual proteins are normalized to their content, obtained by measuring the intensity of colloidal Coomassie Blue or SYPRO ruby stained spots. Such analysis allows comparison of levels of oxidatively modified brain proteins in AD versus control subjects.
Mass Spectrometry and Database Searching
Once a protein spot is identified as significantly involved in AD or MCI, it is digested with trypsin in the gel, and the resulting tryptic peptides undergo mass spectrometry analysis [10, 12, 13, 36-38, 174]. The peptide mass fingerprints obtained are characteristic of a specific protein, which allows correct identification of a particular protein using a suitable database that compares the experimental masses with theoretical masses of trypsin-generated protein sequences.
The mass spectrometric methods that are mostly used include MALDI (matrix assisted laser desorption/ionization) and ESI (electrospray ionization). In MALDI analysis the peptide sample is mixed with a matrix (usually α-cyano-4-hydroxycinnamic acid or 2,5-dihydroxybenzoic acid) and deposited on to a plate that is subjected to laser radiation, which allows the matrix to absorb the energy and then transfer the proton to the peptides in the gas phase. In ESI, MS/MS analysis of peptides leads to sequence information of these tryptic peptides. The exact mechanism for the ionization process in ESI is not well understood; however, a number of hypotheses were put forward to explain the ionization process in ESI. One of such hypothesis resulted from the use of HPLC-coupled to ESI, which suggests that as the liquid leaves the nozzle, the electric field induces a net charge on the small droplets. As the solvent evaporates, the droplet shrinks and the charge density at the surface of the droplet increases. This will eventually lead to the explosion of the droplet producing multiply charge analyte ions.
Surface-enhanced laser desorption /ionization time-of-flight (SELDI-TOF) is another proteomics tool that couples the classical methods of chromatographic sample preparation with mass spectrometry analysis. This method is important for applications in biomarker analysis, but is optimal for proteins below a relatively low molecular weight (<30kDa) [175].
The correct identity of the protein is determined by analysis using peptide mass data to interrogate on-line protein databases. The protein sequence database SwissProt is the most commonly used database for protein identification that is based on computer algorithms [176], and is available gratis through the internet. Most of the various protein search databases that are available online are listed in Table I.
Table I.
Mass spectrometry search engines for peptide mass fingerprinting
| Search engine | URL |
|---|---|
| Mascot | http://www.matrixscience.com |
| MOWSE | http://www.hgmp.mrc.ac.uk/Bioinformatics/Webapp/mowse |
| Profound | http://prowl.rockefeller.edu/profound_bin/WebProFound.exe |
| MS-fit | http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm |
| Peptident | http://ca.expasy.org/tools/peptident.html |
The information obtained from proteomics has significant potential to provide insight into AD pathogenesis, to develop disease markers, and to identify potential targets for drug therapy in AD.
Oxidative Modification of Brain Proteins in AD: Carbonylation and Nitration
Any type of oxidative modification (i.e., nitration, carbonylation, etc.) generally causes the protein to lose of functionality and lower enzyme activity [12]. Oxidatively modified proteins have been identified in AD hippocampus and AD IPL [10, 13, 37, 38, 177, 178]. Previous reports of an increased total protein carbonyls in AD brain [82]initiated studies to determine if specific proteins were more vulnerable or targeted for protein oxidation. Using redox proteomics, we identified specific carbonylated proteins in the hippocampus and inferior parietal lobe (IPL) of the AD brain. . In the IPL region, we reported dihydropyriminidase-related protein 2 (DRP2), α-enolase, heat shock cognate 71(HSC 71), creatine kinase BB (CK BB), glutamine synthase (GS), and ubiquitin carboxyl terminal hydrolase L-1 (UCHL-1) showing a specific increase in protein carbonyls in the AD brain as compared to age-matched control [37, 38]. In a separate study of the AD hippocampus, α-enolase, UCHL-1, DRP2, HSC 71, CK BB, peptidyl prolyl-cis,transisomerase 1 (Pin1), triosephosphate isomerase (TPI), and carbonic anhydrase 2 (CA II) were identified showing an increase in protein carbonyls in the AD brain as compared to age-matched control brain [12]. These data support the notion that protein carbonylation affects energy metabolism, pH regulation, and mitochondrial functions.
Protein nitration has detrimental effects as seen in key proteins such as glutamine synthase [179], ubiquitin [180], tyrosine hydroxylase [181, 182] and Mn superoxide dismutase [183-186], which lose activity upon protein nitration. As noted above, previous research has shown that protein nitration is associated with AD [10, 11, 13, 90, 187]. Our laboratory [10, 13] identified several proteins significantly nitrated in AD hippocampus and IPL compared to control brain. These proteins include: glyceraldehyde-3-phosphate dehydrogenase, ATP synthase α- chain, carbonic anhydrase-II, voltage-dependent anion channel-protein, α-enolase, γ-enolase, β- actin, L-lactate dehydrogenase (LDH), neuropolypeptide h3, and triosephosphate isomerase (TPI) (Table II).
Table II.
Oxidatively modified proteins identified in AD hippocampus and IPL brain regions
| Protein function | Hippocampus | Inferior parietal lobule |
Oxidative modification |
Reference |
|---|---|---|---|---|
| Energy-related enzymes |
α-enolase*, TPI, PGM1, GAPDH |
α-enolase, TPI*, CK, γ-enolase*, LDH |
Carbonylation and nitration |
10, 12, 13,37,38 |
| Proteosome- related proteins |
UCHL-1, HSC-71 |
UCHL-1, HSC- 71 |
Carbonylation | 12, 37,38 |
| Structual proteins | DRP-2 | DRP-2, β-actin* | Carbonylation and nitration |
10, 12, 13, 37 |
| Neurotransmitter- related proteins |
GS | Carbonylation | 37 | |
| Cholinergic function and lipid asymmetry |
Neuropolypeptide h3* |
Nitration | 10 | |
| pH regulation protein |
CAII* | Carbonylation and nitration |
12,13 | |
| Cell cycle; tau phosphorylation; Abeta production |
Pin-1 | Carbonylation | 12 | |
| Synaptic abnormalities and LTP |
γ-SNAP | Carbonylation | 12 | |
| Mitochondrial related protein |
VDAC*, ATP synthase (α- chain)* |
Nitration | 13 |
indicates this protein was found to be significantly nitrated
Functional Classification of Redox Proteomics-Identified Oxidatively Modified Brain Proteins in AD
Energy-related enzymes
α- and γ-enolase, TPI, GAPDH, LDH, PGM1, and CK are seven proteins that were identified to be oxidatively modified in AD brain that affect energy production and use in the cell. α- and γ-enolase are two subunits in the enolase enzyme. This enzyme converts 2-phosphoglycerate to phosphoenolypyruvate in glycolysis. Alpha enolase levels are increased AD brain, but activity is reduced, causing impairment in glucose metabolism [12, 188]. TPI catalyzes the conversion of dihydroxyacetone phosphate (DHAP) to glyceraldehyde-3-phosphate (G3P) in glycolysis. In the previous step, fructose 1,6-bisphosphate is cleaved into DHAP and G3P. Dihydroxyacetone phosphate is not directly involved in glycolysis. The enzyme TPI catalyzes this reaction converting DHAP into G3P (the aldose isomer), and causes an additional molecule of glyceraldehyde 3-phosphate to continue along the glycolytic pathway. TPI has been shown to be oxidatively modified in the AD brain [12]. Another glycolytic protein, PGM1 has been shown to be oxidized in the AD brain, specifically in the hippocampus. PGM 1 converts 3-phosphoglycerate to 2-phosphoglycerate. Glycolysis is especially important to the energy production in the brain, since glucose is the main source of energy. Impaired glycolytic function directly relates to less ATP available to the cells and various cellular processes that require ATP may be impaired, which is consistent with findings of altered glucose metabolism and tolerance in AD patients [189-192].
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is the enzyme responsible for the converting glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate, a high-energy phosphate product. NAD+ and inorganic phosphate are needed to drive the reaction, consequently producing NADH as a byproduct of the reaction. Studies of the AD brain have shown GAPDH to be oxidatively modified with decreased enzyme activity [193, 194]. L- Lactate dehydrogenase (LDH) reduces pyruvate to lactate using NADH as a cofactor. A benefit of this reaction is to generate NAD+, which is used as a co-factor for GAPDH in glycolysis. The oxidation and consequential dysfunction of LDH in AD also leads to reduced GAPDH activity. A reduction in GAPDH activity hinders glycolysis, lowering energy production in the cell. GAPDH also serves as a NO trap, which utilizes a large number of Cys residues [195]. Hence, oxidative modification of GAPDH would lead to a decreased protection of neurons and increased 3-NT formation. CK BB catalyzes the phosphorylation of creatine to creatine phosphate, which is an important high energy storage molecule [196]. The phosphate group is then used in the production of ATP providing immediate energy to the cell [197]. ATP is in constant demand by various ATPases to maintain ion pumps, lipid asymmetry, and cell-cell- communication. The oxidative dysfunction of these enzymes identified by redox proteomics could interrupt glycolysis and disrupt energy metabolism in brain, consistent with reduced energy utilization observed in AD brain [190, 191].
Proteasome-related proteins
The ubiquitin – proteasome pathway has been shown to be dysfunctional in the AD brain [198]. Ubiquitin carboxy terminal hyrdolase-1 (UCHL-1) is crucial to the degradation of damaged or misfolded proteins through proteasomal pathway. UCHL-1 has been found to be oxidized in the hippocampus and IPL regions of the AD brain [12, 37, 199] , which is consistent with AD pathology [200]. The dysfunction of UCHL-1 would be consistent with the dysfunction of the proteasome, accumulation of damaged and aggregated proteins, and excess protein ubiquitinylation observed in AD [198]. Also, recent studies show that UCHL-1 can recover synaptic function and contextual memory formation from Aβ-induced oxidation [201]. Hence, oxidatively dysfunctional UCHL-1 may contribute to altered synaptic function and memory of AD.
The cooperation of proteasome and molecular chaperones are required to control protein-protein interaction and the development of aggregates [202]. The identification of the oxidatively – modified heat shock cognate 71 (HSC 71) is consistent with impairment of the proteasomal pathway in AD, as HSC 71 is involved with the proteasome and protein degradation [38, 203]. HSC 71 also directly binds to the cytoplasmic domain of APP, which conceivably could contribute to alterations in APP processing and Aβ production observed in the AD brain [204].
Structural proteins: β- actin and DRP-2
Maintaining cytoskeletal integrity is important, because shortened dendritic length can be associated with or AD pathology [205]. Actin is a major component of thin microfilaments. β-actin plays a role in preserving cytoskeletal structure. Since β-actin is carbonylated [206] and nitrated in AD brain [10], structural integrity could be compromised and these changes may contribute to neuronal death [22].
DRP-2 is a neuronal repair protein identified as oxidized in AD brain [12, 38]. DRP-2 modulates collapsin activity, which is essential to elongating and directing dendrites to adjacent neurons. Collapsin is also critical in axonal outgrowth. Oxidative modification of DRP-2 may to lead to the observed shortened dendrite length in AD [207], with consequent impairment of neuronal communication.
DRP-2 is downregulated in Down's syndrome and AD patients [208]. Shortened dendritic length observed in AD [207] conceivably could be a result of loss of function of oxidatively modified DRP-2. The protein DRP-2 was also identified as oxidized in the inferior parietal lobule of AD patients and in Aβ-treated synaptosomes [38, 209]. Neuronal miscommunication is a likely contributory factor in the memory loss and cognitive decline in seen in AD patients as demonstrated in those with the ApoE4 allele, a risk factor for AD [210]. These redox proteomics results are consistent with the memory and synapse loss attributed to AD as a consequence of the decreased function of these two structural proteins.
Cholinergic function and lipid asymmetry
Neuropolypeptide h3, also known as RAF kinase inhibitor (RKIP), phosphatidylethanolamine binding protein (PEBP), and hippocampal cholinergic neurostimulating protein (HCNP), is critical to the production of choline acetyltransferase, which is vital in signal transduction. The loss of choline acetyltransferase leads to reduced levels of the neurotransmitter, acetylcholine, causing poor neurotransmission [211]. More specifically, there is decreased expression of HCNP in AD hippocampus [212]. N-methyl-D-aspartic acid (NMDA) receptors activate the production of this enzyme, and alteration of the NMDA receptor mediates cholinergic deficits [213]. Alzheimer's disease is associated with cholinergic neuronal loss [214-216]. Since neuropolypeptide h3 is also PEBP, it is possible that lipid asymmetry is also affected by oxidized PEBP. We showed that lipid asymmetry is lost in synaptic membranes treated with Aβ(1-42) or HNE [217, 218].
Neurotransmitter-related proteins
An excess of glutamate causes toxicity to neurons by over-stimulation of neurons by this excitatory neurotransmitter, e.g., excitotoxicity. A high concentration of glutamate over-activates ionotropic glutamate receptors, which leads to an increase of intracellular Ca2+, resulting in cell death. Synaptic depletion is one of the characteristics of AD brain that could be secondary to glutamate excitotoxicity. Glutamine synthetase (GS) is an enzyme identified by redox proteomics as being oxidatively modified in AD brain [37]. GS catalyzes the reaction of glutamate with ammonia to form glutamine, a non-toxic amino acid. Previous studies have shown GS to be oxidatively modified in the AD brain [37]. Further studies have also revealed reduced GS activity in the AD brain [6, 82]. These results are consistent with those of others, who showed that excessive extracellular glutamate leads to intracellular ROS [219]. Decreased GS activity and excitotoxicity leads to neurodegeneration, consistent with synaptic loss in AD.
