Abstract
We have analyzed conserved domains in t-SNAREs [soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein (SNAP) receptors in the target membrane], proteins that are believed to be involved in the fusion of transport vesicles with their target membrane. By using a sensitive computer method, the generalized profile method, we were able to identify a new homology domain that is common in the two protein families previously identified to act as t-SNAREs, the syntaxin and SNAP-25 (synaptosome-associated protein of 25 kDa) families, which therefore constitute a new superfamily. This homology domain of approximately 60 amino acids is predicted to form a coiled-coil structure. The significance of this homology domain could be demonstrated by a partial suppression of the coiled-coil properties of the domain profile. In proteins belonging to the syntaxin family, a single homology domain is located near the transmembrane domain, whereas the members of the SNAP-25 family possess two homology domains. This domain was also identified in several proteins that have been implicated in vesicular transport but do not belong to any of the t-SNARE protein families. Several new yeast, nematode, and mammalian proteins were identified that belong to the new superfamily. The evolutionary conservation of the SNARE coiled-coil homology domain suggests that this domain has a similar function in different membrane fusion proteins.
Most if not all vesicular membrane fusion events in eukaryotic cells are believed to be mediated by a conserved fusion machinery, the SNARE [soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein (SNAP) receptors] machinery (for review, see refs. 1–4). The components of the SNARE machinery have been identified in recent years and have been characterized biochemically, in most detail in the case of the synaptic vesicle fusion machinery. A mechanism emerges in which, in the process of vesicle docking, proteins present on the vesicle (v-SNAREs) have to bind to their counter parts on the target membrane (t-SNAREs) to form a core complex that can then recruit the soluble proteins NSF and SNAP. This so called fusion complex can then disassemble after ATP hydrolysis mediated by the ATPase NSF in a process that leads to membrane fusion and the release of the vesicle contents. t-SNAREs consist of two different families of proteins: the type II integral membrane proteins syntaxins (5) and SNAP-25 (synaptosome-associated protein of 25 kDa, a protein that is unrelated to the soluble protein SNAP), which is anchored in the plasma membrane by attached lipids and does not span the membrane (6, 7). The v-SNARE VAMP/synaptobrevin and the t-SNAREs syntaxin and SNAP-25 can form a stoichiometric ternary complex that involves protein domains predicted to form coiled-coil domains (8–11).
According to the SNARE hypothesis, a correct pairing of t- and v-SNAREs is required for vesicle fusion to occur thereby providing specificity of membrane trafficking by a final proof reading mechanism (12). This hypothesis postulates that many isoforms of t- and v-SNAREs exist, each specific for a particular membrane compartment or class of transport vesicles, respectively. There appears to be, however, a shortage of known SNARE isoforms in mammalian cells. Several mammalian syntaxin isoforms are known (5), most of which appear to be plasma membrane t-SNAREs. Only two mammalian members of the SNAP-25 family of t-SNAREs have been discovered so far: the neuron-specific SNAP-25 (6) and the ubiquitously expressed SNAP-23 (13). So far, no mammalian t-SNAREs have been identified that would be specific, for instance, for various endosome classes, the lysosome, or the endoplasmic reticulum (ER), compartments that are known or believed to utilize the SNARE machinery (14, 15).
To characterize the coiled-coil domains of SNAP-25 and syntaxin and to identify new homologues of these t-SNAREs, we have used the generalized profile technique (16), a very sensitive computer method that allows the identification of distantly related domains in proteins that would otherwise appear unrelated. Similar to other profile-based database search methods, the increase in sensitivity comes from two major sources: (i) Conserved positions in the initial alignment used for the profile construction receive a higher weight in the search than less conserved positions. (ii) Gap-creation and extension penalties are not evenly distributed over the whole sequence but are lower at positions where gaps or insertions have already been observed in the initial alignment. Compared with classical profile methods, the recently introduced generalized profiles offer an improved treatment of incomplete sequences (17), making this method well adapted to searches in expressed sequence tag (EST) databases.
We report herein that both the N- and C-terminal coiled-coil domains of members of the SNAP-25 family and the most C-terminal coiled-coil domain of the syntaxin family are related to each other and form a new homology domain of approximately 60 amino acids. The homology domain was found also in other known proteins involved in vesicular membrane traffic, some of which belong to different protein families. The protein families of t-SNAREs and related proteins, therefore, form a new protein superfamily. Several uncharacterized hypothetical proteins from yeast and Caenorhabditis elegans containing the homology domain could be identified in sequence databases. In addition, six new mammalian members of the t-SNARE superfamily were discovered and assembled from ESTs. The presence of a conserved domain in various proteins implicated in membrane fusion suggests that this domain might have a similar function in different protein families.