Synaptic abnormalities and LTP
Soluble N-ethylmaleimide-sensitive factor (NSF) attachment proteins are highly conserved and are involved in intracellular membrane fusion and vesicular trafficking , which is critical to the function of the synapse. These proteins are involved in neurotransmitter release, hormone secretion, mitochondrial organization, and vesicular transport. γ -Soluble NSF attachment protein (γ-SNAP), is one of three isoforms and is involved in long-term potentiation, a key process for learning and memory [220]. γ-SNAP was identified as oxidatively modified in the AD brain [12], which has clear implications in AD pathology through impaired neurotransmission [221]. This finding also has implications with respect to LTP learning and memory, the diminution of which is a hallmark of AD.
pH regulation protein
Carbonic anhydrase II (CA II) catalyzes the hydration of CO2 to HCO3−, regulates pH in the cell, and aids in transporting HCO3− and CO2. The main function of CA II is maintaining electrolyte and water balance [222, 223]. The activity of CA II is diminished in AD hippocampus [178, 188]. Nitration of CA II can deregulate pH in the cell. Since so many reactions and enzymes, as well as the mitochondrial proton gradient for ATP synthesis, are pH-dependent, malfunction of this enzyme carbonic anhydrase II may be involved in AD.
Cell cycle, tau phosphorylation, Abeta production
Pin-1 is one of the peptidyl-prolyl isomerases (PPIase) that regulates biological functions of proteins including protein assembly, folding, intracellular transport, intracellular signaling, transcription, cell cycle progression and apoptosis. Of particular relevance to AD, Pin1 regulates enzymes associated with both phosphorylation and dephosphorylation of the critical cytoskeletal protein, tau [224]. Also, Pin1 binding to APP regulates the production of Aβ [225]. Finally, Pin1 is related to neurons in the cell cycle [224] . Since we showed that Pin1 was oxidatively dysfunctional in both MCI and AD brain [12, 36, 177], three major pathological hall marks of this dementing disorder (NFT, SP, and synapse loss due to neurons trapped in the cell cycle) may be related to damaged Pin1. In MCI brain elevation of cell cycle proteins was found [226], consistent with this notion. In addition, previous studies reported down regulation of Pin1 levels and activity in AD brain [36, 178]. Pin1 was reported to co-localize with phosphorylated tau in AD brain, and showed an inverse relationship to the levels of tau. In addition, an in vitro study reported that Pin1 protects neurons against age-related neurodegeneration [227]. Pin1 can also restore the ability of phosphorylated tau to bind microtubules and promote their assembly in vitro, a process that might represent a potential therapeutic use for Pin1 [228]. Studies to test the hypothesis of the critical importance of Pin1 in AD pathogenesis are ongoing in our laboratory.
Mitochondrial-related proteins
Voltage-dependent anion channel-protein (VDAC) is the outer component of the mitochondrial permeability transition pore (MPTP). VDAC is critical to importing and exporting various metabolites (i.e., ATP) into the mitochondria. This protein, in its role as a part of the MPTP, plays a part in the release of cytochrome c [229], caspases [230], and apoptosis inducing factor [231] from the mitochondria, essential for apoptosis. Nitration of VDAC alters MPTP function and can induce apoptosis by preventing Bcl-2/VDAC interaction in favor of increasing the proapoptotic factor, Bax and cytochrome c release [232]. Thus neuronal death could ensue. Recent research suggests that VDAC plays a role in Aβ-mediated neurotoxicity [233].
ATP synthase, α-subunit, is an ATP synthesizing enzyme composed of a coupling factor 1 (F1) head and the hydrophobic F0 component. The F1 head is composed of three catalytic sites for ATP production. Once ADP and inorganic phosphate (Pi) bind to the catalytic site of this enzyme, one ATP is created. This binding of ADP to Pi must be in a precise “tight” conformation for ATP to be produced. If the ADP and Pi are in the “loose” or “open” conformations, their affinity is not high enough for ATP production. Therefore, ATP production through ATP synthase occurs in a “rotary catalysis and proton electrochemical gradient” [234]. The rotation occurs by protons inducing a conformational shift. Loss of function of this protein can be detrimental to ATP production and in energy metabolism, consistent with PET studies of AD.
Redox proteomics Identification of oxidatively modified brain proteins in mci
Recent redox proteomics study from our laboratory identified alpha-enolase, glutamine synthetase, pyruvate kinase M2 and Pin1 in MCI hippocampus as being oxidatively modified [36]. In AD brain, three of these proteins, i.e., Pin1, glutamine synthetase, and enolase, were reported as oxidatively modified [37, 38, 177]. These proteins are crucial for energy metabolism, and neurotransmission. The identification of Pin1 as a common target of oxidation between AD and MCI suggests that Pin1 oxidation might be one of the driving forces for the intitation or progression of AD pathogenesis that may ultimately lead to cell death and neurodegeneration. Research to test this notion is ongoing in our laboratory.
Redox proteomics analysis of In-vivo models of AD centered around human Aβ(1-42)
Human Aβ(1-42) Injected into Rat Cholinergic-Rich Basal Forebrain
A cholinergic animal model of AD was prepared by injecting Aβ(1-42) into the nucleus basalis magnocellarius (NBM) of rat brain to replicate the cholinergic dysfunction reported in AD brain [235, 236]. Degeneration of the basal forebrain cholinergic neurons is pronounced in AD and is associated with cognitive deficits [237, 238]. In this animal model oxidative stress was observed in hippocampus, cortex and nucleus basalis; however, an extensive protein oxidation was observed in hippocampus compared to that of other regions [35, 237].
Using regional redox proteomics, a large number of oxidized proteins were identified in this animal model that include: 14-3-3 zeta, β-synuclein, pyruvate dehydrogenase, GAPDH, and PGM1, GS and tubulin (β- chain 15/alpha) and chaperonin 60 (HSP60) [35]. These proteins play a crucial role in signal transduction, cellular structure, energy metabolism, and stress response. As discussed above, a number of proteins that were identified in this animal model were already reported to be oxidatively modified in AD brain, including PGM1, GAPDH, GS and tubulin [10, 12, 13]. The expression of 14-3-3 proteins were reported to be increased in AD brain and CSF [239, 240] and these proteins also were found to be associated with NFT [241].
Recently, it has been shown that 14-3-3 zeta acts as a scaffolding protein simultaneously binding to tau and glycogen synthase kinase 3 β (GSK3β) in a multiprotein tau phosphorylation complex [242]. The hyperphosphorylation of tau could be associated with the oxidation of 14-3-3 zeta in this rat model following Aβ(1-42) injection into NBM that could unite both the importance of Aβ(1-42), and the hyperphosphorylation of tau. Heat shock proteins 60 is a mitochondrial chaperone protein that is involved in mediating the proper folding and assembly of mitochondrial proteins, especially in response to oxidative stress [243]. Expression of HSP60 is significantly decreased in AD and Aβ(25-35) has been shown to induce oxidation of HSP60 in fibroblasts derived from AD patients compared to age matched controls [203, 244]. The proteomics identification of the oxidation of HSP60 by Aβ(1-42) in vivo [35] could presumably lead to loss of function of HSP60 that may lead to increased protein misfolding and aggregation, as well as an increased vulnerability to oxidative stress. Moreover, as noted above, in AD there is evidence that Aβ(1-42) migrates to mitochondria, where HSP 60 conceivably could be affected.
Human Aβ1-42)-Expressing Caenorhabditis elegans (C. elegans)
Transgenic C. elegans is a worm model in which human Aβ (1–42) is expressed through a body-wall muscle myosin promoter and an Aβ minigene [245]. This model has been used to study Aβ toxicity, fibril formation and oxidative stress in vivo [50, 246, 247]. Increased protein oxidation that preceded fibrillar deposition of human Aβ(1-42) was observed, consistent with the notion that small, soluble oligomers of the peptide are the toxic species [50]. We also showed that C. elegans expressing human Aβ(1-42) exhibited an increased oxidative stress and also suggested association of methionine 35 of Aβ(1-42) in the mechanism of oxidative damage [247]. A redox proteomics study in this animal model identified sixteen proteins that were oxidatively modified [34]. The oxidatively modified proteins in this model of Aβ include: ATP synthase α chain, glutamate dehydrogenase, proteasome α- and β-subunit, glutathione S-transferase (ε-class), medium and short-chain acyl-CoA dehydrogenase, arginine kinase, myosin regulatory light chain, actin, adenosine kinase, malate dehydrogenase, transketolase, translation elongation factor EF-1 γ, lipid binding protein, and RACK1 ortholog. These identified proteins were involved in a variety of cellular functions including energy metabolism, protein degradation, cytoskeletal integrity, antioxidant system, signal transduction, and lipid metabolism. Many of these classes of oxidized proteins are also represented in AD brain as noted above.
Canine Model
Beagle dogs are a good model to study the role of Aβ, since these animals have the same Aβ sequence as humans. In addition, aged beagles deposit Aβ and have cognitive dysfunctions similar to humans [248, 249]. Histopathologically, aged beagle canines show reduced cerebral volume, cortical atrophy, ventricular widening as assessed by in vivo magnetic resonance imaging [250-252]. Beagle dogs absorb dietary nutrients in similar ways as humans. Previous studies in aged canines showed that long-term antioxidants reduce cognitive decline [253-255] and lead to rapid improvements in complex learning ability in aging dogs [256, 257]. These characteristics make the beagle dog a highly relevant for studying human AD considering the similarities in Aβ deposition, histopathology, and nutrition. The histopathological results in aged beagle dog imitate human AD brain pathology before any occurance of amyloid deposition [258-260].
Using redox proteomics, our group identified six brain proteins that showed decreased oxidation in aged beagles that had received behavioral enrichment and an antioxidant-supplemented diet relative to control aged animals. These proteins that were protected from oxidative modification in aged beagles include: glutamate dehydrogenase (GDH), GAPDH, α-enolase, neurofilament triplet L protein, GST and fascin actin bundling protein [174]. In addition, our group also reported increased activity of key antioxidant enzymes such as glutathione-S-transferase (GST) and superoxide dismutase (SOD) in this co-treated animal model of Aβ [174]. These protected proteins play an important role in energy metabolism, maintenance and stabilization of cell structure, and cellular defense. We suggest that these proteins identified by redox proteomics may play key roles in learning and memory in normal physiology, since protection of these proteins from oxidation in aged beagles was correlated with improved learning and memory.. Among the brain proteins identified by proteomics as oxidized in aged beagles, enolase, GAPDH and GST were found to be commonly oxidized between AD and aged beagle dogs.
The in vivo models of Aβ support a critical role of Aβ-induced oxidative stress and consequently the role of Aβ in AD pathology. The beagle studies suggest a possible therapeutic approach to lowering risk of developing AD that combines behavioral enrichment with an antioxidant-rich diet that, similar to the aged beagles with Aβ(1-42) of the same sequence as humans, could delay or significantly modulate AD pathology while improving memory.
Future Research
The increased oxidative stress parameters in MCI brain regions [36, 166, 167, 169] suggest that oxidative stress may be an early event in the progression from normal brain to AD pathology. These results further support the hypothesis that oxidative stress is a mediator of synaptic loss and a presumed factor for the formation of neurofibrillary tangles and senile plaques [10, 12, 13, 37, 38, 98, 199]. A better understanding of MCI could help in delineating the mechanism of AD and in developing effective therapeutics to slow or prevent conversion of MCI to AD and to treat AD. Recent AD clinical research has emphasized early detection of AD with the hope that early treatment will slow or prevent the progression of AD pathogenesis. On-going efforts in our laboratory include testing of promising therapeutic approaches to inhibit oxidative damage in MCI, AD, and models thereof [29, 60, 261-268]. Given the expected 14-16 million Americans and more than 22 million persons worldwide expected to develop AD in the relatively near future in the absence of effective intervention, increased national funding for AD-related research is imperative.
Acknowledgements
This work was supported in part by grants from the National Institute of Health [AG-05119; AG-10836].
Abbreviations
- 3-NT
3-nitrotyrosine
- 5-OHU
5-hydroxyuracil
- 8-OHA
8-hydroxyadenine
- 8-OHdG
8-hydroxy-2-deoxyguanine
- 8-OHG
8-hydroxyguanine
- Aβ
amyloid β-peptide
- AD
Alzheimer's disease
- APP
Amyloid precursor protein
- C. elegans
Caenorhabditis elegans
- CAII
Carbonic anhydrase 2
- CK BB
Creatine kinase BB isoform
- CSF
cerebrospinal fluid
- DHAP
Dihydroxyacetone phosphate
- DNPH
2,4-dinitrophenylhydrazine
- DRP2
Dihydropyrimidinase-related protein 2
- ESI
Electrospray ionization
- G3P
Glyceraldehyde-3-phosphate
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GDH
Glutamate dehydrogenase
- GS
Glutamine synthase
- GSK3β
Glycogen synthase kinase 3 β
- GST
Glutathione–S–transferase
- HNCP
Hippocampal cholinergic neurostimulating protein
- HNE
4-hydroxy-2-trans-nonenal
- HSC 71
Heat shock cognate 71
- HSP60
Chaperonin 60
- ICAT
Isotopically coded affinity tags
- IEF
isoelectric focusing
- iNOS
Inducible nitric oxide synthase
- IPL
Inferior parietal lobe
- LDH
L-lactate dehydrogenase
- MALDI
Matrix assisted laser desorption/ionization
- MCI
Mild cognitive impairment
- mtDNA
Mitochondrial DNA
- MetO
Methionine sulfoxide
- MetS·+
Sulfuranyl radical
- MPTP
mitochondrial permeability transition pore
- MRP-1
Multi-drug resistant protein
- NBM
nucleus basalis magnocellarius
- nDNA
Nuclear DNA
- NFT
Neurofibrillary tangles
- NMDA
N-methyl-D-aspartic acid
- nNOS
Neuronal NOS
- NO•
Nitric oxide
- NSF
N-ethylmaleimide-sensitive factor
- O2•−
Superoxide radical anion
- ONOO−
Peroxynitrite
- PEBP
Phosphatidylethanolamine binding protein
- PGM1
Phosphoglycerate mutase 1
- PHF
Paired helical filaments
- Pin1
Peptidyl prolyl-cis,trans isomerase 1
- RKIP
RAF kinase inhibitor
- RNS
Reactive nitrogen species
- ROS
Reactive oxygen species
- RSNO
Nitrosothiol
- SELDI-TOF
Surface-enhanced laser desorption /ionization time-of-flight
- γ-SNAP
γ -Soluble N-ethylmaleimide-sensitive attachment protein
- SOD
Superoxide dismutase
- SP
Senile plaques
- TBARS
Thiobarbituric acid reactive substance
- TPI
Triosephosphate isomerase
- UCHL-1
Ubiquitin carboxy-terminal hydrolase L-1
- VDAC
Voltage-dependant anion channel protein
- VF
Ventricular cerebrospinal fluid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Butterfield DA. Amyloid beta-peptide (1-42)-induced oxidative stress and neurotoxicity: implications for neurodegeneration in Alzheimer's disease brain. A review. Free Radic Res. 2002;36:1307–1313. doi: 10.1080/1071576021000049890. [DOI] [PubMed] [Google Scholar]
- 2.Butterfield DA, Castegna A, Lauderback CM, Drake J. Evidence that amyloid beta-peptide-induced lipid peroxidation and its sequelae in Alzheimer's disease brain contribute to neuronal death. Neurobiol Aging. 2002;23:655–664. doi: 10.1016/s0197-4580(01)00340-2. [DOI] [PubMed] [Google Scholar]
- 3.Butterfield DA, Drake J, Pocernich C, Castegna A. Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid beta-peptide. Trends Mol Med. 2001;7:548–554. doi: 10.1016/s1471-4914(01)02173-6. [DOI] [PubMed] [Google Scholar]
- 4.Butterfield DA, Lauderback CM. Lipid peroxidation and protein oxidation in Alzheimer's disease brain: potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free Radic Biol Med. 2002;32:1050–1060. doi: 10.1016/s0891-5849(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 5.Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 6.Butterfield DA, Stadtman ER. Protein Oxidation processes in aging brain. Adv Cell Aging Gerontol. 1997;2:161–191. [Google Scholar]
- 7.Lovell MA, Xie C, Markesbery WR. Acrolein is increased in Alzheimer's disease brain and is toxic to primary hippocampal cultures. Neurobiol Aging. 2001;22:187–194. doi: 10.1016/s0197-4580(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 8.Mark RJ, Lovell MA, Markesbery WR, Uchida K, Mattson MP. A role for 4-hydroxynonenal, an aldehydic product of lipid peroxidation, in disruption of ion homeostasis and neuronal death induced by amyloid beta-peptide. J Neurochem. 1997;68:255–264. doi: 10.1046/j.1471-4159.1997.68010255.x. [DOI] [PubMed] [Google Scholar]
- 9.Smith MA, Taneda S, Richey PL, Miyata S, Yan SD, Stern D, Sayre LM, Monnier VM, Perry G. Advanced Maillard reaction end products are associated with Alzheimer disease pathology. Proc Natl Acad Sci U S A. 1994;91:5710–5714. doi: 10.1073/pnas.91.12.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer's disease brain. J Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 11.Smith MA, Richey Harris PL, Sayre LM, Beckman JS, Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Merchant M, Markesbery WR, Butterfield DA. Redox proteomics identification of oxidized proteins in Alzheimer's disease hippocampus and cerebellum: an approach to understand pathological and biochemical alterations in AD. Neurobiol Aging. 2006;27:1564–1576. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 13.Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, Markesbery WR, Butterfield DA. Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 14.Berlett BS, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 15.Dalle-Donne I, Scaloni A, Butterfield DA. Redox Proteomics. Wiley; New York: 2006. [Google Scholar]
- 16.Dalle-Donne I, Scaloni A, Giustarini D, Cavarra E, Tell G, Lungarella G, Colombo R, Rossi R, Milzani A. Proteins as biomarkers of oxidative/nitrosative stress in diseases: the contribution of redox proteomics. Mass Spectrom Rev. 2005;24:55–99. doi: 10.1002/mas.20006. [DOI] [PubMed] [Google Scholar]
- 17.Stadtman ER, Levine RL. Free radical-mediated oxidation of free amino acids and amino acid residues in proteins. Amino Acids. 2003;25:207–218. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 18.Butterfield DA. beta-Amyloid-associated free radical oxidative stress and neurotoxicity: implications for Alzheimer's disease. Chem Res Toxicol. 1997;10:495–506. doi: 10.1021/tx960130e. [DOI] [PubMed] [Google Scholar]
- 19.Levine RL, Williams JA, Stadtman ER, Shacter E. Carbonyl assays for determination of oxidatively modified proteins. Methods Enzymol. 1994;233:346–357. doi: 10.1016/s0076-6879(94)33040-9. [DOI] [PubMed] [Google Scholar]
- 20.Koppenol WH, Moreno JJ, Pryor WA, Ischiropoulos H, Beckman JS. Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem Res Toxicol. 1992;5:834–842. doi: 10.1021/tx00030a017. [DOI] [PubMed] [Google Scholar]
- 21.Murphy MP, Packer MA, Scarlett JL, Martin SW. Peroxynitrite: a biologically significant oxidant. Gen Pharmacol. 1998;31:179–186. doi: 10.1016/s0306-3623(97)00418-7. [DOI] [PubMed] [Google Scholar]
- 22.Beckman JS, Koppenol WH. Nitric oxide, superoxide, and peroxynitrite: the good, the bad, and ugly. Am J Physiol. 1996;271:C1424–1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 23.Ischiropoulos H. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem Biophys Res Commun. 2003;305:776–783. doi: 10.1016/s0006-291x(03)00814-3. [DOI] [PubMed] [Google Scholar]
- 24.Radi R, Beckman JS, Bush KM, Freeman BA. Peroxynitrite oxidation of sulfhydryls. The cytotoxic potential of superoxide and nitric oxide. J Biol Chem. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 25.Broillet MC. S-nitrosylation of proteins. Cell Mol Life Sci. 1999;55:1036–1042. doi: 10.1007/s000180050354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gow AJ, Duran D, Malcolm S, Ischiropoulos H. Effects of peroxynitrite-induced protein modifications on tyrosine phosphorylation and degradation. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 27.Butterfield DA, Kanski J. Brain protein oxidation in age-related neurodegenerative disorders that are associated with aggregated proteins. Mech Ageing Dev. 2001;122:945–962. doi: 10.1016/s0047-6374(01)00249-4. [DOI] [PubMed] [Google Scholar]
- 28.Stadtman ER, Berlett BS. Reactive oxygen-mediated protein oxidation in aging and disease. Chem Res Toxicol. 1997;10:485–494. doi: 10.1021/tx960133r. [DOI] [PubMed] [Google Scholar]
- 29.Abdul HM, Butterfield DA. Involvement of PI3K/PKG/ERK1/2 signaling pathways in cortical neurons to trigger protection by cotreatment of acetyl-L-carnitine and alpha-lipoic acid against HNE-mediated oxidative stress and neurotoxicity: implications for Alzheimer's disease. Free Radic Biol Med. 2007;42:371–384. doi: 10.1016/j.freeradbiomed.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hensley K, Robinson KA, Gabbita SP, Salsman S, Floyd RA. Reactive oxygen species, cell signaling, and cell injury. Free Radic Biol Med. 2000;28:1456–1462. doi: 10.1016/s0891-5849(00)00252-5. [DOI] [PubMed] [Google Scholar]
- 31.Naoi M, Maruyama W, Shamoto-Nagai M, Yi H, Akao Y, Tanaka M. Oxidative stress in mitochondria: decision to survival and death of neurons in neurodegenerative disorders. Mol Neurobiol. 2005;31:81–93. doi: 10.1385/MN:31:1-3:081. [DOI] [PubMed] [Google Scholar]
- 32.Butterfield DA. Proteomics: a new approach to investigate oxidative stress in Alzheimer's disease brain. Brain Res. 2004;1000:1–7. doi: 10.1016/j.brainres.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Butterfield DA, Boyd-Kimball D, Castegna A. Proteomics in Alzheimer's disease: insights into potential mechanisms of neurodegeneration. J Neurochem. 2003;86:1313–1327. doi: 10.1046/j.1471-4159.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- 34.Boyd-Kimball D, Poon HF, Lynn BC, Cai J, Pierce WM, Jr., Klein JB, Ferguson J, Link CD, Butterfield DA. Proteomic identification of proteins specifically oxidized in Caenorhabditis elegans expressing human Abeta(1-42): implications for Alzheimer's disease. Neurobiol Aging. 2006;27:1239–1249. doi: 10.1016/j.neurobiolaging.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 35.Boyd-Kimball D, Sultana R, Poon HF, Lynn BC, Casamenti F, Pepeu G, Klein JB, Butterfield DA. Proteomic identification of proteins specifically oxidized by intracerebral injection of amyloid beta-peptide (1-42) into rat brain: implications for Alzheimer's disease. Neuroscience. 2005;132:313–324. doi: 10.1016/j.neuroscience.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 36.Butterfield DA, Poon HF, Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: Insights into the development of Alzheimer's disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 38.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 39.Perluigi M, Fai Poon H, Hensley K, Pierce WM, Klein JB, Calabrese V, De Marco C, Butterfield DA. Proteomic analysis of 4-hydroxy-2-nonenal-modified proteins in G93A-SOD1 transgenic mice--a model of familial amyotrophic lateral sclerosis. Free Radic Biol Med. 2005;38:960–968. doi: 10.1016/j.freeradbiomed.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 40.Poon HF, Farr SA, Thongboonkerd V, Lynn BC, Banks WA, Morley JE, Klein JB, Butterfield DA. Proteomic analysis of specific brain proteins in aged SAMP8 mice treated with alpha-lipoic acid: implications for aging and age-related neurodegenerative disorders. Neurochem Int. 2005;46:159–168. doi: 10.1016/j.neuint.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Poon HF, Shepherd HM, Reed TT, Calabrese V, Stella AM, Pennisi G, Cai J, Pierce WM, Klein JB, Butterfield DA. Proteomics analysis provides insight into caloric restriction mediated oxidation and expression of brain proteins associated with age-related impaired cellular processes: Mitochondrial dysfunction, glutamate dysregulation and impaired protein synthesis. Neurobiol Aging. 2006;27:1020–1034. doi: 10.1016/j.neurobiolaging.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 42.Morris JC, Storandt M, Miller JP, McKeel DW, Price JL, Rubin EH, Berg L. Mild cognitive impairment represents early-stage Alzheimer disease. Arch Neurol. 2001;58:397–405. doi: 10.1001/archneur.58.3.397. [DOI] [PubMed] [Google Scholar]