METHODS
All database searches were performed with current releases of SwissProt (18), GenPept (19), and dbEST (20). blast searches (21) were executed on the EPFL/ISREC network server. Generalized profile construction and searches were run locally by using programs of the pftools package (by P.B.; available from the authors upon request). Initial profiles were constructed with the parameter optimizations as described (22), applying a blosum45 substitution matrix (23), gap penalties of 2.1, and gap-extension penalties of 0.2. Profile statistics were derived from the analysis of the score distribution of a locally shuffled database as described (24).
Coiled-coil predictions were performed with the computer programs coils (25, 26) and paircoil (27). The generic coiled-coil profile was constructed from the residue frequencies of the mtidk matrix (26) used in the coils program. The generic profile was scaled to the same standard deviation as the original SNAP-25 domain profile and an average match score of 0. For coiled-coil reduction, 30% of the generic coiled-coil profile was subtracted from the original domain profile.
Phylogenetic tree reconstruction was performed with the neighbor-joining method (28). To assess the statistical significance of the clustering, bootstrapping analysis with 100 replicates was applied.
RESULTS AND DISCUSSION
To find sequences related to the SNAP-25 family of proteins, we first performed blast searches (21) with current releases of protein databases. Besides the characterized SNAP-25-like t-SNAREs from various organisms, two new hypothetical proteins from the nematode C. elegans (K02D10.1 and T14G12.2) and one from budding yeast (YMR017w) were identified with high significance (P < 10−6). These three uncharacterized proteins and SNAP-25 share the same domain organization— i.e., two conserved coiled-coil regions of ≈70 residues each separated by a less conserved spacer region. Therefore, they are likely to be SNAP-25-like t-SNAREs and were accepted as new members of the SNAP-25 family. Like Sec9p, the characterized SNAP-25 homologue from yeast (29), these proteins lack the posttranslationally palmitoylated cysteine residues within the spacer region.
Due to the coiled-coil nature of the query sequence and the corresponding atypical amino acid composition, the blast statistics are not very reliable. The highest-scoring irrelevant protein (tropomyosin) still had a score that would normally be considered significant (P < 10−4), which makes the identification of distant homologues virtually impossible.
Construction of a SNAP-25 Profile: Identification of a Common Domain in the SNAP-25 and Syntaxin Families of t-SNAREs.
To specifically find more distantly related members of the SNAP-25 family, generalized profiles for the conserved N- and C-terminal coiled-coil domains were constructed. A database search performed with the profile of the C-terminal domain found significant matches to several proteins involved in vesicular transport, as well as some uncharacterized proteins. The highest scoring sequences were the hypothetical nematode protein C15C7.1 (P < 10−5), the yeast vacuolar maintenance protein Vam7p (P < 0.003), the hypothetical yeast protein YDR468c (P < 0.004), and interestingly syntaxin 1 from Drosophila (P < 0.02). It should be noted that the statistical estimations obtained from profile searches are generally more conservative and reliable than the blast statistics, which assume a constant amino acid composition over all database sequences. Including the high scoring sequences C15C7.1, Vam7p, and YDR468c into the next iteration cycle of profile construction and database search resulted in significant scores for the yeast SNARE Bet1p and several syntaxin isoforms.
The matching region in the syntaxins corresponds to the membrane-proximal coiled-coil domain, previously designated helix 3 or H3 (9) or domain C (8). For syntaxin 1, this domain has been shown to mediate interactions with SNAP-25, VAMP/synaptobrevin, α-SNAP, and synaptotagmin (2), and the equivalent domain of Sed5p (the yeast homologue of syntaxin 5) might be involved in homodimerization (30). The finding that the two classes of t-SNAREs, syntaxin and SNAP-25, which are otherwise not related to each other, share a common homology domain suggests the intriguing possibility that they are evolutionarily derived from a common ancestor and that the homology domain has a similar function in these protein families. Subsignificant but in this biological context conspicuously high scores were obtained for the vesicular fusion protein p115/TAP and interestingly also for a second domain within the SNAP-25 family itself, corresponding to the N-terminal coiled-coil region (see below).
Partial Elimination of Coiled-Coil Properties Improves the Specificity of the Domain Profile.
Subsequent cycles of iterative profile refinement turned out to be problematic. The superposition of very distantly related coiled-coil sequences with identical heptad register led to an excessive amplification of the coiled-coil properties of the resulting profile. As a consequence, evolutionarily unrelated proteins with particularly pronounced coiled-coil regions reached inappropriately high scores, and subtle sequence similarities that are indicators of distant evolutionary relationship were progressively obscured by the strong coiled-coil preference of the profile.