- 43.Petersen RC, Doody R, Kurz A, Mohs RC, Morris JC, Rabins PV, Ritchie K, Rossor M, Thal L, Winblad B. Current concepts in mild cognitive impairment. Arch Neurol. 2001;58:1985–1992. doi: 10.1001/archneur.58.12.1985. [DOI] [PubMed] [Google Scholar]
- 44.Portet F, Ousset PJ, Touchon J. [What is a mild cognitive impairment?] Rev Prat. 2005;55:1891–1894. [PubMed] [Google Scholar]
- 45.Portet F, Ousset PJ, Visser PJ, Frisoni GB, Nobili F, Scheltens P, Vellas B, Touchon J. Mild cognitive impairment (MCI) in medical practice: a critical review of the concept and new diagnostic procedure. Report of the MCI Working Group of the European Consortium on Alzheimer's Disease. J Neurol Neurosurg Psychiatry. 2006;77:714–718. doi: 10.1136/jnnp.2005.085332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Devanand DP, Pradhaban G, Liu X, Khandji A, De Santi S, Segal S, Rusinek H, Pelton GH, Honig LS, Mayeux R, Stern Y, Tabert MH, de Leon MJ. Hippocampal and entorhinal atrophy in mild cognitive impairment: prediction of Alzheimer disease. Neurology. 2007;68:828–836. doi: 10.1212/01.wnl.0000256697.20968.d7. [DOI] [PubMed] [Google Scholar]
- 47.Du AT, Schuff N, Amend D, Laakso MP, Hsu YY, Jagust WJ, Yaffe K, Kramer JH, Reed B, Norman D, Chui HC, Weiner MW. Magnetic resonance imaging of the entorhinal cortex and hippocampus in mild cognitive impairment and Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2001;71:441–447. doi: 10.1136/jnnp.71.4.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A. 1986;83:4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. doi: 10.1152/physrev.2001.81.2.741. [DOI] [PubMed] [Google Scholar]
- 50.Drake J, Link CD, Butterfield DA. Oxidative stress precedes fibrillar deposition of Alzheimer's disease amyloid beta-peptide (1-42) in a transgenic Caenorhabditis elegans model. Neurobiol Aging. 2003;24:415–420. doi: 10.1016/s0197-4580(02)00225-7. [DOI] [PubMed] [Google Scholar]
- 51.Lambert JC, Mann DM, Harris JM, Chartier-Harlin MC, Cumming A, Coates J, Lemmon H, Clair D, Iwatsubo T, Lendon C. The −48 C/T polymorphism in the presenilin 1 promoter is associated with an increased risk of developing Alzheimer's disease and an increased Abeta load in brain. J Med Genet. 2001;38:353–355. doi: 10.1136/jmg.38.6.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oda T, Wals P, Osterburg HH, Johnson SA, Pasinetti GM, Morgan TE, Rozovsky I, Stine WB, Snyder SW, Holzman TF, et al. Clusterin (apoJ) alters the aggregation of amyloid beta-peptide (A beta 1-42) and forms slowly sedimenting A beta complexes that cause oxidative stress. Exp Neurol. 1995;136:22–31. doi: 10.1006/exnr.1995.1080. [DOI] [PubMed] [Google Scholar]
- 53.Walsh DM, Hartley DM, Kusumoto Y, Fezoui, Y, Condron MM, Lomakin A, Benedek GB, Selkoe DJ, Teplow DB. Amyloid beta-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J Biol Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 54.Verdile G, Gandy SE, Martins RN. The role of presenilin and its interacting proteins in the biogenesis of Alzheimer's beta amyloid. Neurochem Res. 2007;32:609–623. doi: 10.1007/s11064-006-9131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isoo N, Sato C, Miyashita H, Shinohara M, Takasugi N, Morohashi Y, Tsuji S, Tomita T, Iwatsubo T. Abeta42 Overproduction Associated with Structural Changes in the Catalytic Pore of {gamma}-Secretase: COMMON EFFECTS OF PEN-2 N-TERMINAL ELONGATION AND FENOFIBRATE. J Biol Chem. 2007;282:12388–12396. doi: 10.1074/jbc.M611549200. [DOI] [PubMed] [Google Scholar]
- 56.Newman M, Musgrave FI, Lardelli M. Alzheimer disease: amyloidogenesis, the presenilins and animal models. Biochim Biophys Acta. 2007;1772:285–297. doi: 10.1016/j.bbadis.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Butterfield DA, Boyd-Kimball D. The critical role of methionine 35 in Alzheimer's amyloid beta-peptide (1-42)-induced oxidative stress and neurotoxicity. Biochim Biophys Acta. 2005;1703:149–156. doi: 10.1016/j.bbapap.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 58.Clementi ME, Pezzotti M, Orsini F, Sampaolese B, Mezzogori D, Grassi C, Giardina B, Misiti F. Alzheimer's amyloid beta-peptide (1-42) induces cell death in human neuroblastoma via bax/bcl-2 ratio increase: an intriguing role for methionine 35. Biochem Biophys Res Commun. 2006;342:206–213. doi: 10.1016/j.bbrc.2006.01.137. [DOI] [PubMed] [Google Scholar]
- 59.Crouch PJ, Barnham KJ, Duce JA, Blake RE, Masters CL, Trounce IA. Copper-dependent inhibition of cytochrome c oxidase by Abeta(1-42) requires reduced methionine at residue 35 of the Abeta peptide. J Neurochem. 2006;99:226–236. doi: 10.1111/j.1471-4159.2006.04050.x. [DOI] [PubMed] [Google Scholar]
- 60.Joshi G, Sultana R, Perluigi M, Butterfield D. In vivo protection of synaptosomes from oxidative stress mediated by Fe2+/H2O2 or 2,2-azobis-(2-amidinopropane) dihydrochloride by the glutathione mimetic tricyclodecan-9-ylxanthogenate. Free Radic Biol Med. 2005;38:1023–1031. doi: 10.1016/j.freeradbiomed.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 61.Murray IV, Sindoni ME, Axelsen PH. Promotion of oxidative lipid membrane damage by amyloid beta proteins. Biochemistry. 2005;44:12606–12613. doi: 10.1021/bi050926p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moskovitz J, Berlett BS, Poston JM, Stadtman ER. Methionine sulfoxide reductase in antioxidant defense. Methods Enzymol. 1999;300:239–244. doi: 10.1016/s0076-6879(99)00130-5. [DOI] [PubMed] [Google Scholar]
- 63.Maher P. Redox control of neural function: background, mechanisms, and significance. Antioxid Redox Signal. 2006;8:1941–1970. doi: 10.1089/ars.2006.8.1941. [DOI] [PubMed] [Google Scholar]
- 64.Lovell MA, Xie C, Gabbita SP, Markesbery WR. Decreased thioredoxin and increased thioredoxin reductase levels in Alzheimer's disease brain. Free Radic Biol Med. 2000;28:418–427. doi: 10.1016/s0891-5849(99)00258-0. [DOI] [PubMed] [Google Scholar]
- 65.Gabbita SP, Aksenov MY, Lovell MA, Markesbery WR. Decrease in peptide methionine sulfoxide reductase in Alzheimer's disease brain. J Neurochem. 1999;73:1660–1666. doi: 10.1046/j.1471-4159.1999.0731660.x. [DOI] [PubMed] [Google Scholar]
- 66.Kanski J, Aksenova M, Schoneich C, Butterfield DA. Substitution of isoleucine-31 by helical-breaking proline abolishes oxidative stress and neurotoxic properties of Alzheimer's amyloid beta-peptide. Free Radic Biol Med. 2002;32:1205–1211. doi: 10.1016/s0891-5849(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 67.Pogocki D, Schoneich C. Redox properties of Met(35) in neurotoxic beta-amyloid peptide. A molecular modeling study. Chem Res Toxicol. 2002;15:408–418. doi: 10.1021/tx0101550. [DOI] [PubMed] [Google Scholar]
- 68.Varadarajan S, Kanski J, Aksenova M, Lauderback C, Butterfield DA. Different mechanisms of oxidative stress and neurotoxicity for Alzheimer's A beta(1--42) and A beta(25--35) J Am Chem Soc. 2001;123:5625–5631. doi: 10.1021/ja010452r. [DOI] [PubMed] [Google Scholar]
- 69.Milller B, Williams T, Schoneich C. Mechanism of Sulfoxide Formation through Reaction of Sulfur Radical Cation Complexes with Superoxide of Hydroxide Ion in Oxygenated Aqueous Solution. J Am Chem Soc. 1996;118:11014–11025. [Google Scholar]
- 70.Brot N, Weissbach H. Biochemistry and physiological role of methionine sulfoxide residues in proteins. Arch Biochem Biophys. 1983;223:271–281. doi: 10.1016/0003-9861(83)90592-1. [DOI] [PubMed] [Google Scholar]
- 71.Vogt W. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radic Biol Med. 1995;18:93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 72.Varadarajan S, Yatin S, Kanski J, Jahanshahi F, Butterfield DA. Methionine residue 35 is important in amyloid beta-peptide-associated free radical oxidative stress. Brain Res Bull. 1999;50:133–141. doi: 10.1016/s0361-9230(99)00093-3. [DOI] [PubMed] [Google Scholar]
- 73.Naslund J, Schierhorn A, Hellman U, Lannfelt L, Roses AD, Tjernberg LO, Silberring J, Gandy SE, Winblad B, Greengard P, et al. Relative abundance of Alzheimer A beta amyloid peptide variants in Alzheimer disease and normal aging. Proc Natl Acad Sci U S A. 1994;91:8378–8382. doi: 10.1073/pnas.91.18.8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nordstedt C, Naslund J, Tjernberg LO, Karlstrom AR, Thyberg J, Terenius L. The Alzheimer A beta peptide develops protease resistance in association with its polymerization into fibrils. J Biol Chem. 1994;269:30773–30776. [PubMed] [Google Scholar]
- 75.Brunelle P, Rauk A. The radical model of Alzheimer's disease: specific recognition of Gly29 and Gly33 by Met35 in a beta-sheet model of Abeta: an ONIOM study. J Alzheimers Dis. 2002;4:283–289. doi: 10.3233/jad-2002-4403. [DOI] [PubMed] [Google Scholar]
- 76.Kanski J, Aksenova M, Butterfield DA. The hydrophobic environment of Met35 of Alzheimer's Abeta(1-42) is important for the neurotoxic and oxidative properties of the peptide. Neurotox Res. 2002;4:219–223. doi: 10.1080/10298420290023945. [DOI] [PubMed] [Google Scholar]
- 77.Kanski J, Varadarajan S, Aksenova M, Butterfield DA. Role of glycine-33 and methionine-35 in Alzheimer's amyloid beta-peptide 1-42-associated oxidative stress and neurotoxicity. Biochim Biophys Acta. 2002;1586:190–198. doi: 10.1016/s0925-4439(01)00097-7. [DOI] [PubMed] [Google Scholar]
- 78.Barnham KJ, Haeffner F, Ciccotosto GD, Curtain CC, Tew D, Mavros C, Beyreuther K, Carrington D, Masters CL, Cherny RA, Cappai R, Bush AI. Tyrosine gated electron transfer is key to the toxic mechanism of Alzheimer's disease beta-amyloid. FASEB J. 2004;18:1427–1429. doi: 10.1096/fj.04-1890fje. [DOI] [PubMed] [Google Scholar]
- 79.Huang X, Atwood CS, Hartshorn MA, Multhaup G, Goldstein LE, Scarpa RC, Cuajungco MP, Gray DN, Lim J, Moir RD, Tanzi RE, Bush AI. The A beta peptide of Alzheimer's disease directly produces hydrogen peroxide through metal ion reduction. Biochemistry. 1999;38:7609–7616. doi: 10.1021/bi990438f. [DOI] [PubMed] [Google Scholar]
- 80.Boyd-Kimball D, Sultana R, Mohmmad-Abdul H, Butterfield DA. Rodent Abeta(1-42) exhibits oxidative stress properties similar to those of human Abeta(1-42): Implications for proposed mechanisms of toxicity. J Alzheimers Dis. 2004;6:515–525. doi: 10.3233/jad-2004-6509. [DOI] [PubMed] [Google Scholar]
- 81.Ritchie CW, Bush AI, Mackinnon A, Macfarlane S, Mastwyk M, MacGregor L, Kiers L, Cherny R, Li QX, Tammer A, Carrington D, Mavros C, Volitakis I, Xilinas M, Ames D, Davis S, Beyreuther K, Tanzi RE, Masters CL. Metal-protein attenuation with iodochlorhydroxyquin (clioquinol) targeting Abeta amyloid deposition and toxicity in Alzheimer disease: a pilot phase 2 clinical trial. Arch Neurol. 2003;60:1685–1691. doi: 10.1001/archneur.60.12.1685. [DOI] [PubMed] [Google Scholar]
- 82.Hensley K, Hall N, Subramaniam R, Cole P, Harris M, Aksenov M, Aksenova M, Gabbita SP, Wu JF, Carney JM, et al. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 83.Markesbery WR, Lovell MA. Four-hydroxynonenal, a product of lipid peroxidation, is increased in the brain in Alzheimer's disease. Neurobiol Aging. 1998;19:33–36. doi: 10.1016/s0197-4580(98)00009-8. [DOI] [PubMed] [Google Scholar]
- 84.Lauderback CM, Hackett JM, Huang FF, Keller JN, Szweda LI, Markesbery WR, Butterfield DA. The glial glutamate transporter, GLT-1, is oxidatively modified by 4-hydroxy-2-nonenal in the Alzheimer's disease brain: the role of Abeta1-42. J Neurochem. 2001;78:413–416. doi: 10.1046/j.1471-4159.2001.00451.x. [DOI] [PubMed] [Google Scholar]