To alleviate this problem, we artificially reduced the coiled-coil characteristics of the sequence profiles. To that end, the generalized profiles were decomposed into a linear combination of two components. The first component was a multiple of a generic coiled-coil profile, constructed from various families of coiled-coil proteins (25, 26), the second component was the residual profile lacking coiled-coil preference. It was, however, not intended to suppress the coiled-coil characteristics of the query profile entirely since protein regions distantly related to the SNAP-25 family would be expected to exist in that protein fold. The degree of coiled-coil suppression was adjusted in a way that scores for clearly unrelated proteins with nearly perfect coiled coils no longer reached a significance threshold of P = 0.1. A coiled-coil reduction of ≥20% was generally sufficient to reach that task, whereas a reduction of >40% was too drastic and prevented any database matches from meeting our stringent significance criterion of P < 0.01. We chose a conservative value of 30% reduction that was sufficient to suppress spurious high scores of extensively coiled-coil cytoskeletal proteins without severely affecting the highest scoring matches.
A new set of database searches, now using the artificially “decoiled” profiles, yielded an improved separation between true and false matches and was also suitable for iterative profile refinement by inclusion of newly accepted family members in subsequent rounds of profile construction. The initial search with the C-terminal domain of the SNAP-25 family found again the nematode protein C15C7.1; the yeast proteins YDR468c, Vam7p, and Bet1p; and two syntaxin isoforms with highly significant scores (P < 10−5 to P < 10−2). Several additional syntaxins had better scores than the highest scoring irrelevant sequence. Inclusion of only the highest scoring matches into the next profiles led to significant matches for the yeast vesicular fusion proteins Sft1p and Ufe1p and for the complete syntaxin family. Finally, inclusion of the syntaxins into the subsequent profile resulted in highly significant matches (P < 10−3) for several other uncharacterized proteins and, most interestingly, also for the N-terminal coiled-coil domain of most SNAP-25-related proteins. This discovery that the N- and C-terminal coiled-coil domains of the SNAP-25 family are related to each other suggests that they originated from an internal duplication.
No obviously unrelated protein ever reached scores better than P < 0.1 in any iteration of the profile search. Searches with the final profile in six-frame translated EST databases led to the identification of several homologous sequences that could be assembled to six new non-orthologous mammalian proteins, herein tentatively named TSL-1 to TSL-6 (for t-SNARE-like). TSL-4 could be assembled to a full-length protein, whereas the others are only known as partial sequences.
The t-SNARE Superfamily.
The final sequence profile describes a homology domain of approximately 60 amino acids with a predicted coiled-coil structure in all subfamilies. This homology domain was found in all currently known t-SNAREs, as well as several proteins of unknown or disputed function that are likely to be structurally and perhaps functionally related to the t-SNAREs (see below). It therefore seems appropriate to refer to this homology domain as the t-SNARE domain. Members of different protein families contain this domain, and therefore, they form a new superfamily.
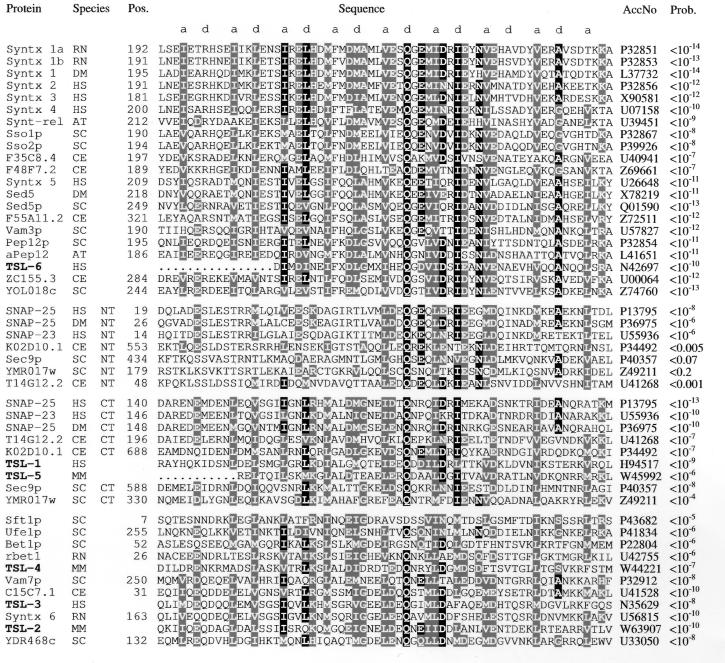
An alignment of the t-SNARE domains of representative members of the t-SNARE superfamily and all newly identified related proteins is shown in Fig. 1. The most likely coiled-coil register is indicated, and it is obvious that positions a and d, which form the protein–protein contact and are usually occupied by hydrophobic residues, are particularly well conserved. An unusual glutamine residue at a d-position in the core of the coiled-coil region is almost invariant. The conserved hydrophobic positions in the coiled-coil show a preference for isoleucine at position a and leucine at position d. An analysis of observed coiled-coil structures, theoretical considerations, and protein design experiments suggest that this distribution favors two-stranded coiled-coils (31). In contrast, three-stranded coiled-coils show a preference for leucine at position a and branched residues at position d.