- 85.Perry G, Cash AD, Smith MA. Alzheimer Disease and Oxidative Stress. J Biomed Biotechnol. 2002;2:120–123. doi: 10.1155/S1110724302203010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tohgi H, Abe T, Yamazaki K, Murata T, Ishizaki E, Isobe C. Alterations of 3-nitrotyrosine concentration in the cerebrospinal fluid during aging and in patients with Alzheimer's disease. Neurosci Lett. 1999;269:52–54. doi: 10.1016/s0304-3940(99)00406-1. [DOI] [PubMed] [Google Scholar]
- 87.Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. Biochim Biophys Acta. 1998;1366:211–223. doi: 10.1016/s0005-2728(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 88.Schopfer FJ, Baker PR, Freeman BA. NO-dependent protein nitration: a cell signaling event or an oxidative inflammatory response? Trends Biochem Sci. 2003;28:646–654. doi: 10.1016/j.tibs.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 89.Lafon-Cazal M, Culcasi M, Gaven F, Pietri S, Bockaert J. Nitric oxide, superoxide and peroxynitrite: putative mediators of NMDA-induced cell death in cerebellar granule cells. Neuropharmacology. 1993;32:1259–1266. doi: 10.1016/0028-3908(93)90020-4. [DOI] [PubMed] [Google Scholar]
- 90.Hensley K, Maidt ML, Yu Z, Sang H, Markesbery WR, Floyd RA. Electrochemical analysis of protein nitrotyrosine and dityrosine in the Alzheimer brain indicates region-specific accumulation. J Neurosci. 1998;18:8126–8132. doi: 10.1523/JNEUROSCI.18-20-08126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Horiguchi T, Uryu K, Giasson BI, Ischiropoulos H, LightFoot R, Bellmann C, Richter-Landsberg C, Lee VM, Trojanowski JQ. Nitration of tau protein is linked to neurodegeneration in tauopathies. Am J Pathol. 2003;163:1021–1031. doi: 10.1016/S0002-9440(10)63462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lovell MA, Ehmann WD, Butler SM, Markesbery WR. Elevated thiobarbituric acid-reactive substances and antioxidant enzyme activity in the brain in Alzheimer's disease. Neurology. 1995;45:1594–1601. doi: 10.1212/wnl.45.8.1594. [DOI] [PubMed] [Google Scholar]
- 93.Lovell MA, Ehmann WD, Mattson MP, Markesbery WR. Elevated 4-hydroxynonenal in ventricular fluid in Alzheimer's disease. Neurobiol Aging. 1997;18:457–461. doi: 10.1016/s0197-4580(97)00108-5. [DOI] [PubMed] [Google Scholar]
- 94.McGrath LT, McGleenon BM, Brennan S, McColl D, Mc IS, Passmore AP. Increased oxidative stress in Alzheimer's disease as assessed with 4-hydroxynonenal but not malondialdehyde. QJM. 2001;94:485–490. doi: 10.1093/qjmed/94.9.485. [DOI] [PubMed] [Google Scholar]
- 95.Sayre LM, Zelasko DA, Harris PL, Perry G, Salomon RG, Smith MA. 4-Hydroxynonenal-derived advanced lipid peroxidation end products are increased in Alzheimer's disease. J Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 96.Boyd-Kimball D, Mohmmad Abdul H, Reed T, Sultana R, Butterfield DA. Role of phenylalanine 20 in Alzheimer's amyloid beta-peptide (1-42)-induced oxidative stress and neurotoxicity. Chem Res Toxicol. 2004;17:1743–1749. doi: 10.1021/tx049796w. [DOI] [PubMed] [Google Scholar]
- 97.Subramaniam R, Roediger F, Jordan B, Mattson MP, Keller JN, Waeg G, Butterfield DA. The lipid peroxidation product, 4-hydroxy-2-trans-nonenal, alters the conformation of cortical synaptosomal membrane proteins. J Neurochem. 1997;69:1161–1169. doi: 10.1046/j.1471-4159.1997.69031161.x. [DOI] [PubMed] [Google Scholar]
- 98.Sultana R, Butterfield DA. Oxidatively modified GST and MRP1 in Alzheimer's disease brain: implications for accumulation of reactive lipid peroxidation products. Neurochem Res. 2004;29:2215–2220. doi: 10.1007/s11064-004-7028-0. [DOI] [PubMed] [Google Scholar]
- 99.Szilard L. On the Nature of the Aging Process. Proc Natl Acad Sci U S A. 1959;45:30–45. doi: 10.1073/pnas.45.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Crawford DR, Suzuki T, Sesay J, Davies KJ. Analysis of gene expression following oxidative stress. Methods Mol Biol. 2002;196:155–162. doi: 10.1385/1-59259-274-0:155. [DOI] [PubMed] [Google Scholar]
- 101.Davies KJ. Oxidative stress: the paradox of aerobic life. Biochem Soc Symp. 1995;61:1–31. doi: 10.1042/bss0610001. [DOI] [PubMed] [Google Scholar]
- 102.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 103.Steenken S. Structure, acid/base properties and transformation reactions of purine radicals. Free Radic Res Commun. 1989;6:117–120. doi: 10.3109/10715768909073445. [DOI] [PubMed] [Google Scholar]
- 104.Zhang J, Dawson VL, Dawson TM, Snyder SH. Nitric oxide activation of poly(ADP-ribose) synthetase in neurotoxicity. Science. 1994;263:687–689. doi: 10.1126/science.8080500. [DOI] [PubMed] [Google Scholar]
- 105.Mecocci P, MacGarvey U, Beal MF. Oxidative damage to mitochondrial DNA is increased in Alzheimer's disease. Ann Neurol. 1994;36:747–751. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 106.Gabbita SP, Lovell MA, Markesbery WR. Increased nuclear DNA oxidation in the brain in Alzheimer's disease. J Neurochem. 1998;71:2034–2040. doi: 10.1046/j.1471-4159.1998.71052034.x. [DOI] [PubMed] [Google Scholar]
- 107.Shan X, Lin CL. Quantification of oxidized RNAs in Alzheimer's disease. Neurobiol Aging. 2006;27:657–662. doi: 10.1016/j.neurobiolaging.2005.03.022. [DOI] [PubMed] [Google Scholar]
- 108.Ding Q, Markesbery WR, Cecarini V, Keller JN. Decreased RNA, and increased RNA oxidation, in ribosomes from early Alzheimer's disease. Neurochem Res. 2006;31:705–710. doi: 10.1007/s11064-006-9071-5. [DOI] [PubMed] [Google Scholar]
- 109.Nunomura A, Perry G, Pappolla MA, Wade R, Hirai K, Chiba S, Smith MA. RNA oxidation is a prominent feature of vulnerable neurons in Alzheimer's disease. J Neurosci. 1999;19:1959–1964. doi: 10.1523/JNEUROSCI.19-06-01959.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shan X, Tashiro H, Lin CL. The identification and characterization of oxidized RNAs in Alzheimer's disease. J Neurosci. 2003;23:4913–4921. doi: 10.1523/JNEUROSCI.23-12-04913.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abe T, Tohgi H, Isobe C, Murata T, Sato C. Remarkable increase in the concentration of 8-hydroxyguanosine in cerebrospinal fluid from patients with Alzheimer's disease. J Neurosci Res. 2002;70:447–450. doi: 10.1002/jnr.10349. [DOI] [PubMed] [Google Scholar]
- 112.Beal MF. Mitochondria take center stage in aging and neurodegeneration. Ann Neurol. 2005;58:495–505. doi: 10.1002/ana.20624. [DOI] [PubMed] [Google Scholar]
- 113.Hirai K, Aliev G, Nunomura A, Fujioka H, Russell RL, Atwood CS, Johnson AB, Kress Y, Vinters HV, Tabaton M, Shimohama S, Cash AD, Siedlak SL, Harris PL, Jones PK, Petersen RB, Perry G, Smith MA. Mitochondrial abnormalities in Alzheimer's disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sullivan PG, Brown MR. Mitochondrial aging and dysfunction in Alzheimer's disease. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:407–410. doi: 10.1016/j.pnpbp.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 115.Blass JP. The mitochondrial spiral. An adequate cause of dementia in the Alzheimer's syndrome. Ann N Y Acad Sci. 2000;924:170–183. doi: 10.1111/j.1749-6632.2000.tb05576.x. [DOI] [PubMed] [Google Scholar]
- 116.Blass JP, Sheu RK, Gibson GE. Inherent abnormalities in energy metabolism in Alzheimer disease. Interaction with cerebrovascular compromise. Ann N Y Acad Sci. 2000;903:204–221. doi: 10.1111/j.1749-6632.2000.tb06370.x. [DOI] [PubMed] [Google Scholar]
- 117.DeKosky ST, Scheff SW. Synapse loss in frontal cortex biopsies in Alzheimer's disease: correlation with cognitive severity. Ann Neurol. 1990;27:457–464. doi: 10.1002/ana.410270502. [DOI] [PubMed] [Google Scholar]
- 118.Minoshima S, Giordani B, Berent S, Frey KA, Foster NL, Kuhl DE. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol. 1997;42:85–94. doi: 10.1002/ana.410420114. [DOI] [PubMed] [Google Scholar]
- 119.Scheff SW, DeKosky ST, Price DA. Quantitative assessment of cortical synaptic density in Alzheimer's disease. Neurobiol Aging. 1990;11:29–37. doi: 10.1016/0197-4580(90)90059-9. [DOI] [PubMed] [Google Scholar]
- 120.Scheff SW, Price DA. Synapse loss in the temporal lobe in Alzheimer's disease. Ann Neurol. 1993;33:190–199. doi: 10.1002/ana.410330209. [DOI] [PubMed] [Google Scholar]
- 121.Scheff SW, Price DA. Alzheimer's disease-related synapse loss in the cingulate cortex. J Alzheimers Dis. 2001;3:495–505. doi: 10.3233/jad-2001-3509. [DOI] [PubMed] [Google Scholar]
- 122.Perry EK, Perry RH, Tomlinson BE, Blessed G, Gibson PH. Coenzyme A-acetylating enzymes in Alzheimer's disease: possible cholinergic ‘compartment’ of pyruvate dehydrogenase. Neurosci Lett. 1980;18:105–110. doi: 10.1016/0304-3940(80)90220-7. [DOI] [PubMed] [Google Scholar]
- 123.Sorbi S, Bird ED, Blass JP. Decreased pyruvate dehydrogenase complex activity in Huntington and Alzheimer brain. Ann Neurol. 1983;13:72–78. doi: 10.1002/ana.410130116. [DOI] [PubMed] [Google Scholar]
- 124.Yates CM, Butterworth J, Tennant MC, Gordon A. Enzyme activities in relation to pH and lactate in postmortem brain in Alzheimer-type and other dementias. J Neurochem. 1990;55:1624–1630. doi: 10.1111/j.1471-4159.1990.tb04948.x. [DOI] [PubMed] [Google Scholar]
- 125.Castellani R, Hirai K, Aliev G, Drew KL, Nunomura A, Takeda A, Cash AD, Obrenovich ME, Perry G, Smith MA. Role of mitochondrial dysfunction in Alzheimer's disease. J Neurosci Res. 2002;70:357–360. doi: 10.1002/jnr.10389. [DOI] [PubMed] [Google Scholar]
- 126.Gibson GE, Blass JP, Beal MF, Bunik V. The alpha-ketoglutarate-dehydrogenase complex: a mediator between mitochondria and oxidative stress in neurodegeneration. Mol Neurobiol. 2005;31:43–63. doi: 10.1385/MN:31:1-3:043. [DOI] [PubMed] [Google Scholar]
- 127.Gibson GE, Park LC, Sheu KF, Blass JP, Calingasan NY. The alpha-ketoglutarate dehydrogenase complex in neurodegeneration. Neurochem Int. 2000;36:97–112. doi: 10.1016/s0197-0186(99)00114-x. [DOI] [PubMed] [Google Scholar]
- 128.Gibson GE, Sheu KF, Blass JP. Abnormalities of mitochondrial enzymes in Alzheimer disease. J Neural Transm. 1998;105:855–870. doi: 10.1007/s007020050099. [DOI] [PubMed] [Google Scholar]
- 129.Gibson GE, Zhang H, Xu H, Park LC, Jeitner TM. Oxidative stress increases internal calcium stores and reduces a key mitochondrial enzyme. Biochim Biophys Acta. 2002;1586:177–189. doi: 10.1016/s0925-4439(01)00091-6. [DOI] [PubMed] [Google Scholar]
- 130.Sheu KF, Blass JP. The alpha-ketoglutarate dehydrogenase complex. Ann N Y Acad Sci. 1999;893:61–78. doi: 10.1111/j.1749-6632.1999.tb07818.x. [DOI] [PubMed] [Google Scholar]
- 131.Sims NR. Energy metabolism, oxidative stress and neuronal degeneration in Alzheimer's disease. Neurodegeneration. 1996;5:435–440. doi: 10.1006/neur.1996.0059. [DOI] [PubMed] [Google Scholar]
- 132.Skulachev VP. Role of uncoupled and non-coupled oxidations in maintenance of safely low levels of oxygen and its one-electron reductants. Q Rev Biophys. 1996;29:169–202. doi: 10.1017/s0033583500005795. [DOI] [PubMed] [Google Scholar]
- 133.Skulachev VP. Uncoupling: new approaches to an old problem of bioenergetics. Biochim Biophys Acta. 1998;1363:100–124. doi: 10.1016/s0005-2728(97)00091-1. [DOI] [PubMed] [Google Scholar]
- 134.Votyakova TV, Reynolds IJ. DeltaPsi(m)-Dependent and -independent production of reactive oxygen species by rat brain mitochondria. J Neurochem. 2001;79:266–277. doi: 10.1046/j.1471-4159.2001.00548.x. [DOI] [PubMed] [Google Scholar]
- 135.Bozner P, Grishko V, LeDoux SP, Wilson GL, Chyan YC, Pappolla MA. The amyloid beta protein induces oxidative damage of mitochondrial DNA. J Neuropathol Exp Neurol. 1997;56:1356–1362. doi: 10.1097/00005072-199712000-00010. [DOI] [PubMed] [Google Scholar]
- 136.Pappolla MA, Chyan YJ, Omar RA, Hsiao K, Perry G, Smith MA, Bozner P. Evidence of oxidative stress and in vivo neurotoxicity of beta-amyloid in a transgenic mouse model of Alzheimer's disease: a chronic oxidative paradigm for testing antioxidant therapies in vivo. Am J Pathol. 1998;152:871–877. [PMC free article] [PubMed] [Google Scholar]
- 137.Busciglio J, Pelsman A, Wong C, Pigino G, Yuan M, Mori H, Yankner BA. Altered metabolism of the amyloid beta precursor protein is associated with mitochondrial dysfunction in Down's syndrome. Neuron. 2002;33:677–688. doi: 10.1016/s0896-6273(02)00604-9. [DOI] [PubMed] [Google Scholar]
- 138.Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, Xu HW, Stern D, McKhann G, Yan SD. Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. Faseb J. 2005;19:2040–2041. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- 139.Manczak M, Anekonda TS, Henson E, Park BS, Quinn J, Reddy PH. Mitochondria are a direct site of A beta accumulation in Alzheimer's disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum Mol Genet. 2006;15:1437–1449. doi: 10.1093/hmg/ddl066. [DOI] [PubMed] [Google Scholar]
- 140.Chen X, Yan SD. Mitochondrial Abeta: a potential cause of metabolic dysfunction in Alzheimer's disease. IUBMB Life. 2006;58:686–694. doi: 10.1080/15216540601047767. [DOI] [PubMed] [Google Scholar]
- 141.Harjai KJ. Potential new cardiovascular risk factors: left ventricular hypertrophy, homocysteine, lipoprotein(a), triglycerides, oxidative stress, and fibrinogen. Ann Intern Med. 1999;131:376–386. doi: 10.7326/0003-4819-131-5-199909070-00009. [DOI] [PubMed] [Google Scholar]
- 142.Kojda G, Harrison D. Interactions between NO and reactive oxygen species: pathophysiological importance in atherosclerosis, hypertension, diabetes and heart failure. Cardiovasc Res. 1999;43:562–571. doi: 10.1016/s0008-6363(99)00169-8. [DOI] [PubMed] [Google Scholar]
- 143.Maytin M, Leopold J, Loscalzo J. Oxidant stress in the vasculature. Curr Atheroscler Rep. 1999;1:156–164. doi: 10.1007/s11883-999-0012-z. [DOI] [PubMed] [Google Scholar]
- 144.Zalba G, Beaumont J, San Jose G, Fortuno A, Fortuno MA, Diez J. Vascular oxidant stress: molecular mechanisms and pathophysiological implications. J Physiol Biochem. 2000;56:57–64. doi: 10.1007/BF03179777. [DOI] [PubMed] [Google Scholar]
- 145.Fischer P, Jungwirth S, Zehetmayer S, Weissgram S, Hoenigschnabl S, Gelpi E, Krampla W, Tragl KH. Conversion from subtypes of mild cognitive impairment to Alzheimer dementia. Neurology. 2007;68:288–291. doi: 10.1212/01.wnl.0000252358.03285.9d. [DOI] [PubMed] [Google Scholar]
- 146.de la Torre JC. Alzheimer disease as a vascular disorder: nosological evidence. Stroke. 2002;33:1152–1162. doi: 10.1161/01.str.0000014421.15948.67. [DOI] [PubMed] [Google Scholar]
- 147.Wolozin B, Bednar MM. Interventions for heart disease and their effects on Alzheimer's disease. Neurol Res. 2006;28:630–636. doi: 10.1179/016164106X130515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mohmmad Abdul H, Wenk GL, Gramling M, Hauss-Wegrzyniak B, Butterfield DA. APP and PS-1 mutations induce brain oxidative stress independent of dietary cholesterol: implications for Alzheimer's disease. Neurosci Lett. 2004;368:148–150. doi: 10.1016/j.neulet.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 149.de la Torre JC. How do heart disease and stroke become risk factors for Alzheimer's disease? Neurol Res. 2006;28:637–644. doi: 10.1179/016164106X130362. [DOI] [PubMed] [Google Scholar]
- 150.Siuda J, Gorzkowska A, Opala G, Ochudlo S. Vascular risk factors and intensity of cognitive dysfunction in MCI. J Neurol Sci. 2007 doi: 10.1016/j.jns.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 151.Pasquier F, Boulogne A, Leys D, Fontaine P. Diabetes mellitus and dementia. Diabetes Metab. 2006;32:403–414. doi: 10.1016/s1262-3636(07)70298-7. [DOI] [PubMed] [Google Scholar]
- 152.Pilz H, Oguogho A, Chehne F, Lupattelli G, Palumbo B, Sinzinger H. Quitting cigarette smoking results in a fast improvement of in vivo oxidation injury (determined via plasma, serum and urinary isoprostane) Thromb Res. 2000;99:209–221. doi: 10.1016/s0049-3848(00)00249-8. [DOI] [PubMed] [Google Scholar]
- 153.de la Torre JC, Stefano GB. Evidence that Alzheimer's disease is a microvascular disorder: the role of constitutive nitric oxide. Brain Res Brain Res Rev. 2000;34:119–136. doi: 10.1016/s0165-0173(00)00043-6. [DOI] [PubMed] [Google Scholar]
- 154.Helmer C, Pasquier F, Dartigues JF. [Epidemiology of Alzheimer disease and related disorders] Med Sci (Paris) 2006;22:288–296. doi: 10.1051/medsci/2006223288. [DOI] [PubMed] [Google Scholar]
- 155.Pansari K, Gupta A, Thomas P. Alzheimer's disease and vascular factors: facts and theories. Int J Clin Pract. 2002;56:197–203. [PubMed] [Google Scholar]
- 156.Skoog I, Kalaria RN, Breteler MM. Vascular factors and Alzheimer disease. Alzheimer Dis Assoc Disord. 1999;13(Suppl 3):S106–114. doi: 10.1097/00002093-199912003-00016. [DOI] [PubMed] [Google Scholar]
- 157.Chiappelli M, Borroni B, Archetti S, Calabrese E, Corsi MM, Franceschi M, Padovani A, Licastro F. VEGF gene and phenotype relation with Alzheimer's disease and mild cognitive impairment. Rejuvenation Res. 2006;9:485–493. doi: 10.1089/rej.2006.9.485. [DOI] [PubMed] [Google Scholar]
- 158.Solfrizzi V, Panza F, Colacicco AM, D'Introno A, Capurso C, Torres F, Grigoletto F, Maggi S, Del Parigi A, Reiman EM, Caselli RJ, Scafato E, Farchi G, Capurso A. Vascular risk factors, incidence of MCI, and rates of progression to dementia. Neurology. 2004;63:1882–1891. doi: 10.1212/01.wnl.0000144281.38555.e3. [DOI] [PubMed] [Google Scholar]
- 159.Morris JC. Mild cognitive impairment and preclinical Alzheimer's disease. Geriatrics. 2005;(Suppl):9–14. [PubMed] [Google Scholar]
- 160.Petersen RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–308. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 161.Chertkow H, Bergman H, Schipper HM, Gauthier S, Bouchard R, Fontaine S, Clarfield AM. Assessment of suspected dementia. Can J Neurol Sci. 2001;28(Suppl 1):S28–41. doi: 10.1017/s0317167100001189. [DOI] [PubMed] [Google Scholar]
- 162.Guidi I, Galimberti D, Lonati S, Novembrino C, Bamonti F, Tiriticco M, Fenoglio C, Venturelli E, Baron P, Bresolin N, Scarpini E. Oxidative imbalance in patients with mild cognitive impairment and Alzheimer's disease. Neurobiol Aging. 2006;27:262–269. doi: 10.1016/j.neurobiolaging.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 163.Migliore L, Fontana I, Trippi F, Colognato R, Coppede F, Tognoni G, Nucciarone B, Siciliano G. Oxidative DNA damage in peripheral leukocytes of mild cognitive impairment and AD patients. Neurobiol Aging. 2005;26:567–573. doi: 10.1016/j.neurobiolaging.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 164.Wang J, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in mild cognitive impairment. J Neurochem. 2006;96:825–832. doi: 10.1111/j.1471-4159.2005.03615.x. [DOI] [PubMed] [Google Scholar]
- 165.Markesbery WR, Kryscio RJ, Lovell MA, Morrow JD. Lipid peroxidation is an early event in the brain in amnestic mild cognitive impairment. Ann Neurol. 2005;58:730–735. doi: 10.1002/ana.20629. [DOI] [PubMed] [Google Scholar]
- 166.Butterfield DA, Reed T, Perluigi M, De Marco C, Coccia R, Cini C, Sultana R. Elevated protein-bound levels of the lipid peroxidation product, 4-hydroxy-2-nonenal, in brain from persons with mild cognitive impairment. Neurosci Lett. 2006;397:170–173. doi: 10.1016/j.neulet.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 167.Keller JN, Schmitt FA, Scheff SW, Ding Q, Chen Q, Butterfield DA, Markesbery WR. Evidence of increased oxidative damage in subjects with mild cognitive impairment. Neurology. 2005;64:1152–1156. doi: 10.1212/01.WNL.0000156156.13641.BA. [DOI] [PubMed] [Google Scholar]
- 168.Williams TI, Lynn BC, Markesbery WR, Lovell MA. Increased levels of 4-hydroxynonenal and acrolein, neurotoxic markers of lipid peroxidation, in the brain in Mild Cognitive Impairment and early Alzheimer's disease. Neurobiol Aging. 2006;27:1094–1099. doi: 10.1016/j.neurobiolaging.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 169.Butterfield DA, Reed TT, Perluigi M, De Marco C, Coccia R, Keller JN, Markesbery WR, Sultana R. Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: Implications for the role of nitration in the progression of Alzheimer's disease. Brain Res. 2007;1148:243–248. doi: 10.1016/j.brainres.2007.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rabilloud T. Two-dimensional gel electrophoresis in proteomics: old, old fashioned, but it still climbs up the mountains. Proteomics. 2002;2:3–10. [PubMed] [Google Scholar]
- 171.Tilleman K, Stevens I, Spittaels K, Haute CV, Clerens S, Van Den Bergh G, Geerts H, Van Leuven F, Vandesande F, Moens L. Differential expression of brain proteins in glycogen synthase kinase-3 transgenic mice: a proteomics point of view. Proteomics. 2002;2:94–104. [PubMed] [Google Scholar]
- 172.Santoni V, Molloy M, Rabilloud T. Membrane proteins and proteomics: un amour impossible? Electrophoresis. 2000;21:1054–1070. doi: 10.1002/(SICI)1522-2683(20000401)21:6<1054::AID-ELPS1054>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 173.Smolka MB, Zhou H, Purkayastha S, Aebersold R. Optimization of the isotope-coded affinity tag-labeling procedure for quantitative proteome analysis. Anal Biochem. 2001;297:25–31. doi: 10.1006/abio.2001.5318. [DOI] [PubMed] [Google Scholar]
- 174.Opii WO, Sultana R, Abdul HM, Ansari MA, Nath A, Butterfield DA. Oxidative stress and toxicity induced by the nucleoside reverse transcriptase inhibitor (NRTI)-2′,3′-dideoxycytidine (ddC): Relevance to HIV-dementia. Exp Neurol. 2007;204:29–38. doi: 10.1016/j.expneurol.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Poon TC. Opportunities and limitations of SELDI-TOF-MS in biomedical research: practical advices. Expert Rev Proteomics. 2007;4:51–65. doi: 10.1586/14789450.4.1.51. [DOI] [PubMed] [Google Scholar]
- 176.Hoogland C, Sanchez JC, Tonella L, Binz PA, Bairoch A, Hochstrasser DF, Appel RD. The 1999 SWISS-2DPAGE database update. Nucleic Acids Res. 2000;28:286–288. doi: 10.1093/nar/28.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Markesbery WR, Zhou XZ, Lu KP, Butterfield DA. Oxidative modification and down-regulation of Pin1 in Alzheimer's disease hippocampus: A redox proteomics analysis. Neurobiol Aging. 2005 doi: 10.1016/j.neurobiolaging.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 178.Sultana R, Perluigi M, Butterfield DA. Redox proteomics identification of oxidatively modified proteins in Alzheimer's disease brain and in vivo and in vitro models of AD centered around Abeta(1-42) J Chromatogr B Analyt Technol Biomed Life Sci. 2006;833:3–11. doi: 10.1016/j.jchromb.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 179.Berlett BS, Friguet B, Yim MB, Chock PB, Stadtman ER. Peroxynitrite-mediated nitration of tyrosine residues in Escherichia coli glutamine synthetase mimics adenylylation: relevance to signal transduction. Proc Natl Acad Sci U S A. 1996;93:1776–1780. doi: 10.1073/pnas.93.5.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Yi D, Perkins PD. Identification of ubiquitin nitration and oxidation using a liquid chromatography/mass selective detector system. J Biomol Tech. 2005;16:364–370. [PMC free article] [PubMed] [Google Scholar]
- 181.Blanchard-Fillion B, Souza JM, Friel T, Jiang GC, Vrana K, Sharov V, Barron L, Schoneich C, Quijano C, Alvarez B, Radi R, Przedborski S, Fernando GS, Horwitz J, Ischiropoulos H. Nitration and inactivation of tyrosine hydroxylase by peroxynitrite. J Biol Chem. 2001;276:46017–46023. doi: 10.1074/jbc.M105564200. [DOI] [PubMed] [Google Scholar]