Figure 1.
Alignment of the coiled-coil t-SNARE homology domain. The header line shows the register of the predicted coiled-coil. The t-SNARE domains of representative members of the syntaxin family (first sequence block), the SNAP-25 family (second and third blocks), and members of other families or of uncertain family relation (fourth block) are shown with all newly identified mammalian homologous proteins (TSLs, boldface type). Residues conserved in more than 50% of all sequences are printed inversely; positions with conservative substitutions in more than 50% of all sequences are printed on a shaded background. Protein or gene names are indicated, and two-letter species abbreviations is as follows: RN, Rattus norvegicus; HS, Homo sapiens; DM, Drosophila melanogaster; CE, C. elegans; SC, Saccharomyces cerevisiae; AT, Arabidopsis thaliana; MM, Mus musculus. NT and CT refer to the N-terminal and C-terminal homology domain of SNAP-25-related proteins, respectively. Numbers to the left of the alignment indicate the position of the homology domain in the sequence (if known). The sequence accession numbers (AccNo) are shown. Codes starting with the letters P or Q are from SwissProt; others are from GenBank/EMBL. The statistical significance of the sequences matching to the final domain profile are expressed as error probabilities (Prob.). Each individual sequence was optimally aligned against the profile by using the pftools program package. Coiled-coil prediction with the program coils 2.1, using standard parameters (Window width, 21/28; MTIDK matrix) gave coiled-coil probabilities of P > 0.7 over a range of at least 35 residues for all domains listed with two exceptions. TSL-4 and YDR468c reached only maximal values of 0.45 and 0.3, respectively. If the problems in coiled-coil detection for short segments (26) are taken into account, it is likely that all domains adopt a coiled-coil conformation with similar geometry.
A sequence similarity dendrogram of the t-SNARE domains of representative known proteins and the newly identified proteins, constructed by the neighbor-joining method (28), is shown in Fig. 2A. The domain structures of these proteins are shown schematically in Fig. 2B. The members of the t-SNARE superfamily can be clearly divided into subgroups based both on the sequence homologies and the domain structures. All syntaxins and syntaxin-related proteins (domain structure A, A′) contain a C-terminal hydrophobic membrane anchor as do some less closely related members (domain structure B) such as Bet1 (see below). All of these proteins contain one copy of the t-SNARE domain in a distance of 10 to 15 amino acids from the transmembrane domain.
Figure 2.
(A) Nearest-neighbor dendrogram of the t-SNARE homology domain. The same set of sequences as in Fig. 1 is shown. Bifurcation points confirmed in 80–100 bootstrap replicates (out of 100) are marked by solid triangles, in 50–80 replicates are marked by solid circles, in 20–50 replicates are marked by open circles, and in less than 20 replicates are marked by no label. Boldface type on the right refers to the domain structures in B. Species abbreviations are as in Fig. 1. The newly identified SNARE homologues (TSLs) are indicated in boldface type. (B) Representative domain structures of t-SNARE superfamily members. Shaded boxes indicate coiled-coil regions. Open boxes labeled TM indicate putative transmembrane domains. The position of the t-SNARE coiled-coil homology domain is indicated by a solid bar at the bottom of the sequence representation. The chosen representative proteins are: A, syntaxin 1A; A′, Pep12p; B, Bet1p; C, SNAP-25; C′, Sec9; D, Vam7p.
In contrast, the superfamily members that are more closely related to SNAP-25 do not possess an obvious transmembrane domain but instead contain two copies of the t-SNARE domain interrupted by a less conserved spacer region. A relationship between syntaxins and SNAP-25 has been proposed before on the basis of their weak similarity (32, 33). However, the coiled-coil conformation of the region in question makes it impossible to demonstrate a significant similarity in pairwise sequence comparisons. In fact, several clearly unrelated coiled-coil proteins give much better scores when aligned to either syntaxins or SNAP-25. Our analysis provides a statistically valid basis for this homology and additionally detects an ancient domain duplication event in the SNAP-25 family.
The Syntaxin Protein Family.
In the dendrogram, the syntaxins form a single cluster that can be divided into two subgroups. The first group contains mammalian isoforms 1 to 4, yeast Sso1p and Sso2p, and two hypothetical nematode proteins. The t-SNARE domain sequence similarity between the members of this group correlates well with the function of these t-SNAREs. Syntaxins 1 to 4 (5, 34, 35) and Sso1p and Sso2p (29, 36) have been shown to localize to the plasma membrane and are likely to be involved in the fusion of transport vesicles with this membrane. The two uncharacterized proteins from C. elegans (F35C8.4 and F48F7.2), which clearly belong to this group, therefore, possibly also function as t-SNAREs at the plasma membrane of this species.
The second syntaxin subgroup contains the mammalian syntaxin 5; the yeast proteins Sed5p, Pep12p, and Vam3p; a plant syntaxin; and several uncharacterized proteins including the newly identified human TSL-6. Syntaxin 5 and Sed5p have been localized to the ER–Golgi intermediate compartment or the cis-Golgi in mammalian cells and yeast, respectively, and are involved in vesicle traffic between the ER and Golgi (30, 37). The uncharacterized C. elegans protein F55A11.2 is closely related to syntaxin 5 and Sed5p and might, therefore, serve the equivalent function.
Vam3p and Pep12p are involved in vacuolar assembly and vacuolar protein targeting in yeast (38, 39) and the most likely intracellular localization of Pep12p is an intermediate endosomal compartment for the transport of vacuolar hydrolases (39). The plant syntaxin homologue aPep12 has been identified by functional complementation of a yeast pep12 mutant (40) and is indeed most closely related to Pep12p. The newly identified human TSL-6 is most closely related to Pep12p and aPep12. Although the N-terminal half of TSL-6 is unknown, it is clearly a member of the syntaxin family since it contains a C-terminal transmembrane domain close to the t-SNARE domain. The close proximity to Pep12p suggests that TSL-6 is its mammalian homologue and might function as an endosomal t-SNARE responsible for the transport of lysosomal proteins since lysosomes are believed to be functionally equivalent to the vacuole of yeast. This is a potentially important finding. To our knowledge, no endosome-specific t-SNARE has been identified in mammalian cells although membrane traffic between different endosome classes, including fusion with late endosomes, appears to involve the SNARE machinery (41).
Two uncharacterized proteins from C. elegans (ZC155.3) and yeast (YOL018c) appear to form an additional third syntaxin cluster. YOL018c shares the same domain structure with the other syntaxins which suggests that it plays a yet unidentified role in membrane traffic as a potential t-SNARE. A distinct feature of the predicted sequence of YOL018c is that it contains a large luminal domain of 62 amino acids, which is atypical for syntaxins. The nematode protein ZC155.3 seems to have a very unusual domain structure. It should be noted, however, that the sequence of this hypothetical protein has been predicted from genomic sequences. The distances between the exons contributing to the database sequence ZC155.3 suggest that it might be an artifactual fusion of two different coding regions.
Very recently, a human protein has been identified by a database search for homologues to Pep12p and named syntaxin 6 because of its weak overall homology with other syntaxins, its domain structure, and the finding that it binds α-SNAP in vitro (33). From Fig. 2A, it is clear that syntaxin 6 is only distantly related to the other syntaxins, including Pep12p, and rather forms a separate cluster with the two newly identified mammalian proteins TSL-2 and TSL-3 and the nematode protein C15C7.1. This suggests that syntaxin 6 is not the mammalian homologue of Pep12p (we have identified TSL-6 as a likely candidate for the Pep12p homologue, see above) but is a member of a new family of syntaxin-like proteins. Syntaxin 6 has been localized to the Golgi region and proposed to function there as a t-SNARE. An interesting speculation would be that the closely related TSL-2 and -3 might reside in the Golgi as well but in different subcompartments. The presence of multiple syntaxin isoforms on different Golgi cisternae would constitute a strong argument in support of the SNARE hypothesis.
The SNAP-25 Protein Family.
The SNAP-25-like proteins are characterized by a domain structure that is very different from the syntaxin-like proteins. They contain two t-SNARE domains but no hydrophobic membrane anchoring domain (structures C and C′ in Fig. 2B). The C-terminal t-SNARE domains of the various SNAP-25-like proteins form a cluster in the dendrogram as do the N-terminal domains. The relationship between these two clusters is, however, rather distant, suggesting that a possible internal duplication which could be responsible for the presence of the two related domains in a single protein must have been a very early evolutionary event.