- 182.Souza JM, Chen Q, Blanchard-Fillion B, Lorch SA, Hertkorn C, Lightfoot R, Weisse M, Friel T, Paxinou E, Themistocleous M, Chov S, Ischiropoulos H. Reactive nitrogen species and proteins: biological significance and clinical relevance. Adv Exp Med Biol. 2001;500:169–174. doi: 10.1007/978-1-4615-0667-6_22. [DOI] [PubMed] [Google Scholar]
- 183.Anantharaman M, Tangpong J, Keller JN, Murphy MP, Markesbery WR, Kiningham KK, Clair DK. Beta-amyloid mediated nitration of manganese superoxide dismutase: implication for oxidative stress in a APPNLH/NLH X PS-1P264L/P264L double knock-in mouse model of Alzheimer's disease. Am J Pathol. 2006;168:1608–1618. doi: 10.2353/ajpath.2006.051223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. Peroxynitrite-mediated tyrosine nitration catalyzed by superoxide dismutase. Arch Biochem Biophys. 1992;298:431–437. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 185.Tangpong J, Cole MP, Sultana R, Estus S, Vore M, Clair W, Ratanachaiyavong S, Clair DK, Butterfield DA. Adriamycin-mediated nitration of manganese superoxide dismutase in the central nervous system: insight into the mechanism of chemobrain. J Neurochem. 2007;100:191–201. doi: 10.1111/j.1471-4159.2006.04179.x. [DOI] [PubMed] [Google Scholar]
- 186.Yamakura F, Taka H, Fujimura T, Murayama K. Inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J Biol Chem. 1998;273:14085–14089. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]
- 187.Good PF, Werner P, Hsu A, Olanow CW, Perl DP. Evidence of neuronal oxidative damage in Alzheimer's disease. Am J Pathol. 1996;149:21–28. [PMC free article] [PubMed] [Google Scholar]
- 188.Meier-Ruge W, Iwangoff P, Reichlmeier K. Neurochemical enzyme changes in Alzheimer's and Pick's disease. Arch Gerontol Geriatr. 1984;3:161–165. doi: 10.1016/0167-4943(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 189.Beal MF. Energy, oxidative damage, and Alzheimer's disease: clues to the underlying puzzle. Neurobiol Aging. 1994;15(Suppl 2):S171–174. doi: 10.1016/0197-4580(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 190.Hoyer S. Oxidative energy metabolism in Alzheimer brain. Studies in early-onset and late-onset cases. Mol Chem Neuropathol. 1992;16:207–224. doi: 10.1007/BF03159971. [DOI] [PubMed] [Google Scholar]
- 191.Messier C, Gagnon M. Glucose regulation and brain aging. J Nutr Health Aging. 2000;4:208–213. [PubMed] [Google Scholar]
- 192.Vanhanen M, Soininen H. Glucose intolerance, cognitive impairment and Alzheimer's disease. Curr Opin Neurol. 1998;11:673–677. doi: 10.1097/00019052-199812000-00011. [DOI] [PubMed] [Google Scholar]
- 193.Kish SJ, Lopes-Cendes I, Guttman M, Furukawa Y, Pandolfo M, Rouleau GA, Ross BM, Nance M, Schut L, Ang L, DiStefano L. Brain glyceraldehyde-3-phosphate dehydrogenase activity in human trinucleotide repeat disorders. Arch Neurol. 1998;55:1299–1304. doi: 10.1001/archneur.55.10.1299. [DOI] [PubMed] [Google Scholar]
- 194.Mazzola JL, Sirover MA. Reduction of glyceraldehyde-3-phosphate dehydrogenase activity in Alzheimer's disease and in Huntington's disease fibroblasts. J Neurochem. 2001;76:442–449. doi: 10.1046/j.1471-4159.2001.00033.x. [DOI] [PubMed] [Google Scholar]
- 195.Hara MR, Cascio MB, Sawa A. GAPDH as a sensor of NO stress. Biochim Biophys Acta. 2006;1762:502–509. doi: 10.1016/j.bbadis.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 196.Bessman SP, Carpenter CL. The creatine-creatine phosphate energy shuttle. Annu Rev Biochem. 1985;54:831–862. doi: 10.1146/annurev.bi.54.070185.004151. [DOI] [PubMed] [Google Scholar]
- 197.Aksenov MY, Aksenova MV, Payne RM, Smith CD, Markesbery WR, Carney JM. The expression of creatine kinase isoenzymes in neocortex of patients with neurodegenerative disorders: Alzheimer's and Pick's disease. Exp Neurol. 1997;146:458–465. doi: 10.1006/exnr.1997.6550. [DOI] [PubMed] [Google Scholar]
- 198.Keller JN, Hanni KB, Markesbery WR. Impaired proteasome function in Alzheimer's disease. J Neurochem. 2000;75:436–439. doi: 10.1046/j.1471-4159.2000.0750436.x. [DOI] [PubMed] [Google Scholar]
- 199.Choi J, Levey AI, Weintraub ST, Rees HD, Gearing M, Chin LS, Li L. Oxidative modifications and down-regulation of ubiquitin carboxyl-terminal hydrolase L1 associated with idiopathic Parkinson's and Alzheimer's diseases. J Biol Chem. 2004;279:13256–13264. doi: 10.1074/jbc.M314124200. [DOI] [PubMed] [Google Scholar]
- 200.Mirra SS, Heyman A, McKeel D, Sumi SM, Crain BJ, Brownlee LM, Vogel FS, Hughes JP, van Belle G, Berg L. The Consortium to Establish a Registry for Alzheimer's Disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer's disease. Neurology. 1991;41:479–486. doi: 10.1212/wnl.41.4.479. [DOI] [PubMed] [Google Scholar]
- 201.Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O. Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory. Cell. 2006;126:775–788. doi: 10.1016/j.cell.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 202.McClellan AJ, Tam S, Kaganovich D, Frydman J. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol. 2005;7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- 203.Yoo BC, Kim SH, Cairns N, Fountoulakis M, Lubec G. Deranged expression of molecular chaperones in brains of patients with Alzheimer's disease. Biochem Biophys Res Commun. 2001;280:249–258. doi: 10.1006/bbrc.2000.4109. [DOI] [PubMed] [Google Scholar]
- 204.Kouchi Z, Sorimachi H, Suzuki K, Ishiura S. Proteasome inhibitors induce the association of Alzheimer's amyloid precursor protein with Hsc73. Biochem Biophys Res Commun. 1999;254:804–810. doi: 10.1006/bbrc.1998.9977. [DOI] [PubMed] [Google Scholar]
- 205.Spires TL, Meyer-Luehmann M, Stern EA, McLean PJ, Skoch J, Nguyen PT, Bacskai BJ, Hyman BT. Dendritic spine abnormalities in amyloid precursor protein transgenic mice demonstrated by gene transfer and intravital multiphoton microscopy. J Neurosci. 2005;25:7278–7287. doi: 10.1523/JNEUROSCI.1879-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Aksenov MY, Aksenova MV, Butterfield DA, Geddes JW, Markesbery WR. Protein oxidation in the brain in Alzheimer's disease. Neuroscience. 2001;103:373–383. doi: 10.1016/s0306-4522(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 207.Coleman PD, Flood DG. Neuron numbers and dendritic extent in normal aging and Alzheimer's disease. Neurobiol Aging. 1987;8:521–545. doi: 10.1016/0197-4580(87)90127-8. [DOI] [PubMed] [Google Scholar]
- 208.Lubec G, Nonaka M, Krapfenbauer K, Gratzer M, Cairns N, Fountoulakis M. Expression of the dihydropyrimidinase related protein 2 (DRP-2) in Down syndrome and Alzheimer's disease brain is downregulated at the mRNA and dysregulated at the protein level. J Neural Transm Suppl. 1999;57:161–177. doi: 10.1007/978-3-7091-6380-1_10. [DOI] [PubMed] [Google Scholar]
- 209.Boyd-Kimball D, Castegna A, Sultana R, Poon HF, Petroze R, Lynn BC, Klein JB, Butterfield DA. Proteomic identification of proteins oxidized by Abeta(1-42) in synaptosomes: implications for Alzheimer's disease. Brain Res. 2005;1044:206–215. doi: 10.1016/j.brainres.2005.02.086. [DOI] [PubMed] [Google Scholar]
- 210.Ji Y, Gong Y, Gan W, Beach T, Holtzman DM, Wisniewski T. Apolipoprotein E isoform-specific regulation of dendritic spine morphology in apolipoprotein E transgenic mice and Alzheimer's disease patients. Neuroscience. 2003;122:305–315. doi: 10.1016/j.neuroscience.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 211.Ojika K. [Hippocampal cholinergic neurostimulating peptide] Seikagaku. 1998;70:1175–1180. [PubMed] [Google Scholar]
- 212.Maki M, Matsukawa N, Yuasa H, Otsuka Y, Yamamoto T, Akatsu H, Okamoto T, Ueda R, Ojika K. Decreased expression of hippocampal cholinergic neurostimulating peptide precursor protein mRNA in the hippocampus in Alzheimer disease. J Neuropathol Exp Neurol. 2002;61:176–185. doi: 10.1093/jnen/61.2.176. [DOI] [PubMed] [Google Scholar]
- 213.Jouvenceau A, Dutar P, Billard JM. Alteration of NMDA receptor-mediated synaptic responses in CA1 area of the aged rat hippocampus: contribution of GABAergic and cholinergic deficits. Hippocampus. 1998;8:627–637. doi: 10.1002/(SICI)1098-1063(1998)8:6<627::AID-HIPO5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 214.Coyle JT, Price DL, DeLong MR. Alzheimer's disease: a disorder of cortical cholinergic innervation. Science. 1983;219:1184–1190. doi: 10.1126/science.6338589. [DOI] [PubMed] [Google Scholar]
- 215.Perry EK, Curtis M, Dick DJ, Candy JM, Atack JR, Bloxham CA, Blessed G, Fairbairn A, Tomlinson BE, Perry RH. Cholinergic correlates of cognitive impairment in Parkinson's disease: comparisons with Alzheimer's disease. J Neurol Neurosurg Psychiatry. 1985;48:413–421. doi: 10.1136/jnnp.48.5.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wevers A, Witter B, Moser N, Burghaus L, Banerjee C, Steinlein OK, Schutz U, de Vos RA, Steur EN, Lindstrom J, Schroder H. Classical Alzheimer features and cholinergic dysfunction: towards a unifying hypothesis? Acta Neurol Scand Suppl. 2000;176:42–48. doi: 10.1034/j.1600-0404.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- 217.Abdul HM, Butterfield DA. Protection against amyloid beta-peptide (1-42)-induced loss of phospholipid asymmetry in synaptosomal membranes by tricyclodecan-9-xanthogenate (D609) and ferulic acid ethyl ester: implications for Alzheimer's disease. Biochim Biophys Acta. 2005;1741:140–148. doi: 10.1016/j.bbadis.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 218.Castegna A, Thongboonkerd V, Klein J, Lynn BC, Wang YL, Osaka H, Wada K, Butterfield DA. Proteomic analysis of brain proteins in the gracile axonal dystrophy (gad) mouse, a syndrome that emanates from dysfunctional ubiquitin carboxyl-terminal hydrolase L-1, reveals oxidation of key proteins. J Neurochem. 2004;88:1540–1546. doi: 10.1046/j.1471-4159.2003.02288.x. [DOI] [PubMed] [Google Scholar]
- 219.Lafon-Cazal M, Fagni L, Guiraud MJ, Mary S, Lerner-Natoli M, Pin JP, Shigemoto R, Bockaert J. mGluR7-like metabotropic glutamate receptors inhibit NMDA-mediated excitotoxicity in cultured mouse cerebellar granule neurons. Eur J Neurosci. 1999;11:663–672. doi: 10.1046/j.1460-9568.1999.00475.x. [DOI] [PubMed] [Google Scholar]
- 220.Lledo PM, Zhang X, Sudhof TC, Malenka RC, Nicoll RA. Postsynaptic membrane fusion and long-term potentiation. Science. 1998;279:399–403. doi: 10.1126/science.279.5349.399. [DOI] [PubMed] [Google Scholar]
- 221.Stenbeck G. Soluble NSF-attachment proteins. Int J Biochem Cell Biol. 1998;30:573–577. doi: 10.1016/s1357-2725(97)00064-2. [DOI] [PubMed] [Google Scholar]
- 222.Bertram G, Zierold K, Wessing A. Carbonic anhydrase supports electrolyte transport in Drosophila Malpighian tubules. Evidence by X-ray microanalysis of cryosections. J Insect Physiol. 1997;43:17–28. doi: 10.1016/s0022-1910(96)00078-9. [DOI] [PubMed] [Google Scholar]
- 223.Lucas JM, Knapp LW. A physiological evaluation of carbon sources for calcification in the octocoral Leptogorgia virgulata (Lamarck) J Exp Biol. 1997;200:2653–2662. doi: 10.1242/jeb.200.20.2653. [DOI] [PubMed] [Google Scholar]
- 224.Butterfield DA, Abdul HM, Opii W, Newman SF, Joshi G, Ansari MA, Sultana R. Pin1 in Alzheimer's disease. J Neurochem. 2006;98:1697–1706. doi: 10.1111/j.1471-4159.2006.03995.x. [DOI] [PubMed] [Google Scholar]
- 225.Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X, Xia W, Nicholson LK, Lu KP. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-beta production. Nature. 2006;440:528–534. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 226.Sultana R, Butterfield DA. Regional expression of key cell cycle proteins in brain from subjects with amnestic mild cognitive impairment. Neurochem Res. 2007;32:655–662. doi: 10.1007/s11064-006-9123-x. [DOI] [PubMed] [Google Scholar]
- 227.Liou MJ, Lu MC, Chen JN. Oxidation of explosives by Fenton and photo-Fenton processes. Water Res. 2003;37:3172–3179. doi: 10.1016/S0043-1354(03)00158-1. [DOI] [PubMed] [Google Scholar]
- 228.Thorpe JR, Morley SJ, Rulten SL. Utilizing the peptidyl-prolyl cis-trans isomerase pin1 as a probe of its phosphorylated target proteins. Examples of binding to nuclear proteins in a human kidney cell line and to tau in Alzheimer's diseased brain. J Histochem Cytochem. 2001;49:97–108. doi: 10.1177/002215540104900110. [DOI] [PubMed] [Google Scholar]
- 229.Madesh M, Hajnoczky G. VDAC-dependent permeabilization of the outer mitochondrial membrane by superoxide induces rapid and massive cytochrome c release. J Cell Biol. 2001;155:1003–1015. doi: 10.1083/jcb.200105057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Shimizu H, Banno Y, Sumi N, Naganawa T, Kitajima Y, Nozawa Y. Activation of p38 mitogen-activated protein kinase and caspases in UVB-induced apoptosis of human keratinocyte HaCaT cells. J Invest Dermatol. 1999;112:769–774. doi: 10.1046/j.1523-1747.1999.00582.x. [DOI] [PubMed] [Google Scholar]
- 231.Crompton M. The mitochondrial permeability transition pore and its role in cell death. Biochem J. 1999;341:233–249. [PMC free article] [PubMed] [Google Scholar]
- 232.Tsujimoto Y, Shimizu S. VDAC regulation by the Bcl-2 family of proteins. Cell Death Differ. 2000;7:1174–1181. doi: 10.1038/sj.cdd.4400780. [DOI] [PubMed] [Google Scholar]
- 233.Marin R, Ramirez CM, Gonzalez M, Gonzalez-Munoz E, Zorzano A, Camps M, Alonso R, Diaz M. Voltage-dependent anion channel (VDAC) participates in amyloid beta-induced toxicity and interacts with plasma membrane estrogen receptor alpha in septal and hippocampal neurons. Mol Membr Biol. 2007;24:148–160. doi: 10.1080/09687860601055559. [DOI] [PubMed] [Google Scholar]
- 234.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 235.Kasa P, Rakonczay Z, Gulya K. The cholinergic system in Alzheimer's disease. Prog Neurobiol. 1997;52:511–535. doi: 10.1016/s0301-0082(97)00028-2. [DOI] [PubMed] [Google Scholar]
- 236.Roberson MR, Harrell LE. Cholinergic activity and amyloid precursor protein metabolism. Brain Res Brain Res Rev. 1997;25:50–69. doi: 10.1016/s0165-0173(97)00016-7. [DOI] [PubMed] [Google Scholar]
- 237.Giovannini MG, Scali C, Prosperi C, Bellucci A, Vannucchi MG, Rosi S, Pepeu G, Casamenti F. Beta-amyloid-induced inflammation and cholinergic hypofunction in the rat brain in vivo: involvement of the p38MAPK pathway. Neurobiol Dis. 2002;11:257–274. doi: 10.1006/nbdi.2002.0538. [DOI] [PubMed] [Google Scholar]
- 238.Whitehouse PJ, Price DL, Clark AW, Coyle JT, DeLong MR. Alzheimer disease: evidence for selective loss of cholinergic neurons in the nucleus basalis. Ann Neurol. 1981;10:122–126. doi: 10.1002/ana.410100203. [DOI] [PubMed] [Google Scholar]
- 239.Burkhard PR, Sanchez JC, Landis T, Hochstrasser DF. CSF detection of the 14-3-3 protein in unselected patients with dementia. Neurology. 2001;56:1528–1533. doi: 10.1212/wnl.56.11.1528. [DOI] [PubMed] [Google Scholar]
- 240.Fountoulakis M, Cairns N, Lubec G. Increased levels of 14-3-3 gamma and epsilon proteins in brain of patients with Alzheimer's disease and Down syndrome. J Neural Transm Suppl. 1999;57:323–335. doi: 10.1007/978-3-7091-6380-1_23. [DOI] [PubMed] [Google Scholar]
- 241.Layfield R, Fergusson J, Aitken A, Lowe J, Landon M, Mayer RJ. Neurofibrillary tangles of Alzheimer's disease brains contain 14-3-3 proteins. Neurosci Lett. 1996;209:57–60. doi: 10.1016/0304-3940(96)12598-2. [DOI] [PubMed] [Google Scholar]
- 242.Agarwal-Mawal A, Qureshi HY, Cafferty PW, Yuan Z, Han D, Lin R, Paudel HK. 14-3-3 connects glycogen synthase kinase-3 beta to tau within a brain microtubule-associated tau phosphorylation complex. J Biol Chem. 2003;278:12722–12728. doi: 10.1074/jbc.M211491200. [DOI] [PubMed] [Google Scholar]
- 243.Bozner P, Wilson GL, Druzhyna NM, Bryant-Thomas TK, LeDoux SP, Wilson GL, Pappolla MA. Deficiency of chaperonin 60 in Down's syndrome. J Alzheimers Dis. 2002;4:479–486. doi: 10.3233/jad-2002-4604. [DOI] [PubMed] [Google Scholar]
- 244.Choi J, Malakowsky CA, Talent JM, Conrad CC, Carroll CA, Weintraub ST, Gracy RW. Anti-apoptotic proteins are oxidized by Abeta25-35 in Alzheimer's fibroblasts. Biochim Biophys Acta. 2003;1637:135–141. doi: 10.1016/s0925-4439(02)00227-2. [DOI] [PubMed] [Google Scholar]
- 245.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Link CD, Johnson CJ, Fonte V, Paupard M, Hall DH, Styren S, Mathis CA, Klunk WE. Visualization of fibrillar amyloid deposits in living, transgenic Caenorhabditis elegans animals using the sensitive amyloid dye, X-34. Neurobiol Aging. 2001;22:217–226. doi: 10.1016/s0197-4580(00)00237-2. [DOI] [PubMed] [Google Scholar]
- 247.Yatin SM, Varadarajan S, Link CD, Butterfield DA. In vitro and in vivo oxidative stress associated with Alzheimer's amyloid beta-peptide (1-42) Neurobiol Aging. 1999;20:325–330. doi: 10.1016/s0197-4580(99)00056-1. discussion 339-342. [DOI] [PubMed] [Google Scholar]
- 248.Head E, McCleary R, Hahn FF, Milgram NW, Cotman CW. Region-specific age at onset of beta-amyloid in dogs. Neurobiol Aging. 2000;21:89–96. doi: 10.1016/s0197-4580(00)00093-2. [DOI] [PubMed] [Google Scholar]
- 249.Johnstone EM, Chaney MO, Norris FH, Pascual R, Little SP. Conservation of the sequence of the Alzheimer's disease amyloid peptide in dog, polar bear and five other mammals by cross-species polymerase chain reaction analysis. Brain Res Mol Brain Res. 1991;10:299–305. doi: 10.1016/0169-328x(91)90088-f. [DOI] [PubMed] [Google Scholar]
- 250.Su MY, Head E, Brooks WM, Wang Z, Muggenburg BA, Adam GE, Sutherland R, Cotman CW, Nalcioglu O. Magnetic resonance imaging of anatomic and vascular characteristics in a canine model of human aging. Neurobiol Aging. 1998;19:479–485. doi: 10.1016/s0197-4580(98)00081-5. [DOI] [PubMed] [Google Scholar]
- 251.Tapp PD, Siwak CT, Estrada J, Head E, Muggenburg BA, Cotman CW, Milgram NW. Size and reversal learning in the beagle dog as a measure of executive function and inhibitory control in aging. Learn Mem. 2003;10:64–73. doi: 10.1101/lm.54403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Tapp PD, Siwak CT, Gao FQ, Chiou JY, Black SE, Head E, Muggenburg BA, Cotman CW, Milgram NW, Su MY. Frontal lobe volume, function, and beta-amyloid pathology in a canine model of aging. J Neurosci. 2004;24:8205–8213. doi: 10.1523/JNEUROSCI.1339-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Head E, Callahan H, Muggenburg BA, Cotman CW, Milgram NW. Visual-discrimination learning ability and beta-amyloid accumulation in the dog. Neurobiol Aging. 1998;19:415–425. doi: 10.1016/s0197-4580(98)00084-0. [DOI] [PubMed] [Google Scholar]
- 254.Milgram NW, Head E, Zicker SC, Ikeda-Douglas C, Murphey H, Muggenberg BA, Siwak CT, Tapp PD, Lowry SR, Cotman CW. Long-term treatment with antioxidants and a program of behavioral enrichment reduces age-dependent impairment in discrimination and reversal learning in beagle dogs. Exp Gerontol. 2004;39:753–765. doi: 10.1016/j.exger.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 255.Milgram NW, Head E, Zicker SC, Ikeda-Douglas CJ, Murphey H, Muggenburg B, Siwak C, Tapp D, Cotman CW. Learning ability in aged beagle dogs is preserved by behavioral enrichment and dietary fortification: a two-year longitudinal study. Neurobiol Aging. 2005;26:77–90. doi: 10.1016/j.neurobiolaging.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 256.Cotman CW, Head E, Muggenburg BA, Zicker S, Milgram NW. Brain aging in the canine: a diet enriched in antioxidants reduces cognitive dysfunction. Neurobiol Aging. 2002;23:809–818. doi: 10.1016/s0197-4580(02)00073-8. [DOI] [PubMed] [Google Scholar]
- 257.Milgram NW, Zicker SC, Head E, Muggenburg BA, Murphey H, Ikeda-Douglas CJ, Cotman CW. Dietary enrichment counteracts age-associated cognitive dysfunction in canines. Neurobiol Aging. 2002;23:737–745. doi: 10.1016/s0197-4580(02)00020-9. [DOI] [PubMed] [Google Scholar]
- 258.Faulstich ME. Brain imaging in dementia of the Alzheimer type. Int J Neurosci. 1991;57:39–49. doi: 10.3109/00207459109150345. [DOI] [PubMed] [Google Scholar]
- 259.Gosche KM, Mortimer JA, Smith CD, Markesbery WR, Snowdon DA. Hippocampal volume as an index of Alzheimer neuropathology: findings from the Nun Study. Neurology. 2002;58:1476–1482. doi: 10.1212/wnl.58.10.1476. [DOI] [PubMed] [Google Scholar]
- 260.Mungas D, Reed BR, Jagust WJ, DeCarli C, Mack WJ, Kramer JH, Weiner MW, Schuff N, Chui HC. Volumetric MRI predicts rate of cognitive decline related to AD and cerebrovascular disease. Neurology. 2002;59:867–873. doi: 10.1212/wnl.59.6.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Abdul HM, Calabrese V, Calvani M, Butterfield DA. Acetyl-L-carnitine-induced up-regulation of heat shock proteins protects cortical neurons against amyloid-beta peptide 1-42-mediated oxidative stress and neurotoxicity: implications for Alzheimer's disease. J Neurosci Res. 2006;84:398–408. doi: 10.1002/jnr.20877. [DOI] [PubMed] [Google Scholar]
- 262.Ansari MA, Joshi G, Huang Q, Opii WO, Abdul HM, Sultana R, Butterfield DA. In vivo administration of D609 leads to protection of subsequently isolated gerbil brain mitochondria subjected to in vitro oxidative stress induced by amyloid beta-peptide and other oxidative stressors: relevance to Alzheimer's disease and other oxidative stress-related neurodegenerative disorders. Free Radic Biol Med. 2006;41:1694–1703. doi: 10.1016/j.freeradbiomed.2006.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Boyd-Kimball D, Sultana R, Abdul HM, Butterfield DA. Gamma-glutamylcysteine ethyl ester-induced up-regulation of glutathione protects neurons against Abeta(1-42)-mediated oxidative stress and neurotoxicity: implications for Alzheimer's disease. J Neurosci Res. 2005;79:700–706. doi: 10.1002/jnr.20394. [DOI] [PubMed] [Google Scholar]
- 264.Drake J, Kanski J, Varadarajan S, Tsoras M, Butterfield DA. Elevation of brain glutathione by gamma-glutamylcysteine ethyl ester protects against peroxynitrite-induced oxidative stress. J Neurosci Res. 2002;68:776–784. doi: 10.1002/jnr.10266. [DOI] [PubMed] [Google Scholar]
- 265.Farr SA, Poon HF, Dogrukol-Ak D, Drake J, Banks WA, Eyerman E, Butterfield DA, Morley JE. The antioxidants alpha-lipoic acid and N-acetylcysteine reverse memory impairment and brain oxidative stress in aged SAMP8 mice. J Neurochem. 2003;84:1173–1183. doi: 10.1046/j.1471-4159.2003.01580.x. [DOI] [PubMed] [Google Scholar]
- 266.Perluigi M, Joshi G, Sultana R, Calabrese V, De Marco C, Coccia R, Butterfield DA. In vivo protection by the xanthate tricyclodecan-9-yl-xanthogenate against amyloid beta-peptide (1-42)-induced oxidative stress. Neuroscience. 2006 doi: 10.1016/j.neuroscience.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 267.Sultana R, Newman S, Mohmmad-Abdul H, Keller JN, Butterfield DA. Protective effect of the xanthate, D609, on Alzheimer's amyloid beta-peptide (1-42)-induced oxidative stress in primary neuronal cells. Free Radic Res. 2004;38:449–458. doi: 10.1080/1071576042000206478. [DOI] [PubMed] [Google Scholar]
- 268.Sultana R, Ravagna A, Mohmmad-Abdul H, Calabrese V, Butterfield DA. Ferulic acid ethyl ester protects neurons against amyloid beta- peptide(1-42)-induced oxidative stress and neurotoxicity: relationship to antioxidant activity. J Neurochem. 2005;92:749–758. doi: 10.1111/j.1471-4159.2004.02899.x. [DOI] [PubMed] [Google Scholar]