The mammalian isoforms SNAP-25 and SNAP-23 and the Drosophila SNAP-25 are very closely related to each other, and also Sec9p, the known yeast homologue of SNAP-25, clearly belongs to this family. In addition, we identified three uncharacterized hypothetical proteins, K02D10.1 and T14G12.2 from C. elegans and the yeast protein YMR017w, as new members of the SNAP-25 family. Particularly interesting is YMR017w which is closely related to Sec9p and is the first yeast isoform of Sec9p identified so far. Sec9p functions at the plasma membrane in yeast, probably analogous to SNAP-25 in mammals (29). No SNAP-25-like t-SNARE has been identified so far, neither in yeast nor in mammals, which would be specific for a membrane compartment other than the plasma membrane. The finding of a Sec9p homologue clearly merits a further investigation of the role of this new protein in membrane traffic. Both Sec9p and YMR017w share the same domain structure (C′) which differs from the other members of this family because of their large N-terminal extension. It has been shown that this extension is not necessary for the essential function of Sec9p (29). The C-terminal t-SNARE domains of Sec9p and YMR017w are closely related to each other but do not form a cluster with the other members of the SNAP-25 family.
The t-SNARE domains of the newly discovered mammalian proteins TSL-1 and TSL-5 cluster with the C-terminal t-SNARE domains of the SNAP-25 family (Fig. 2A). Only the C-terminal parts of these two proteins are known and they clearly do not contain a hydrophobic membrane anchor. They are therefore likely to be new members of the SNAP-25 family although the existence of a second, N-terminal, t-SNARE domain remains to be established. The discovery of new mammalian SNAP-25-related proteins will make it possible to address the question of whether the members of this t-SNARE family are differentially localized to different membrane compartments in the cell, as has been demonstrated before for members of the syntaxin family (34, 35). Such a differential localization would indicate a function in providing specificity of membrane fusion.
Other Members of the t-SNARE Superfamily.
Other proteins like the yeast proteins Sft1p, Ufe1p, Bet1p, Vam7p, and YDR468c and the newly identified mammalian TSL-4 cannot unambiguously be assigned to a subgroup. However, on the basis of the significant domain similarity, they are clearly members of the t-SNARE superfamily established herein.
The two yeast proteins Sft1p and Ufe1p form a distinct cluster that is most closely related to the syntaxins. Ufe1p is believed to be a t-SNARE responsible for the retrograde transport to the ER in yeast (42) and according to its domain structure is clearly a member of the syntaxin family. The domain structure of the very small Sft1p is different, however: it consists only of the t-SNARE domain and a C-terminal membrane anchor. Sft1p has been identified as a multicopy suppressor of a yeast Sed5p mutant and has been proposed to be required for traffic between Golgi compartments (43). It has been suggested that Sft1p acts as a v-SNARE mostly because of its typical v-SNARE-like structure (i.e., a very small type II membrane protein with a cytoplasmic domain predicted to form a coiled-coil). However, Sft1p shows no sequence homology to the known v-SNARE VAMP/synaptobrevin or its related proteins. In light of our finding of a domain typical for t-SNAREs in Sft1p, it seems worthwhile to consider the possibility that Sft1p acts as a t-SNARE rather than a v-SNARE.
A similar conflict is encountered with Bet1p. Bet1p is a small protein with a C-terminal membrane anchor and consists mostly of the t-SNARE domain in its cytoplasmic part (domain structure B). Yeast Bet1p is required for ER-to-Golgi transport and resides on the ER. It has been proposed to be a v-SNARE but its presence on transport vesicles is controversial (44). Although Bet1p displays a structure reminiscent of the v-SNAREs of the VAMP/synaptobrevin family, it shares no sequence homology with this family. Our finding that Bet1p belongs to the t-SNARE superfamily might, therefore, suggest a different function for this protein. The newly identified mouse TSL-4, whose complete sequence is known, is as closely related to yeast Bet1p as is the recently cloned rat homologue rbet1 (45). Since we could also identify EST sequences of the mouse rbet1, which differ from TSL-4 (data not shown), we therefore conclude that at least two different homologues of Bet1p exist in mammalian cells. Interestingly, rbet1 has been localized to the Golgi apparatus in fibroblasts, whereas the yeast counterpart resides on the ER membrane. It should be most interesting to determine the subcellular localization of TSL-4 in comparison.
The yeast Vam7p, believed to be involved in vacuole assembly, has a unique domain structure resembling the SNAP-25 structure but contains only one copy of the t-SNARE domain.
A protein that was consistently detected in the iteration cycles of our database searches is p115/TAP. The scores for this protein never reached significant levels, although they were always above the scores calculated for the next best unrelated protein. This lack of improvement in the score argues against an evolutionary relationship. p115/TAP was therefore not included in the profiles and it remains unclear whether it is indeed a true member of the t-SNARE superfamily. It is, however, functionally related to SNAREs. p115/TAP is required for the docking or fusion step of intra-Golgi vesicular transport (46), for the fusion of transcytotic vesicles with their target membrane (47), and, in concert with NSF and SNAP, for the reassembly of Golgi fragments (48). Uso1p, the yeast homologue of p115/TAP, was found to be required for the formation of the ER-to-Golgi v-SNARE/t-SNARE complex (49). The domain structure of p115/TAP is entirely different from the SNAREs: it forms a homodimer with a globular N-terminal half and an extended C-terminal coiled-coil-rich domain of approximately 250 residues (47, 50). Only the region between residues 637 and 699 in this C-terminal part shows homology to the t-SNARE domain. In light of their functional relationship, even a very weak sequence similarity between the t-SNAREs and p115/TAP might suggest an evolutionary relationship and points to a possible important role of the featured domain in p115/TAP.
The evolutionary relationship of various proteins implicated in membrane fusion sheds a new light on the mechanism of this process, and the existence of a conserved coiled-coil domain present in different membrane fusion proteins suggests a similar function of this domain in these proteins. The discovery of new potential t-SNARE homologues should facilitate the testing of the central prediction of the SNARE hypothesis, namely, that different SNARE isoforms function specifically in distinct membrane trafficking steps.
Acknowledgments
This work was supported by a Feodor–Lynen Fellowship of the Alexander von Humboldt Foundation to T.W., a postdoctoral fellowship from the Irvington Institute for Immunology to S.H.L., a National Institutes of Health institutional National Research Service Award (T32HL07731) to S.C., National Institutes of Health Grants R01 AI25144 and R01 AI36953 and an American Heart Association Established Investigator Award to K.M., and a grant from the Swiss National Research Foundation to P.B.
ABBREVIATIONS
- EST
expressed sequence tag
- NSF
N-ethylmaleimide-sensitive factor
- SNAP-25
synaptosome-associated protein of 25 kDa
- SNAP
soluble NSF attachment protein
- SNARE
SNAP receptor
- TSL
t-SNARE-like
- t-SNARE
SNARE in the target membrane
- v-SNARE
SNARE in the vesicle membrane
References
- 1.Rothman J E. Nature (London) 1994;372:55–63. doi: 10.1038/372055a0. [DOI] [PubMed] [Google Scholar]
- 2.Bennett M K. Curr Opin Cell Biol. 1995;7:581–586. doi: 10.1016/0955-0674(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 3.Südhof T C. Nature (London) 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 4.Calakos N, Scheller R H. Physiol Rev. 1996;76:1–29. doi: 10.1152/physrev.1996.76.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Bennett M K, Garcia-Arraras J E, Elferink L A, Peterson K, Fleming A M, Hazuka C D, Scheller R H. Cell. 1993;74:863–873. doi: 10.1016/0092-8674(93)90466-4. [DOI] [PubMed] [Google Scholar]
- 6.Oyler G A, Higgins G A, Hart R A, Battenberg E, Billingsley M, Bloom F E, Wilson M C. J Cell Biol. 1989;109:3039–3052. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Söllner T, Whiteheart S W, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman J E. Nature (London) 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 8.Chapman E R, An S, Barton N, Jahn R. J Biol Chem. 1994;269:27427–27432. [PubMed] [Google Scholar]
- 9.Kee Y, Lin R C, Hsu S C, Scheller R H. Neuron. 1995;14:991–998. doi: 10.1016/0896-6273(95)90337-2. [DOI] [PubMed] [Google Scholar]
- 10.McMahon H T, Südhof T C. J Biol Chem. 1995;270:2213–2217. doi: 10.1074/jbc.270.5.2213. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi T, McMahon H, Yamasaki S, Binz T, Hata Y, Südhof T C, Niemann H. EMBO J. 1994;13:5051–5061. doi: 10.1002/j.1460-2075.1994.tb06834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothman J E, Warren G. Curr Biol. 1994;4:220–233. doi: 10.1016/s0960-9822(00)00051-8. [DOI] [PubMed] [Google Scholar]
- 13.Ravichandran V, Chawla A, Roche P A. J Biol Chem. 1996;271:13300–13303. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 14.Haas A, Wickner W. EMBO J. 1996;15:3296–3305. [PMC free article] [PubMed] [Google Scholar]
- 15.Diaz R, Mayorga L S, Weidman P J, Rothman J E, Stahl P D. Nature (London) 1989;339:398–400. doi: 10.1038/339398a0. [DOI] [PubMed] [Google Scholar]
- 16.Bucher P, Bairoch A. Intell Syst Mol Biol. 1994;2:53–61. [PubMed] [Google Scholar]
- 17.Bucher P, Karplus K, Moeri N, Hofmann K. Computers Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 18.Bairoch A, Apweiler R. Nucleic Acids Res. 1996;24:21–25. doi: 10.1093/nar/24.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benson D A, Boguski M, Lipman D J, Ostell J. Nucleic Acids Res. 1996;24:1–5. doi: 10.1093/nar/24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boguski M S, Lowe T M, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Luthy R, Xenarios I, Bucher P. Protein Sci. 1994;3:139–146. doi: 10.1002/pro.5560030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henikoff S, Henikoff J G. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofmann K, Bucher P. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- 25.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 26.Lupas A. Methods Enzymol. 1996;266:513–524. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 27.Berger B, Wilson D B, Wolf E, Tonchev T, Milla M, Kim P S. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Brennwald P, Kearns B, Champion K, Keranen S, Bankaitis V, Novick P. Cell. 1994;79:245–258. doi: 10.1016/0092-8674(94)90194-5. [DOI] [PubMed] [Google Scholar]
- 30.Banfield D K, Lewis M J, Rabouille C, Warren G, Pelham H R. J Cell Biol. 1994;127:357–371. doi: 10.1083/jcb.127.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupas A. Trends Biochem Sci. 1996;21:375–382. [PubMed] [Google Scholar]
- 32.Risinger C, Blomqvist A G, Lundell I, Lambertsson A, Nassel D, Pieribone V A, Brodin L, Larhammar D. J Biol Chem. 1993;268:24408–24414. [PubMed] [Google Scholar]
- 33.Bock J B, Lin R C, Scheller R H. J Biol Chem. 1996;271:17961–17965. doi: 10.1074/jbc.271.30.17961. [DOI] [PubMed] [Google Scholar]
- 34.Low S H, Chapin S J, Weimbs T, Kömüves L G, Bennett M K, Mostov K E. Mol Biol Cell. 1996;7:2007–2018. doi: 10.1091/mbc.7.12.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaisano H Y, Ghai M, Malkus P N, Sheu L, Bouquillon A, Bennett M K, Trimble W S. Mol Biol Cell. 1996;7:2019–2027. doi: 10.1091/mbc.7.12.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aalto M K, Ronne H, Keranen S. EMBO J. 1993;12:4095–4104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dascher C, Matteson J, Balch W E. J Biol Chem. 1994;269:29363–29366. [PubMed] [Google Scholar]
- 38.Wada Y, Ohsumi Y, Anraku Y. J Biol Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- 39.Becherer K A, Rieder S E, Emr S D, Jones E W. Mol Biol Cell. 1996;7:579–594. doi: 10.1091/mbc.7.4.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bassham D C, Gal S, da Silva Conceicao A, Raikhel N V. Proc Natl Acad Sci USA. 1995;92:7262–7266. doi: 10.1073/pnas.92.16.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aniento F, Gruenberg J. Cold Spring Harbor Symp Quant Biol. 1995;60:205–209. doi: 10.1101/sqb.1995.060.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Lewis M J, Pelham H R B. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- 43.Pelham H R B, Banfield D K, Lewis M J. Cold Spring Harbor Symp Quant Biol. 1995;60:105–111. doi: 10.1101/sqb.1995.060.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Jiang Y, Sacher M, Singer-Krüger B, Lian J P, Stone S, Ferro-Novick S. Cold Spring Harbor Symp Quant Biol. 1995;60:119–126. doi: 10.1101/sqb.1995.060.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Hay J C, Hirling H, Scheller R H. J Biol Chem. 1996;271:5671–5679. doi: 10.1074/jbc.271.10.5671. [DOI] [PubMed] [Google Scholar]
- 46.Waters M G, Clary D O, Rothman J E. J Cell Biol. 1992;118:1015–1026. doi: 10.1083/jcb.118.5.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barroso M, Nelson D S, Sztul E. Proc Natl Acad Sci USA. 1995;92:527–531. doi: 10.1073/pnas.92.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabouille C, Levine T P, Peters J M, Warren G. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- 49.Sapperstein S K, Lupashin V V, Schmitt H D, Waters M G. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sapperstein S K, Walter D M, Grosvenor A R, Heuser J E, Waters M G. Proc Natl Acad Sci USA. 1995;92:522–526. doi: 10.1073/pnas.92.2.522. [DOI] [PMC free article] [PubMed] [Google Scholar]